Correlation of the Effect of Native Bioagents on Soil Properties and Their Influence on Stem Rot Disease of Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Pathogen and Bio-Control Agents
2.2. Antagonism Assay
2.3. In Vivo Application of S. oryzae and Its Severity in Plants
2.4. Seed and Soil Treatment
2.5. Sample Collection and Estimation of Defense-Related Enzymes
2.5.1. Phenylalanine Ammonia-Lyase (PAL) Assay
2.5.2. Peroxidase (PO) Assay
2.5.3. Polyphenol Oxidase (PPO) Assay
2.5.4. Total Phenol Content (TPC) Assay
2.6. Soil Analysis
2.6.1. Estimation of Soil Chemical Properties
2.6.2. Estimation of Soil Enzymes
Assay of Dehydrogenase Enzyme
Assay of Phosphatase Enzyme
Assay of Urease Enzyme
2.7. Estimation of Plant Nutrient Analysis
2.8. Statistical Analysis
3. Results
3.1. Antagonism Assay
3.2. In Vivo Application of S. oryzae and Its Severity in Plants
3.3. Plant Growth Promotion and Yield Attributes
3.4. Biochemical Parameters
3.5. Soil Chemical Properties
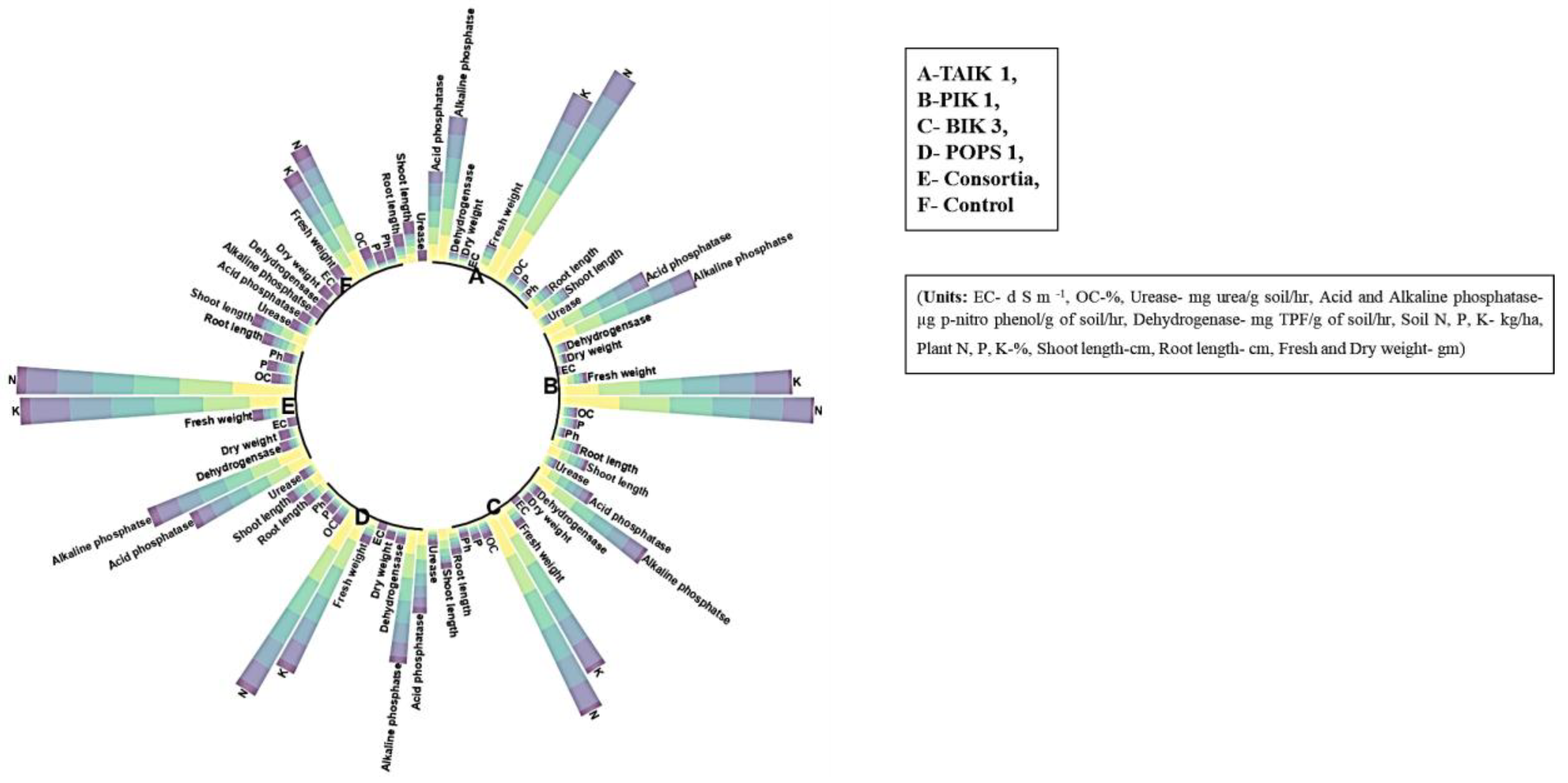
3.6. Soil Enzymes
3.7. Plant Nutrients
3.8. Correlation Analysis
3.9. Stepwise Regression Analysis
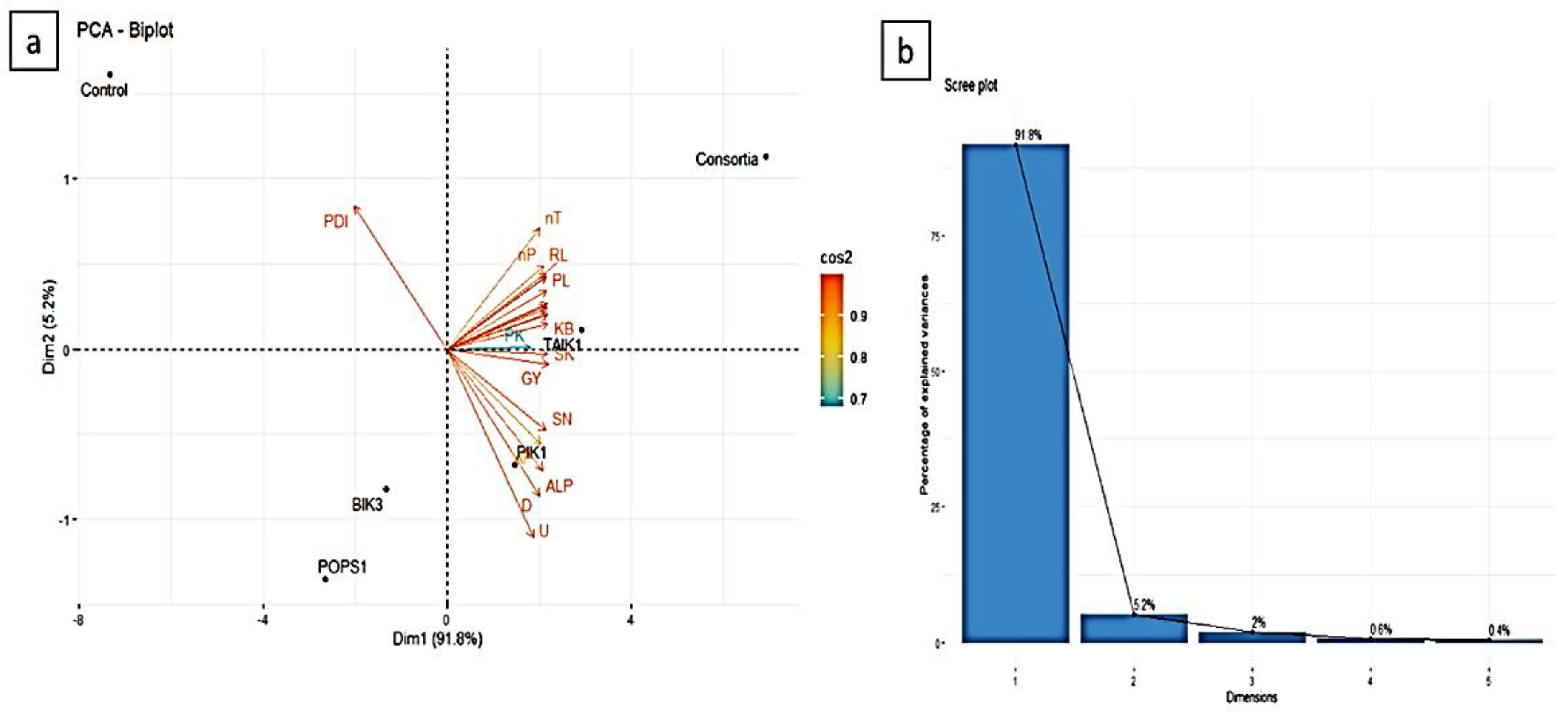
3.10. PCA
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Neher, D.A. Soil community composition and ecosystem processes: Comparing agricultural ecosystems with natural ecosystems. Agrofor. Syst. 1999, 45, 159–185. [Google Scholar] [CrossRef]
- Raj, A.; Jhariya, M.K.; Yadav, D.K.; Banerjee, A.; Meena, R.S. Soil for sustainable environment and ecosystems management. In Sustainable Agriculture, Forest and Environmental Management; Springer: Berlin/Heidelberg, Germany, 2019; pp. 189–221. [Google Scholar]
- Fan, K.; Chu, H.; Eldridge, D.J.; Gaitan, J.J.; Liu, Y.R.; Sokoya, B.; Wang, J.T.; Hu, H.W.; He, J.Z.; Sun, W.; et al. Soil biodiversity supports the delivery of multiple ecosystem functions in urban greenspaces. Nat. Ecol. Evol. 2023, 7, 113–126. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Else, K.B.; Giulia, B.; Zhanguo, B.; Rachel, E.C.; Gerlinde, D.D.; Ron, G.; Luuk, F.; Violette, G.; Thom Erdel, E.; Simsek, U.; et al. Effects of Fungi on Soil Organic Carbon and Soil Enzyme Activity under Agricultural and Pasture Land of Eastern Türkiye. Sustainability 2023, 15, 1765. [Google Scholar]
- Nikita, B.; Puneet, S.C. Excessive and Disproportionate Use of Chemicals Cause Soil Contamination and Nutritional. In Soil Contamination-Threats and Sustainable Solutions; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- Jagaba, A.H.; Kutty, S.R.M.; Isa, M.H.; Ghaleb, A.A.S.; Lawal, I.M.; Usman, A.K.; Birniwa, A.H.; Noor, A.; Abubakar, S.; Umaru, I.; et al. Toxic effects of xenobiotic compounds on the microbial community of activated sludge. ChemBioEng Rev. 2022, 9, 497–535. [Google Scholar] [CrossRef]
