Transcriptome Analysis of Spirulina platensis sp. at Different Salinity and Nutrient Compositions for Sustainable Cultivation in Vietnam
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Culture Media and Experimental Conditions
2.3. Determination of Microalgae Growth
2.4. Determining the Specific Growth Rate
2.5. Determination of Pigments
2.6. Determination of Lipid, Protein, Carbohydrate Contents
2.7. Measurement of the Net Photosynthetic Rate (Pn)
2.8. Determination of Chlorophyll a Fluorescence Photosynthetic Activity
2.9. Isolation of RNA and cDNA Synthesis
2.10. Library Construction and Illumina Sequencing
2.11. Gene Expression Quantification and Differential Expression Analysis
2.12. Gene Ontology and KEGG Analyses
2.13. Statistical Analysis
3. Results and Discussion
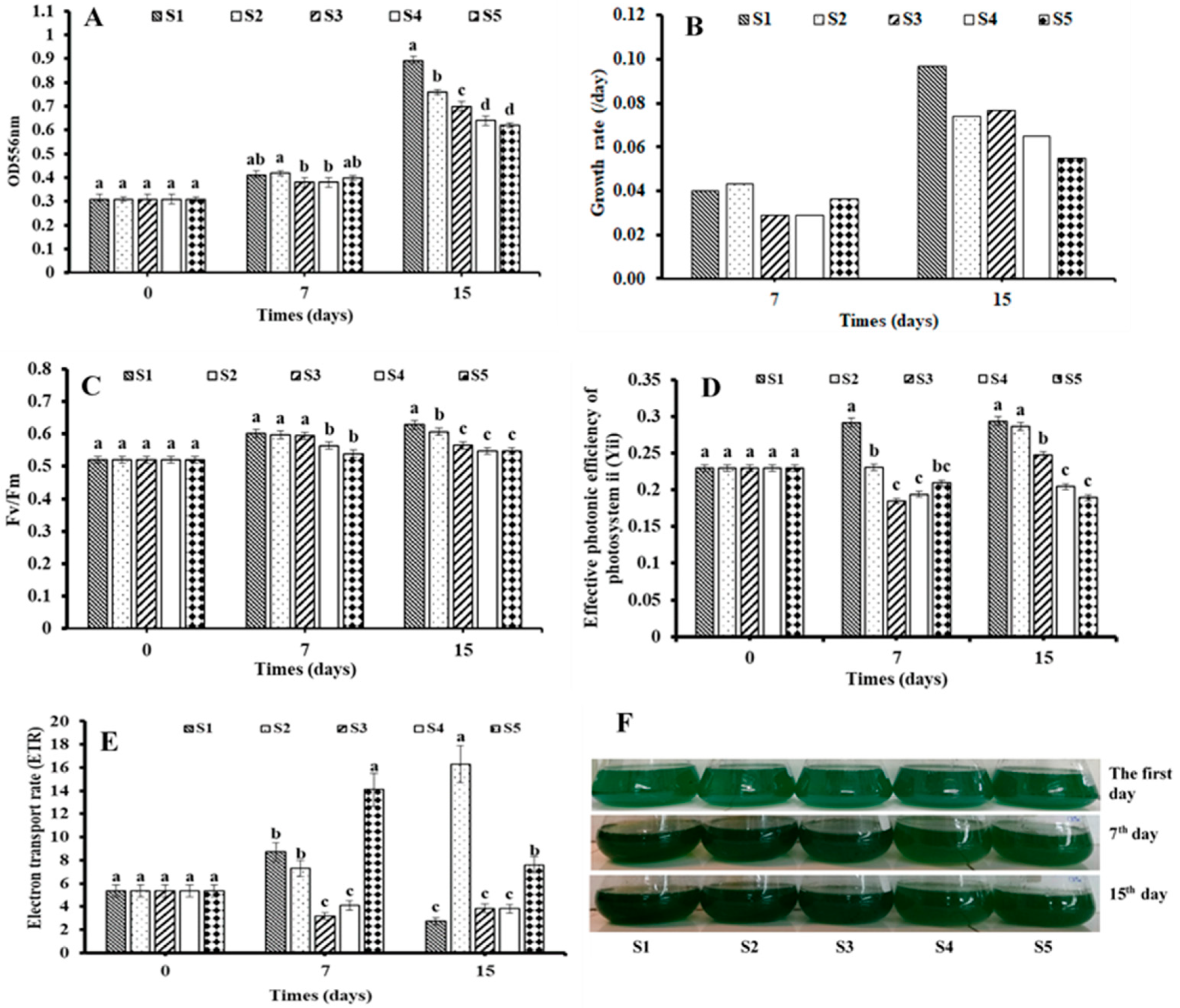
3.1. Effect of SOT Medium Containing Salinity of 30‰ with Different Nutrition Concentration on the Growth of Salt Tolerant S. platensis ST under Laboratory Condition