- Jianhua, Q.; Zhuoran, L.; Zhihuan, W.; Fuxuan, B.; Shuqi, W.; Min, D.; Qi, H.; Yifan, W.; Hui, Y.; Ying, Z. Cyclodextrin-functionalized magnetic alginate microspheres for synchronous removal of lead and bisphenol a from contaminated Soil. Chem. Eng. J. 2023, 461, 142079. [Google Scholar]
- Evangelia, E.G. The impact of heavy metal contamination on soil quality and plant nutrition. Sustainable management of moderate contaminated agricultural and urban soils, using low cost materials and promoting circular economy. Sustain. Chem. Pharm. 2023, 33, 101046. [Google Scholar]
- Jianhua, Q.; Ruixin, L.; Xuewei, B.; Zhuoran, L.; Kaige, L.; Qi, H.; Xinru, Z.; Guangshan, Z.; Shouyi, M.; Ying, Z. Remediation of atrazine contaminated soil by microwave activated persulfate system: Performance, mechanism and DFT calculation. J. Clean. Prod. 2023, 399, 136546. [Google Scholar]
- Jianhua, Q.; Yihang, Y.; Xinmiao, Z.; Lei, W.; Yue, T.; Zhao, J.; Hui, Y.; Min, D.; Ying, Z. Stabilization of lead and cadmium in soil by sulfur-iron functionalized biochar: Performance, mechanisms and microbial community evolution. J. Hazard. Mater. 2022, 425, 127876. [Google Scholar]
- Punam, K.; Mukesh, M.; Pooja, G.; Manish, K.D.; Gopal, N.; Upadhyay, R.S. Plant growth promoting rhizobacteria and their biopriming for growth promotion in mung bean (Vigna radiata (L.) R. Wilczek). Biocatal. Agric. Biotechnol. 2018, 16, 163–171. [Google Scholar]
- Kumar, S.; Chandra, R.; Behera, L.; Keswani, C.; Sansinenea, E. Dual Trichoderma consortium mediated elevation of systemic defense response against early blight in potato. Eur. J. Plant Pathol. 2021, 162, 681–696. [Google Scholar] [CrossRef]
- de Andrade, L.A.; Santos, C.H.B.; Frezarin, E.T.; Sales, L.R.; Rigobelo, E.C. Plant growth-promoting rhizobacteria for sustainable agricultural production. Microorganisms 2023, 11, 1088. [Google Scholar] [CrossRef]
- Kumari, S.; Bharat, N.K.; Thakur, A.K. Role of plant growth-promoting rhizobacteria (PGPR) and bio-control agents (BCAs) in crop production. Int. J. Econ. Plants 2020, 7, 144–150. [Google Scholar] [CrossRef]
- Mishra, R.K.; Sahu, P.K.; Mishra, V.; Jamal, H.; Varma, A.; Tripathi, S. Isolation and characterization of halotolerant plant growth promoting rhizobacteria from mangrove region of Sundarbans, India for enhanced crop productivity. Front. Plant Sci. 2023, 14, 1122347. [Google Scholar]
- Neina, D. The role of soil pH in plant nutrition and soil remediation. Appl. Environ. Soil Sci. 2019, 2019, 5794869. [Google Scholar] [CrossRef] [Green Version]
- Shah, K.K.; Tripathi, S.; Tiwari, I.; Shrestha, J.; Modi, B.; Paudel, N.; Das, B.D. Role of soil microbes in sustainable crop production and soil health: A review. Agric. Sci. Technol. 2021, 13, 1313–1336. [Google Scholar] [CrossRef]
- Pooja, S.; Rajesh, K. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar]
- Chandran, H.; Meena, M.; Swapnil, P. Plant growth-promoting rhizobacteria as a green alternative for sustainable agriculture. Sustainability 2021, 13, 10986. [Google Scholar] [CrossRef]
- Muhammad, I.R.; Liyakat, H.M.; Tanvir, S.; Talal, A.; Iqbal, M.I.; Ismail, M.O. Bacteria and fungi can contribute to nutrients bioavailability and aggregate formation in degraded soils. Microbiol. Res. 2016, 183, 26–41. [Google Scholar]
- Singh, D.P.; Prabha, R.; Renu, S.; Sahu, P.K.; Singh, V. Agrowaste bioconversion and microbial fortification have prospects for soil health, crop productivity, and eco-enterprising. Int. J. Recycl. Org. Waste Agric. 2019, 8, 457–472. [Google Scholar] [CrossRef] [Green Version]
- Shen, H.; He, X.; Liu, Y.; Chen, Y.; Tang, J.; Guo, T. A Complex Inoculant of N2-Fixing, P- and K-Solubilizing Bacteria from a Purple Soil Improves the Growth of Kiwifruit (Actinidia chinensis) Plantlets. Front. Microbiol. 2016, 7, 841. [Google Scholar] [CrossRef]
- Meena, V.S.; Meena, S.K.; Verma, J.P.; Kumar, A.; Aeron, A.; Mishra, P.K.; Bisht, J.K.; Pattanayak, A.; Naveed, M.; Dotaniya, M.L. Plant beneficial rhizospheric microorganism (PBRM) strategies to improve nutrients use efficiency: A review. Ecol. Eng. 2017, 107, 8–32. [Google Scholar] [CrossRef]
- Gopika, K.; Jagadeeshwar, R.; Rao, V.K.; Vijayalakshmi, K. An overview of stem rot disease of rice (Sclerotium oryzae catt.) and its comprehensive management. Int. J. Appl. Biol. Pharml. Technol. 2016, 7, 111–124. [Google Scholar]
- Pramesh, D.; Alase, S.; Muniraju, K.M.; Kumara, M.K. A combination fungicide for the management of sheath blight, sheath rot and stem rot diseases of paddy. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 3500–3509. [Google Scholar]
- Bashyal, B.M.; Parmar, P.; Zaidi, N.W.; Sunil, K.S.; Prakash, G.; Rashmi, A. Improved methodology for the isolation of false smut pathogen Ustilaginoidea virens of rice. Indian Phytopathol. 2021, 74, 249–252. [Google Scholar] [CrossRef]
- Kannan, C.; Mishra, D.; Miriyala, A.; Prakasam, V.; Basavaraj, K.; Jasudasu, G.; Srinivas, P.M.; Sundaram, R.M. Native isolates of Trichoderma as bio-suppressants against sheath blight and stem rot pathogens of rice. Egypt. J. Biol. Pest Control. 2021, 31, 12. [Google Scholar]
- Sinclair, J.B.; Dhingra, O.D. Basic Plant Pathology Methods; CRC Press: Boca Raton, FL, USA, 2017; pp. 287–300. [Google Scholar]
- Gangwar, G.P.; Sinha, A.P. Comparative antagonistic potential of Trichoderma spp. against Xanthomonas oryzae pv. oryzae. Ann. Plant Sci. 2010, 8, 458–463. [Google Scholar]
- Kumar, V.; Singh, R.; Jalali, B.L. Evaluation of resistance to stem rot and yield losses caused by the disease in rice. Indian Phytopathol. 2003, 4, 403–407. [Google Scholar]