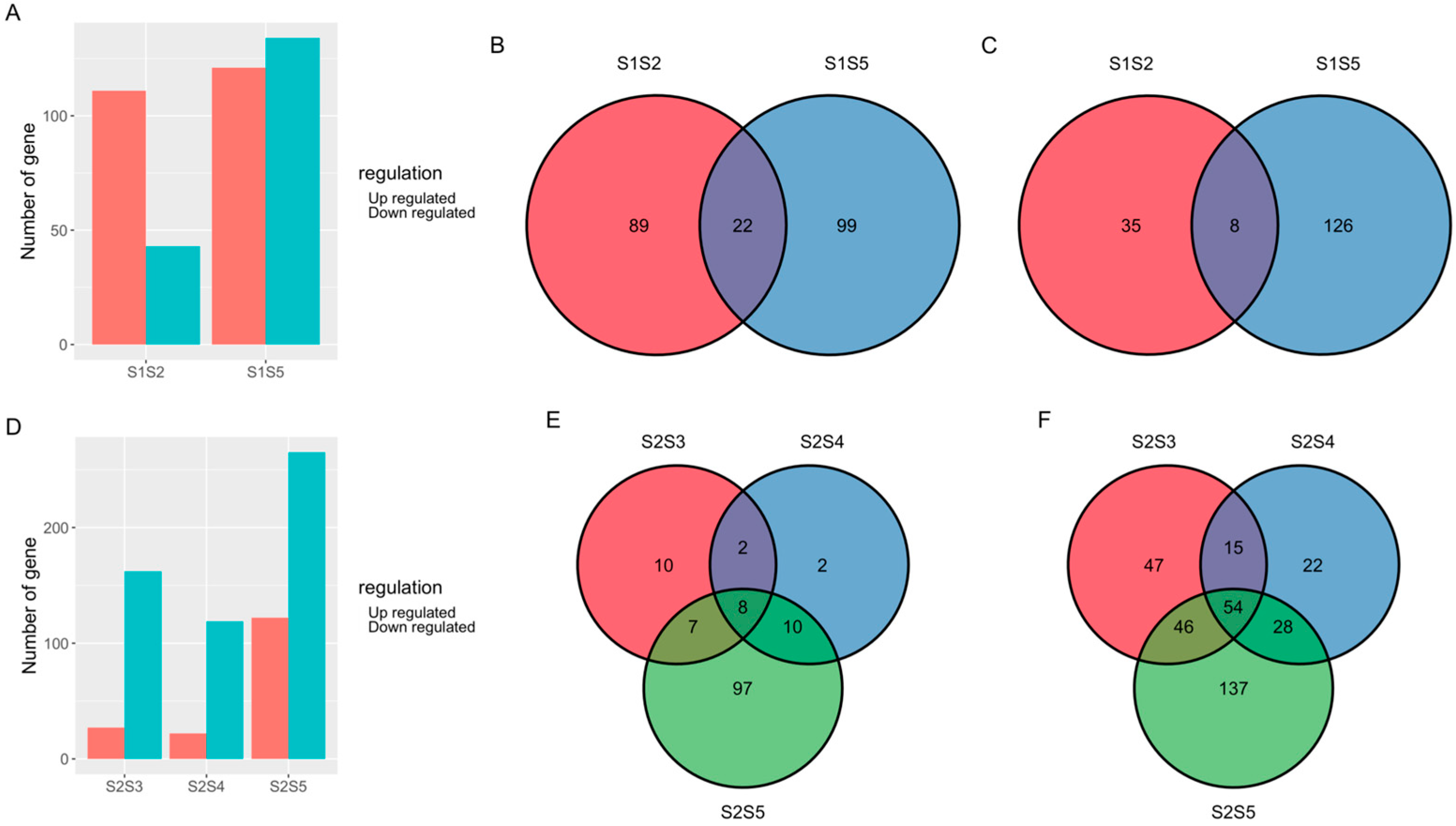
3.2. Transcriptome Analysis
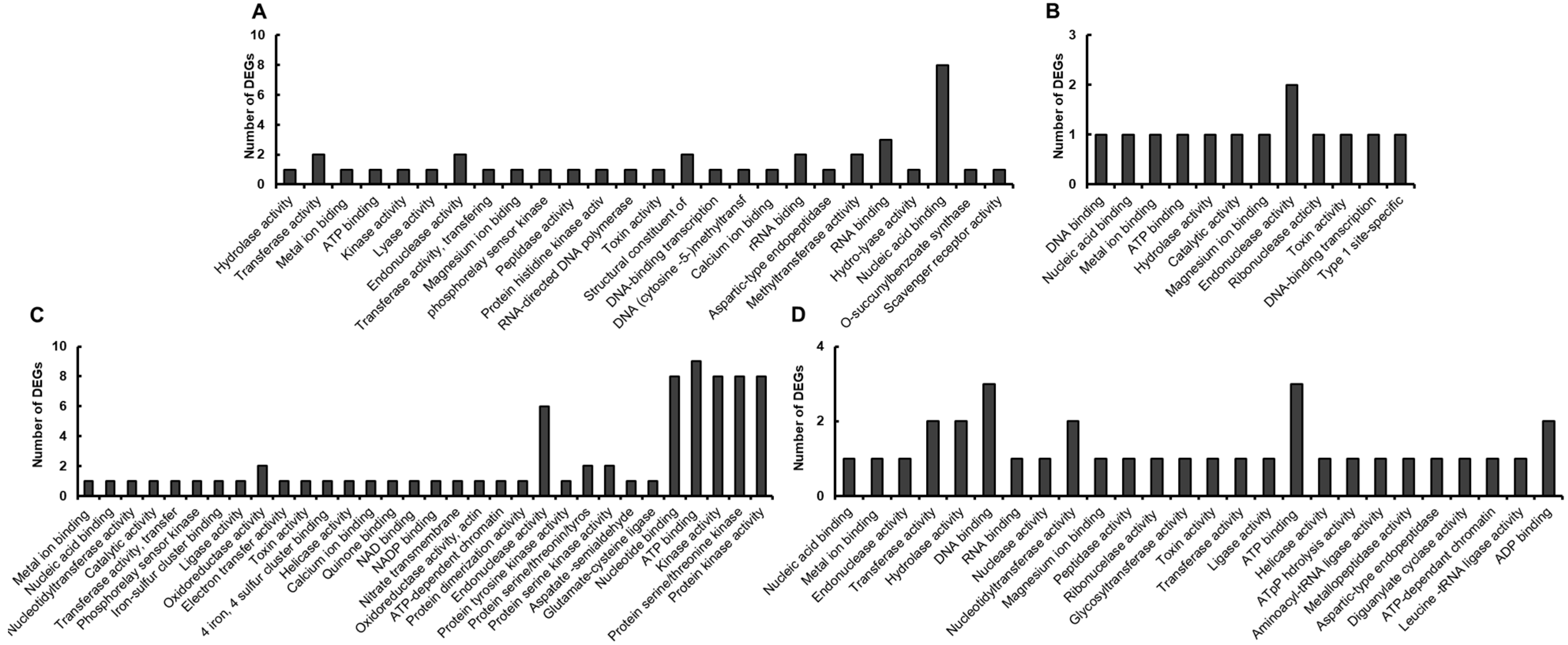
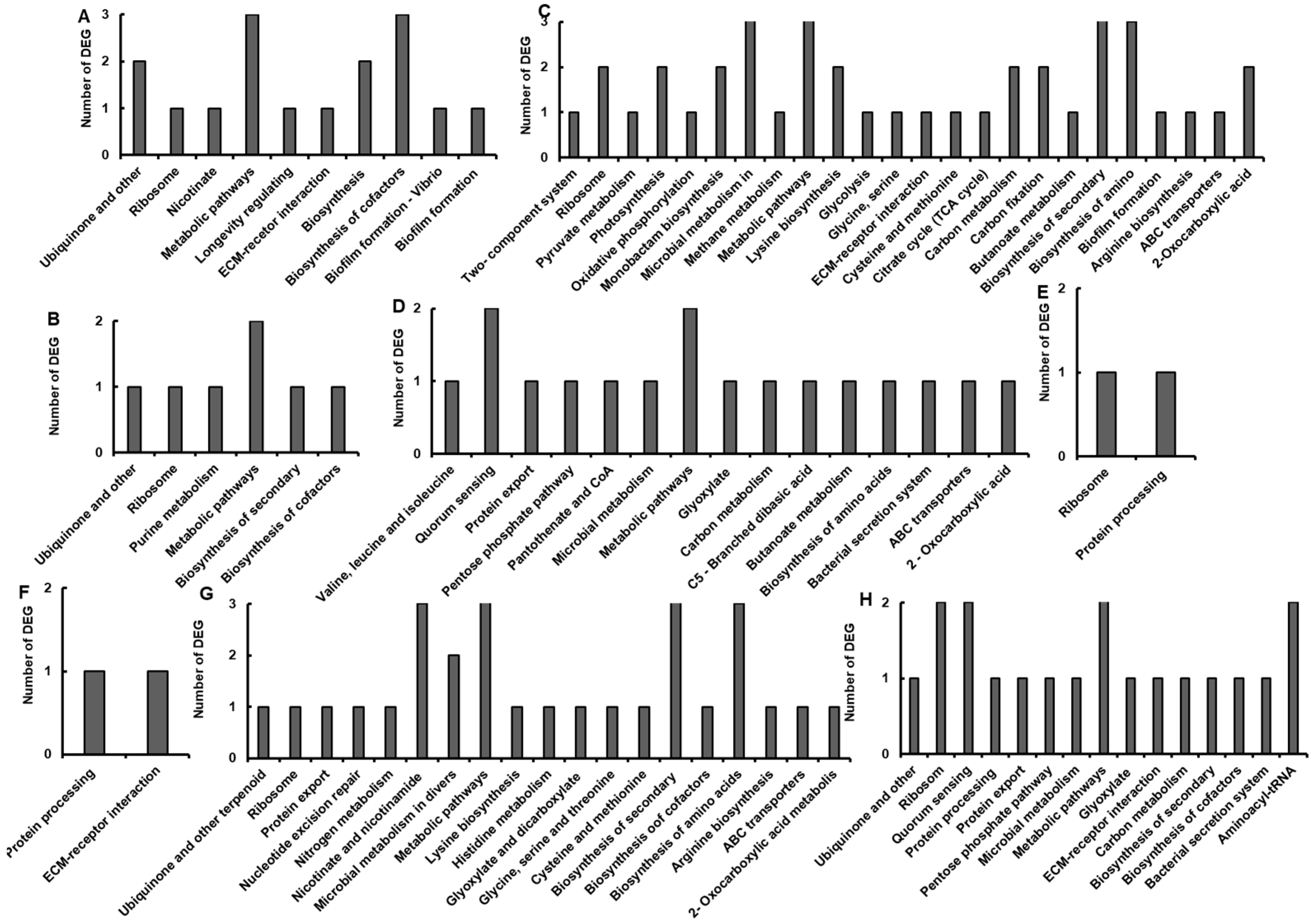
3.3. GO and KEGG Analysis of DEGs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nhung, T.T.; Vo, P.L.; Nghi, V.V.; Bang, H.Q. Salt intrusion adaptation measures for sustainable agricultural development under climate change effects: A case of Ca Mau Peninsula, Vietnam. Clim. Risk Manag. 2019, 23, 88–100. [Google Scholar] [CrossRef]
- Toan, T.L.; Huu, N.; Simioni, M.; Phan, H.; Arai, H.; Mermoz, S.; Bouvet, A.; de Eccher, I.; Diallo, Y.; Duong, T.H.; et al. Agriculture in Viet Nam under the impact of climate change. In Climate Change in Vietnam. Impacts and Adaptation; A COP26 Assessment Report of the GEMMES Viet Nam Project; HAL: Glasgow, UK, 2021; Available online: https://hal.inrae.fr/hal-03456472/document (accessed on 30 July 2023).
- Karkos, P.D.; Leong, S.C.; Karkos, C.D.; Sivaji, N.; Assimakopoulos, D.A. Spirulina in Clinical Practice: Evidence-Based Human Applications. Evid. Based Complement. Altern. Med. 2011, 2011, 531053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brito, A.d.F.; Silva, A.S.; de Oliveira, C.V.; de Souza, A.A.; Ferreira, P.B.; de Souza, I.L.; da Cunha, A.L.C.; da Silva, F.G.; de Souza, S.R.; Tavares, R.L.; et al. Spirulina platensis prevents oxidative stress and inflammation promoted by strength training in rats: Dose-response relation study. Sci. Rep. 2020, 10, 6382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ai, X.; Yu, P.; Li, X.; Lai, X.; Yang, X.; Yang, M.; Liu, F.; Luan, F.; Meng, X. Polysaccharides from Spirulina platensis: Extraction methods, structural features and bioactivities diversity. Int. J. Biol. Macromol. 2023, 231, 123211. [Google Scholar] [CrossRef]
- Lafarga, T.; Fernández-Sevilla, J.M.; González-López, C.; Acién-Fernández, F.G. Spirulina for the food and functional food industries. Food Res. Int. 2020, 137, 109356. [Google Scholar] [CrossRef]
- Liu, Q.; Huang, Y.; Zhang, R.; Cai, T.; Cai, Y. Medical Application of Spirulina platensis Derived C-Phycocyanin. Evid. Based Complement. Altern. Med. 2016, 2016, 7803846. [Google Scholar]
- Jung, F.; Krüger-Genge, A.; Waldeck, P.; Küpper, J.H. Spirulina platensis, a super food? J. Cell. Biotechnol. 2019, 5, 43–54. [Google Scholar] [CrossRef]
- Wang, F.; Yu, X.; Cui, Y.; Xu, L.; Huo, S.; Ding, Z.; Hu, Q.; Xie, W.; Xiao, H.; Zhang, D. Efficient extraction of phycobiliproteins from dry biomass of Spirulina platensis using sodium chloride as extraction enhancer. Food Chem. 2023, 406, 135005. [Google Scholar] [CrossRef]
- Salama, A.F.; Abdel-Hamid, N.M.; El-sheekh, M.; Tosson, E.; Gabr, A.M. Spirulina platensis microalgae protects against diethyl nitrosamine carcinogenic effect on female albino rats. Alex. J. Vet. Sci. 2017, 53, 167–179. [Google Scholar] [CrossRef]
- Tredici, M.R. Mass Production of Microalgae: Photobioreactors. In Handbook of Microalgal Culture; Wiley-Blackwell: Hoboken, NJ, USA, 2003; pp. 178–214. [Google Scholar]
- Moisander, P.H.; McClinton Lii, E.; Paerl, H.W. Salinity Effects on Growth, Photosynthetic Parameters, and Nitrogenase Activity in Estuarine Planktonic Cyanobacteria. Microb. Ecol. 2002, 43, 432–442. [Google Scholar] [CrossRef]
- Mary Leema, J.T.; Kirubagaran, R.; Vinithkumar, N.V.; Dheenan, P.S.; Karthikayulu, S. High value pigment production from Arthrospira (Spirulina) platensis cultured in seawater. Bioresour. Technol. 2010, 101, 9221–9227. [Google Scholar] [CrossRef] [PubMed]
- Ciferri, O. Spirulina, the edible microorganism. Microbiol. Rev. 1983, 47, 551–578. [Google Scholar] [CrossRef]
- Al Mahrouqi, H.; Naqqiuddin, M.A.; Achankunju, J.; Omar, H.; Ismail, A. Different salinity effects on the mass cultivation of Spirulina (Arthrospira platensis) under sheltered outdoor conditions in Oman and Malaysia. J. Algal. Biomass. Util. 2015, 6, 1–14. [Google Scholar]
- Li, Q.; Chang, R.; Sun, Y.; Li, B. iTRAQ-Based Quantitative Proteomic Analysis of Spirulina platensis in Response to Low Temperature Stress. PLoS ONE 2018, 13, e0196442. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, X.; Ki, F.; Dahlgen, R.; Wang, H. Identification of microRNA-size sRNAs Related to Salt Tolerance in Spirulina platensis. Plant Mol. Biol. Rep. 2016, 34, 539–555. [Google Scholar] [CrossRef]
- Shalaby, E.A.; Shanab, S.M.M.; Singh, V. Salt stress enhancement of antioxidant and antiviral efficiency of Spirulina platensis. J. Med. Plants Res. 2010, 4, 2622–2632. [Google Scholar]