- International Rice Research Institute (IRRI). Standard Evaluation System for Rice; International Rice Research Institute (IRRI): Los Banos, Manila, Philippines, 2002; p. 24. Available online: http://www.knowledgebank.irri.org/images/docs/rice-standard-evaluation-system.pdf (accessed on 10 June 2023).
- Brueske, C.H. Phenylalanine ammonia lyase activity in tomato roots infected and resistant to the root-knot nematode, Meloidogyne incognita. Physiol. Plant Pathol. 1980, 16, 409–414. [Google Scholar] [CrossRef]
- Hammerschmidt, R.; Nuckles, E.M.; Kuc, J. Association of enhanced peroxidase activity with induced systemic resistance of cucumber to Colletotrichum lagenarium. Physiol. Plant Pathol. 1982, 20, 73–82. [Google Scholar] [CrossRef]
- Gauillard, F.; Richardforget, F.; Nicolas, J. New spectrophotometric assay for polyphenol oxidase active ity. Anal. Biochem. 1993, 215, 59–65. [Google Scholar] [CrossRef]
- Malick, C.P.; Singh, M.B. Plant Enzymology and Histo Enzymology; Kalyani Publishers: New Delhi, India, 1980; p. 286. [Google Scholar]
- McLean, E.O. Soil pH and lime requirement. In Methods of Soil Analysis: Part. 2 Chemical and Microbiological Properties; Wiley: Hoboken, NJ, USA, 1983; Volume 9, pp. 199–224. [Google Scholar]
- Rhoades, J.D. Soluble Salts. In Methods of Soil Analysis: Part. 2 Chemical and Microbiological Properties; Agronomy Monographs; American Society of Agronomy: Madison, WI, USA, 1983; pp. 167–179. [Google Scholar]
- Walkley, A.; Black, L.A. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Subbaiah, B.V. A rapid procedure for estimation of available nitrogen in Soil. Curr. Sci. 1956, 25, 259–260. [Google Scholar]
- Watanabe, F.S.; Olsen, S.R. Test of an ascorbic acid method for determining phosphorus in water and NaHCO3 extracts from Soil. Soil Sci. Soc. Am. J. 1965, 29, 677–678. [Google Scholar] [CrossRef]
- Muhr, G.R. Soil testing in India. In USAID Mission to India; USAID: New Delhi, India, 1965; p. 120. [Google Scholar]
- Casida, J.L.E.; Klein, D.A.; Santoro, T. Soil dehydrogenase activity. Soil Sci. 1964, 98, 371–376. [Google Scholar] [CrossRef]
- Tabatabai, M.A.; Bremner, J.M. Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biol. Biochem. 1969, 1, 301–307. [Google Scholar] [CrossRef]
- Eivazi, F.; Tabatabai, M.A. Phosphatases in soils. Soil Biol. Biochem. 1977, 9, 167–172. [Google Scholar] [CrossRef]
- Piper, C.S. Vanado-Molybdo-Phosphoric Yellow Colour Method. Soil Sci. Plant Anal. 1966, 10, 368. [Google Scholar]
- Chapman, H.D.; Pratt, P.F. Methods of Analysis for Soils, Plants and Waters; University of California, Division of Agricultural Sciences: Oakland, CA, USA, 1961. [Google Scholar]
- Husson, F.; Josse, J.; Le, S.; Mazet, J. FactoMineR: Multivariate Exploratory Data Analysis and Data Mining. R Package Version 2.3. 2020. Available online: https://CRAN.R-project.org/package=FactoMineR (accessed on 3 June 2023).
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses, R Package v. 1.0.7. 2020. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 3 June 2023).
- Kannan, C.; Divya, M.; Rekha, G.; Maruthi, P.; Hajira, S.; Sundaram, R.M. Diversity analysis of antagonistic microbes against bacterial leaf and fungal sheath blight diseases of rice. Egypt. J. Biol. Pest Cont. 2021, 31, 115. [Google Scholar] [CrossRef]
- Nawaz, M.E.; Malik, K.; Hassan, M.N. Rice-associated antagonistic bacteria suppress the Fusarium fujikoroi causing rice bakanae disease. Biol. Cont. 2022, 67, 101–109. [Google Scholar] [CrossRef]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the Antimicrobial Compounds Produced by Members of the Bacillus subtilis group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef] [Green Version]
- Adrien, B.; Martin, F. Phenazines in plant-beneficial Pseudomonas spp. biosynthesis, regulation, function and genomics. Environ. Microbiol. 2018, 20, 3905–3917. [Google Scholar]
- Pandit, M.A.; Kumar, J.; Gulati, S.; Bhandari, N.; Mehta, P.; Katyal, R.; Rawat, C.D.; Mishra, V.; Kaur, J. Major Biological Control Strategies for Plant Pathogens. Pathogens 2022, 11, 273. [Google Scholar] [CrossRef]
- Moubark, M.Y.; Abdel-Monaim, M.F. Effect of bio-control agents on yield, yield components and root rot control in two wheat cultivars at New Valley region. Not. Sci. Biol. 2011, 3, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Ivica, D.; Tamara, J.; Marija, P.; Giuliano, D.; Djordje, F. Plant-associated Bacillus and Pseudomonas antimicrobial activities in plant disease suppression via biological control mechanisms—A review. Physiol. Mol. Plant Pathol. 2022, 117, 101754. [Google Scholar]
- Stuti, S.; Shweena, K.; Rajni, S. Pseudomonas mediated nutritional and growth promotional activities for sustainable food security. Microbiol. Curr. Res. 2021, 2, 2666–5174. [Google Scholar]
- Tian, J.; Ge, F.; Zhang, D.; Deng, S.; Liu, X. Roles of Phosphate Solubilizing Microorganisms from Managing Soil Phosphorus Deficiency to Mediating Biogeochemical P Cycle. Biol. J. 2021, 10, 158. [Google Scholar] [CrossRef]
- Sowmya, V.; Raja Gopalan, N.S.; Maruthi, P.; Baskar, M.; Umadevi, G.; Ladha Lakshmi, D.; Koteshwar, P.; Jesudasu, G.; Santhosha, R.; Srinivas, P.; et al. Native bio-control agents from the rice fields of Telangana, India: Characterization and unveiling the potential against stem rot and false smut diseases of rice. Res. Sq. 2023. [Google Scholar] [CrossRef]
- Burns, R.G. Enzyme activity in soil: Location and a possible role in microbial ecology. Soil Biol. Biochem. 1982, 14, 423–427. [Google Scholar] [CrossRef]
- Mukherjee, M.; Mukherjee, P.K.; Horwitz, B.A.; Zachow, C.; Berg, G.; Zeilinger, S. Trichoderma-plant-pathogen interactions: Advances in genetics of biological control. Indian J. Microbiol. 2012, 52, 522–529. [Google Scholar] [CrossRef] [Green Version]
- Singh, J.S.; Koushal, S.; Kumar, A.; Vimal, S.R.; Gupta, V.K. Book review: Microbial inoculants in sustainable agricultural Productivity—Vol. II: Functional application. Front. Microbiol. 2016, 7, 7. [Google Scholar] [CrossRef] [Green Version]
- Halifu, S.; Deng, X.; Song, X.; Song, R.; Liang, X. Inhibitory mechanism of Trichoderma virens ZT05 on Rhizoctonia solani. Plants 2020, 9, 912. [Google Scholar] [CrossRef]