- Sandeep, K.P.; Shukla, S.P.; Harikrishna, V.; Muralidhar, A.P.; Vennila, A.; Purushothaman, C.S.; Kumar, R.R. Utilization of inland saline water for Spirulina cultivation. J. Water Reuse Desalination 2013, 3, 346–356. [Google Scholar] [CrossRef]
- Tam, L.T.; Le, T.T.; Nguyen, C.H.; Hoang, T.M.H.; Ngo, T.H.T.; Dang, D.H. Assessing the salt tolerance of Spirulina platensis freshwater strains and examining cheap culture media for cultivation of the potential strain. Vietnam. J. Biotechnol. 2021, 19, 381–392. [Google Scholar] [CrossRef]
- Dang, D.H.; Le, T.T.; Nguyen, C.H.; Le, A.H.; Hoang, T.M.H.; Luu, T.T.; Ngo, T.H.T. Selection of suitable medium for the culture of the salt tolerant Spirulina platensis ST strain under laboratory and pilot conditions. In Proceedings of the National Biotechnology Conference 2022, Dak Lak, Vietnam, 4 November 2022. [Google Scholar]
- Zarrouk, C. Contribution à L’éTude D’Une Cyanophycée Influencée de Divers Facteurs Physiques ET Chimiques sur la Croissance ET la Photo-Synthèse de Spirulina Maxima (Setch. ET Gardner) Geitler. Ph.D. Thesis, University of Paris, Paris, France, 1966. [Google Scholar]
- Guillard, R.R.L.; Sieracki, M.S. Counting cells in cultures with the light microscope. In Algal Culturing Techniques; Elsevier: Amsterdam, The Netherlands, 2005; pp. 239–252. [Google Scholar]
- Dang, D.H. Cultivation of Nutrient-Rich Microalgae for Application as Functional Food for Humans and Domestic Animals in Vietnam; Natural Science and Technology: Hanoi, Vietnam, 2019. [Google Scholar]
- Wellburn, A.R.; Lichtenthaler, H. Formulae and program to determine total carotenoids and chlorophylls a and b of leaf extracts in different solvents. In Advances in Photosynthesis Research, Proceedings of the VIth International Congress on Photosynthesis, Brussels, Belgium, 1–6 August 1983; Sybesma, C., Ed.; Springer: Dordrecht, The Netherlands, 1984; pp. 9–12. [Google Scholar]
- Hadiyanto, H.; Suttrisnorhadi, S. Response surface optimization of ultrasound assisted extraction (UAE) of phycocyanin from microalgae Spirulina platensis. Emir. J. Food Agric. 2016, 28, 227–234. [Google Scholar] [CrossRef] [Green Version]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Soni, B.; Kalavadia, B.; Trivedi, U.; Madamwar, D. Extraction, purification and characterization of phycocyanin from Oscillatoria quadripunctulata—Isolated from the rocky shores of Bet-Dwarka, Gujarat, India. Process. Biochem. 2006, 41, 2017–2023. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Sun, L.; Ren, L.; Zhuang, X.; Ji, X.; Yan, J.; Huang, H. Differential effects of nutrient limitations on biochemical constituents and docosahexaenoic acid production of Schizochytrium sp. Bioresour. Technol. 2014, 159, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Tam, L.T.; Ha, N.C.; Thom, L.T.; Zhu, J.; Wakisaka, M.; Hong, D.D. Ferulic acid extracted from rice bran as a growth promoter for the microalga Nannochloropsis oculata. J. Appl. Phycol. 2021, 33, 37–45. [Google Scholar] [CrossRef]
- Kitajima, M.; Butler, W.L. Quenching of chlorophyll fluorescence and primary photochemistry in chloroplasts by dibromothymoquinone. Biochim. Biophys. Acta Bioenerg. 1975, 376, 105–115. [Google Scholar] [CrossRef]
- Qiu, Z.; Wang, L.; Zhou, Q. Effects of bisphenol A on growth, photosynthesis and chlorophyll fluorescence in above-ground organs of soybean seedlings. Chemosphere 2013, 90, 1274–1280. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Chen, Y.H.; Liao, Y.C.; Huang, X.W.; Lu, T.J. Effect of hot water extracts of Arthrospira maxima (Spirulina) against respiratory syncytial virus. Phytomedicine 2023, 110, 154611. [Google Scholar] [CrossRef]