- Rais, A.; Jabeen, Z.; Shair, F.; Hafeez, F.; Hassan, M. Bacillus spp. a bio-control agent enhances the activity of antioxidant defense enzymes in rice against Pyricularia oryzae. PLoS ONE 2017, 12, e0187412. [Google Scholar] [CrossRef] [Green Version]
- Wiesel, L.; Newton, A.C.; Elliott, I.; Booty, D.; Gilroy, E.M.; Birch, P.R.J.; Hein, I. Molecular effects of resistance elicitors from biological origin and their potential for crop protection. Front. Plant Sci. 2014, 5, 655. [Google Scholar] [CrossRef] [Green Version]
- Deice Raasch-Fernandes, L.; Bonaldo, S.M.; Jesus Rodrigues, D.; Magela Vieira-Junior, G.; Regina Freitas Schwan-Estrada, K.; Rocco da Silva, C.; Gabriela Araújo Verçosa, A.; Lopes de Oliveira, D.; Wender Debiasi, B. Induction of phytoalexins and proteins related to pathogenesis in plants treated with extracts of cutaneous secretions of southern Amazonian Bufonidae amphibians. PLoS ONE 2019, 14, e0211020. [Google Scholar] [CrossRef] [Green Version]
- Zandi, P.; Schnug, E. Reactive Oxygen Species, Antioxidant Responses and Implications from a Microbial Modulation Perspective. Biology 2022, 11, 155. [Google Scholar] [CrossRef]
- Kaur, S.; Samota, M.K.; Choudhary, M.; Abhay, K.P.; Anshu, S.; Julie, T. How do plants defend themselves against pathogens-Biochemical mechanisms and genetic interventions. Physiol. Mol. Biol. Plants 2022, 28, 485–504. [Google Scholar] [CrossRef]
- Aryal, S.; Baniya, M.K.; Danekhu, K.; Kunwar, P.; Gurung, R.; Koirala, N. Total Phenolic Content, Flavonoid Content and Antioxidant Potential of Wild Vegetables from Western Nepal. Plants 2019, 8, 96. [Google Scholar] [CrossRef] [Green Version]
- Jacoby, R.; Peukert, M.; Succurro, A.; Koprivova, A.; Kopriva, S. The Role of Soil Microorganisms in Plant Mineral Nutrition-Current Knowledge and Future Directions. Front. Plant Sci. 2017, 8, 1617. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, M.I.; Biswas, A.K.; Sultana, M.S.; Sarker, B.C.; Mondal, C. Effect of Different Media and pH on Mycelial Growth of Sclerotium oryzae Causing Sheath Rot of Rice. South Asian J. Agric. 2019, 18, 67–72. [Google Scholar] [CrossRef]
- Du, J.X.; Li, Y.; Ur-Rehman, S.; Mukhtar, I.; Yin, Z.; Dong, H.; Wang, H.; Zhang, X.; Gao, Z.; Zhao, X.; et al. Synergistically promoting plant health by harnessing synthetic microbial communities and prebiotics. iScience 2021, 24, 102918. [Google Scholar] [CrossRef]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Keim, R.; Webster, R.K. Fungistasis of sclerotia of Sclerotium oryzae. Phytopathology 1975, 65, 283–287. [Google Scholar] [CrossRef]
- Yadav, R.L.; Shukla, S.K.; Suman, A.; Singh, P.N. Trichoderma inoculation and trash management effects on soil microbial biomass, soil respiration, nutrient uptake and yield of ratoon sugarcane under subtropical conditions. Biol. Fertil. Soils 2009, 45, 461–468. [Google Scholar] [CrossRef]
- Kapri, A.; Tewari, L. Phosphate solubilization potential and phosphatase activity of rhizospheric Trichoderma spp. Braz. J. Microbiol. 2010, 41, 787–795. [Google Scholar] [CrossRef] [Green Version]
- Kucuk, C.; Kivanc, M.; Kinaci, E.; Kinaci, G. Determination of the growth and solubilization capabilities of Trichoderma harzianum T1. Biologia 2008, 63, 167–170. [Google Scholar] [CrossRef]
- Nannipieri, P.; Kandeler, E.; Ruggiero, P. Enzyme activities and microbiological and biochemical processes in Soil. In Enzymes in the Environment; Marcel Dekker Inc.: New York, NY, USA, 2002; pp. 1–33. [Google Scholar]
- Gil-Sotres, F.; Trasar-Cepeda, C.; Leirós, M.C.; Seoane, S. Different approaches to evaluating soil quality using biochemical properties. Soil Biol. Biochem. 2005, 37, 877–887. [Google Scholar] [CrossRef]
- Ng, C.W.W.; Yan, W.H.; Tsim, K.W.K.; So, P.S.; Xia, Y.T.; To, C.T. Effects of Bacillus subtilis and Pseudomonas fluorescens as the soil amendment. Heliyon 2022, 8, e11674. [Google Scholar] [CrossRef]
- Qiu, L.P.; Liu, J.; Wang, Y.Q.; Sun, H.M.; He, W.X. Research on relationship between soil enzyme activities and soil fertility. Plant Nutr. Fert. Sci. 2004, 10, 277–280. [Google Scholar]
- Zenichi, M.; Fumie, I.; Konomi, T.; Masao, T. The Effects of Phosphorus and Magnesium on Sclerotium formation in Rhizoctonia solani Kühn. Jpn. J. Phytopathol. 1991, 57, 649–656. [Google Scholar]
- Ghasemi, S.; Safaie, N.; Shahbazi, S.; Shams-Bakhsh, M.; Askari, H. The Role of Cell Wall Degrading Enzymes in Antagonistic Traits of Trichoderma virens Against Rhizoctonia solani. Iran. J. Biotechnol. 2020, 18, e2333. [Google Scholar]






| Bioagents | Shoot Length (cm) | Root Length (cm) | Fresh Weight (g) | Dry Weight (g) | No. of Tillers | No. of Panicles | Kernel Length (mm) | Kernel Breadth (mm) | Panicle Length (cm) | Test Weight (g) | Grain Yield/Hill (g) | PDI (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PIK1 | 48.77 c | 31.20 c | 32.20 c | 10.50 c | 19.82 b | 8.44 c | 5.80 bc | 2.60 ab | 22.80 c | 21.50 c | 16.10 c | 26.38 d |
| POPS1 | 41.60 e | 25.20 d | 23.30 e | 7.83 e | 18.99 c | 7.99 d | 5.60 cd | 2.30 bc | 20.00 e | 18.50 e | 14.00 e | 38.73 b |
| BIK3 | 44.07 d | 25.77 d | 26.50 d | 8.93 d | 19.01 c | 8.11 d | 5.70 c | 2.40 bc | 21.20 d | 20.00 d | 15.00 d | 36.88 c |
| TAIK1 | 50.90 b | 33.87 b | 35.67 b | 12.00 b | 20.11 d | 9.63 b | 6.00 ab | 2.70 ab | 23.80 b | 22.20 b | 17.10 b | 24.06 e |
| Consortium | 58.33 a | 39.03 a | 41.53 a | 13.57 a | 22.32 a | 10.50 a | 6.20 a | 2.90 a | 25.80 a | 24.10 a | 19.10 a | 19.32 f |
| Control | 36.90 f | 22.33 e | 18.00 f | 6.97 f | 18.54 d | 7.32 e | 5.40 d | 2.10 c | 19.20 f | 17.70 f | 11.20 f | 75.24 a |
| CD | 0.358 | 0.605 | 0.196 | 0.181 | 0.361 | 0.285 | 0.067 | 0.149 | 0.088 | 0.096 | 0.306 | 2.006 |
| CV | 0.647 | 1.267 | 0.647 | 1.830 | 0.663 | 0.687 | 0.661 | 3.461 | 0.231 | 0.368 | 0.871 | 3.290 |
| SEm | 0.116 | 0.197 | 0.064 | 0.059 | 0.034 | 0.025 | 0.022 | 0.049 | 0.029 | 0.031 | 0.018 | 0.023 |
| Bioagents | Soil pH | EC | OC | Urease | Acid Phosphatase | Alkaline Phosphatase | Dehydrogenase | Soil N | Soil P | Soil K | Plant N | Plant P | Plant K |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PIK1 | 8.24 a | 0.75 a | 0.64 a | 6.18 b | 1.25 ab | 1.66 ab | 12.96 c | 283.15 c | 17.06 c | 216.35 c | 0.41 b | 0.07 ab | 1.23 ab |
| POPS1 | 8.15 a | 0.73 a | 0.60 a | 5.85 c | 0.85 cd | 1.42 b | 11.35 d | 235.21 e | 12.39 e | 181.72 e | 0.27 c | 0.04 b | 1.01 ab |
| BIK3 | 8.20 a | 0.74 a | 0.61 a | 6.10 b | 0.64 d | 1.46 b | 11.58 d | 247.63 d | 14.01 d | 195.27 d | 0.38 b | 0.05 ab | 1.10 ab |
| TAIK1 | 8.25 a | 0.75 a | 0.65 a | 6.32 b | 1.00 bc | 1.67 ab | 14.60 b | 297.67 b | 19.67 b | 249.42 b | 0.47 b | 0.08 ab | 0.93 b |
| Consortium | 8.28 a | 0.76 a | 0.68 a | 7.33 a | 1.39 a | 1.78 a | 16.44 a | 315.82 a | 24.91 a | 288.28 a | 0.58 a | 0.11 a | 1.58 a |