- Gagneux-Moreaux, S.; Moreau, C.; Gonzalez, J.L.; Cosson, R.P. Diatom artificial medium (DAM): A new artificial medium for the diatom Haslea ostrearia and other marine microalgae. J. Appl. Phycol. 2007, 19, 549–556. [Google Scholar] [CrossRef] [Green Version]
- Vonshak, A.; Guy, R.; Guy, M. The response of the filamentous cyanobacterium Spirulina platensis to salt stress. Arch. Microbiol. 1988, 150, 417–420. [Google Scholar] [CrossRef]
- Bolsunovskii, A.Y.; Kosinenko, S.V. Intracellular phosphorus pool of the cyanobacterium Spirulina platensis. Microbiology 2000, 69, 116–118. [Google Scholar] [CrossRef]
- Moussa, H.R.; Hassan, M.A.E.F. Growth Enhancers to Mitigate Salinity Stress in Vicia faba. Int. J. Veg. Sci. 2016, 22, 243–250. [Google Scholar] [CrossRef]
- Tester, M.; Davenport, R. Na+ Tolerance and Na+ Transport in Higher Plants. Ann. Bot. 2003, 91, 503–527. [Google Scholar] [CrossRef]
- Bezerra, P.Q.M.; Moraes, L.; Cardoso, L.G.; Druzian, J.I.; Morais, M.G.; Nunes, I.L.; Costa, J.A.V. Spirulina sp. LEB 18 cultivation in seawater and reduced nutrients: Bioprocess strategy for increasing carbohydrates in biomass. Bioresour. Technol. 2020, 316, 123883. [Google Scholar] [CrossRef]
- Sili, C.; Torzillo, G.; Vonshak, A. Arthrospira (Spirulina). In Ecology of Cyanobacteria II: Their Diversity in Space and Time; Whitton, B.A., Ed.; Springer: Dordrecht, The Netherlands, 2012; pp. 677–705. [Google Scholar]
- Ravelonandro, P.H.; Ratianarivo, D.H.; Joannis-Cassan, C.; Isambertc, A.; Raherimandimby, M. Improvement of the growth of Arthrospira (Spirulina) platensis from Toliara (Madagascar): Effect of agitation, salinity and CO2 addition. Food Bioprod. Process. 2011, 89, 209–216. [Google Scholar] [CrossRef] [Green Version]
- Blumwald, E.; Tel-Or, E. Salt Adaptation of the Cyanobacterium Synechococcus 6311 Growing in a Continuous Culture (Turbidostat). Plant Physiol. 1984, 74, 183–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.; Vonshak, A. Effects of salinity stress on photosystem II function in cyanobacterial Spirulina platensis cells. Physiol. Plant 2002, 114, 405–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheevadhanarak, S.; Paithoonrangsarid, K.; Prommeenate, P.; Kaewngam, W.; Musigkain, A.; Tragoonrung, S.; Tabata, S.; Kaneko, T.; Chaijaruwanich, J.; Sangsrakru, D.; et al. Draft genome sequence of Arthrospira platensis C1 (PCC9438). Stand. Genomic Sci. 2012, 6, 43–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumaresan, V.; Nizama, F.; Ravichandran, G.; Viswanathan, K.; Palanisamy, R.; Bhatt, P.; Arasu, M.V.; Al-Dhabi, N.A.; Mala, K.; Arockiaraj, J. Transcriptome changes of blue-green algae, Arthrospira sp. in response to sulfate stress. Algal Res. 2017, 23, 96–103. [Google Scholar] [CrossRef]


| Nutrition Composition (g/L) | SOT Medium | ||||
|---|---|---|---|---|---|
| 100% (S1) | 25% (S2) | 50% (S3) | 75% (S4) | 100% (S5) | |
| Distilled water (L) | 1.000 | 0 | 0 | 0 | 0 |
| Seawater 30‰ (L) | 0 | 1.000 | 1.000 | 1.000 | 1.000 |
| NaHCO3 | 16.800 | 4.200 | 8.400 | 12.600 | 16.800 |
| K2HPO4·3H2O | 0.655 | 0.164 | 0.328 | 0.493 | 0.655 |
| NaNO3 | 2.500 | 0.625 | 1.250 | 1.875 | 2.500 |
| K2SO4 | 1.000 | 0.250 | 0.500 | 0.750 | 1.000 |
| NaCl | 1.000 | 0.250 | 0.500 | 0.750 | 1.000 |
| MgSO4·7H2O | 0.200 | 0.050 | 0.100 | 0.150 | 0.200 |
| CaCl2·2H2O | 0.040 | 0.010 | 0.020 | 0.030 | 0.040 |
| FeSO4·7H2O | 0.010 | 0.0025 | 0.005 | 0.0075 | 0.010 |
| Na2EDTA | 0.080 | 0.020 | 0.040 | 0.060 | 0.080 |
| A5 (mL) | 1.000 | 0.250 | 0.500 | 0.750 | 1.000 |
| Final salinity (‰) | 13 | 42 | 45 | 48 | 52 |
| Parameters (SOT Medium) | Protein (% DCW) | Carbohydrate (% DCW) | Lipid (% DCW) | Phycocyanin (% DCW) | Chlorophyll a (%DCW) | Carotenoid (%DCW) | |
|---|---|---|---|---|---|---|---|
| 13‰ salinity | S1 | 57.75 ± 0.10 | 21.63 ± 0.46 | 6.81 ± 0.40 | 10.90 ± 2.37 | 0.59 ± 0.04 | 0.23 ± 0.00 |
| 42‰ salinity | S2 | 48.73 ± 0.81 | 22.14 ± 0.11 | 7.32 ± 0.61 | 10.23 ± 0.25 | 0.53 ± 0.04 | 0.12 ± 0.01 |
| 45‰ salinity | S3 | 47.17 ± 0.02 | 18.22 ± 0.15 | 8.70 ± 0.25 | 10.17 ± 0.81 | 0.56 ± 0.06 | 0.21 ± 0.00 |
| 48‰ salinity | S4 | 41.16 ± 0.06 | 17.47 ± 0.18 | 11.14 ± 0.33 | 11.18 ± 0.25 | 0.58 ± 0.05 | 0.30 ± 0.04 |
| 52‰ salinity | S5 | 38.94 ± 0.29 | 16.55 ± 0.38 | 10.35 ± 0.55 | 5.90 ± 0.37 | 0.32 ± 0.03 | 0.18 ± 0.04 |
| Sample | Total Reads | Total Bases | Mapped Reads | Uniq Mapped Reads | Multiple Map Reads | GC (%) | Q30 (%) |
|---|---|---|---|---|---|---|---|
| S1 | 18,240,104 | 1,842,250,504 | 4,279,358 (23.46%) | 3,770,540 (20.67%) | 508,818 (2.79%) | 45.50 | 93.40 |
| S2 | 18,163,070 | 1,834,470,070 | 6,616,267 (36.43%) | 4,962,102 (27.32%) | 1,654,165 (9.11%) | 45.00 | 94.10 |
| S3 | 18,187,025 | 1,836,889,525 | 5,681,799 (31.24%) | 4,906,667 (26.98%) | 775,132 (4.26%) | 45.00 | 93.10 |
| S4 | 18,241,587 | 1,842,400,287 | 6,485,723 (35.56%) | 5,707,645 (31.29%) | 778,078 (4.27%) | 46.80 | 93.70 |
| S5 | 18,313,624 | 1,849,676,024 | 2,806,231 (15.33%) | 2,414,712 (13.19%) | 391,519 (2.14%) | 47.30 | 93.30 |
| Total | 91,145,410 | 9,205,686,410 | 25,869,378 | 21,761,666 | 4,107,712 |
| Sample | Total Protein-Coding Genes | Number of Transcripts | Average Number of Transcripts per Genes | Lowest Transcript Count | Highest Transcript Count |
|---|---|---|---|---|---|
| S1 | 5717 | 2,752,902 | 482 | 1 | 8664 |
| S2 | 5757 | 3,720,160 | 646 | 1 | 8286 |
| S3 | 5741 | 3,554,502 | 619 | 1 | 7068 |
| S4 | 5737 | 4,225,425 | 737 | 1 | 8835 |
| S5 | 5692 | 1,767,359 | 310 | 1 | 4775 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, D.D.; Hien, H.T.M.; Thom, L.T.; Ha, N.C.; Huy, L.A.; Thu, N.T.H.; Cuong, N.; Tang, D.Y.Y.; Show, P.L. Transcriptome Analysis of Spirulina platensis sp. at Different Salinity and Nutrient Compositions for Sustainable Cultivation in Vietnam. Sustainability 2023, 15, 11906. https://doi.org/10.3390/su151511906
Hong DD, Hien HTM, Thom LT, Ha NC, Huy LA, Thu NTH, Cuong N, Tang DYY, Show PL. Transcriptome Analysis of Spirulina platensis sp. at Different Salinity and Nutrient Compositions for Sustainable Cultivation in Vietnam. Sustainability. 2023; 15(15):11906. https://doi.org/10.3390/su151511906
Chicago/Turabian StyleHong, Dang Diem, Hoang Thi Minh Hien, Le Thi Thom, Nguyen Cam Ha, Le Anh Huy, Ngo Thi Hoai Thu, Nguyen Cuong, Doris Ying Ying Tang, and Pau Loke Show. 2023. "Transcriptome Analysis of Spirulina platensis sp. at Different Salinity and Nutrient Compositions for Sustainable Cultivation in Vietnam" Sustainability 15, no. 15: 11906. https://doi.org/10.3390/su151511906
APA StyleHong, D. D., Hien, H. T. M., Thom, L. T., Ha, N. C., Huy, L. A., Thu, N. T. H., Cuong, N., Tang, D. Y. Y., & Show, P. L. (2023). Transcriptome Analysis of Spirulina platensis sp. at Different Salinity and Nutrient Compositions for Sustainable Cultivation in Vietnam. Sustainability, 15(15), 11906. https://doi.org/10.3390/su151511906