| Control | 8.08 a | 0.72 a | 0.57 b | 0.73 d | 0.09 e | 0.90 c | 0.14 e | 155.18 f | 7.87 f | 121.29 f | 0.21 c | 0.02 b | 0.76 c |
| CD | 0.023 | 0.028 | 0.268 | 0.057 | 0.070 | 0.074 | 0.111 | 0.024 | 0.002 | 0.772 | 0.009 | 0.001 | 1.952 |
| CV | 1.578 | 1.926 | 1.360 | 2.727 | 1.335 | 1.960 | 9.548 | 0.352 | 1.993 | 2.430 | 1.342 | 5.914 | 2.564 |
| SEm | 0.075 | 0.008 | 0.127 | 0.018 | 0.022 | 0.024 | 0.035 | 0.008 | 0.001 | 0.252 | 0.003 | 0.001 | 0.638 |
| Soil and Plant Characteristics | Parameters | Estimates | Standard Error | Probability | Partial R-Square | Model R-Square |
|---|---|---|---|---|---|---|
| pH | POPS1 | 0.97 | 0.05 | 0.0624 | 0.04 | 0.95 |
| BIK3 | 0.62 | 0.06 | <0.0001 | 0.91 | ||
| EC | BIK3 | 0.65 | 0.08 | <0.0001 | 0.96 | 0.9552 |
| OC | PIK1 | 0.68 | 0.06 | <0.0001 | 0.94 | 0.9414 |
| Available N | TAIK1 | 0.39 | 0.03 | <0.0001 | 0.94 | 0.9403 |
| Available P | PIK1 | 0.69 | 0.05 | <0.0001 | 0.95 | 0.9527 |
| Available K | PIK1 | 0.69 | 0.05 | 0.0003 | 0.33 | 0.9462 |
| Consortium | 0.89 | 0.1 | <0.0001 | 0.61 | ||
| Urease | Consortium | 0.9 | 0.03 | <0.0001 | 0.96 | 0.959 |
| Acid Phosphatase | POPS1 | 0.63 | 0.01 | <0.0001 | 0.98 | 0.9783 |
| Alkaline Phosphatase | POPS1 | 0.4 | 0.05 | 0.0037 | 0.05 | 0.9702 |
| BIK3 | 0.14 | 0.02 | 0.0025 | 0.84 | ||
| TAIK1 | 0.55 | 0.07 | 0.0073 | 0.08 | ||
| Dehydrogenase | POPS1 | 0.42 | 0.08 | <0.0001 | 0.96 | 0.9633 |
| Plant N | BIK3 | 0.44 | 0.03 | 0.0098 | 0.59 | 0.76 |
| Consortium | 0.57 | 0.07 | 0.055 | 0.18 | ||
| Plant P | POPS1 | 0.99 | 0.05 | 0.0287 | 0.02 | 0.9785 |
| BIK3 | 0.49 | 0.09 | <0.0001 | 0.96 | ||
| Plant K | PIK1 | 0.7 | 0.09 | 0.0994 | 0.03 | 0.9836 |
| POPS1 | 0.51 | 0.02 | 0.0548 | 0.03 | ||
| BIK3 | 0.48 | 0 | 0.0992 | 0.01 | ||
| Consortium | 0.35 | 0.05 | <0.0001 | 0.91 |
| Soil and Plant Characteristics | Parameters | Estimates | Standard Error | Probability | Partial R-Square | Model R-Square |
|---|---|---|---|---|---|---|
| pH | POPS1 | 0.67 | 0.08 | <0.0001 | 0.92 | 0.96 |
| BIK3 | 0.5 | 0.03 | 0.0367 | 0.04 | ||
| EC | BIK3 | 0.69 | 0.09 | <0.0001 | 0.96 | 0.9571 |
| OC | PIK1 | 0.77 | 0.01 | <0.0001 | 0.96 | 0.9608 |
| Available N | TAIK1 | 0.26 | 0.02 | <0.0001 | 0.96 | 0.9643 |
| Available P | PIK1 | 0.49 | 0.03 | <0.0001 | 0.97 | 0.973 |
| Available K | PIK1 | 0.25 | 0.06 | 0.0011 | 0.58 | 0.9169 |
| Consortium | 0.97 | 0.05 | 0.0104 | 0.34 | ||
| Urease | TAIK1 | 0.65 | 0.1 | <0.0001 | 0.97 | 0.9685 |
| Acid Phosphatase | POPS1 | 0.12 | 0.06 | <0.0001 | 0.98 | 0.9785 |
| Alkaline Phosphatase | POPS1 | 0.96 | 0.02 | 0.1215 | 0.04 | 0.9753 |
| BIK3 | 0.57 | 0.03 | 0.0001 | 0.86 | ||
| TAIK1 | 0.43 | 0.03 | 0.0059 | 0.07 | ||
| Dehydrogenase | POPS1 | 0.68 | 0.03 | <0.0001 | 0.97 | 0.9726 |
| Plant N | BIK3 | 0.43 | 0.02 | 0.0191 | 0.52 | 0.7677 |
| Consortium | 0.59 | 0.06 | 0.0285 | 0.25 | ||
| Plant P | PIK1 | 0.57 | 0.04 | <0.0001 | 0.94 | 0.9412 |
| Plant K | PIK1 | 0.55 | 0.05 | 0.0527 | 0.03 | 0.9757 |
| TAIK1 | 0.7 | 0.05 | 0.0752 | 0.02 | ||
| Consortium | 0.49 | 0.06 | <0.0001 | 0.92 |
| Soil and Plant Characteristics | Parameters | Estimates | Standard Error | Probability | Partial R-Square | Model R-Square |
|---|---|---|---|---|---|---|
| pH | POPS1 | 0.88 | 0.05 | <0.0001 | 0.9 | 0.9383 |
| BIK3 | 0.67 | 0.05 | 0.0748 | 0.04 | ||
| EC | BIK3 | 0.82 | 0.03 | <0.0001 | 0.94 | 0.9403 |
| OC | PIK1 | 0.65 | 0.07 | <0.0001 | 0.93 | 0.9332 |
| Available N | TAIK1 | 0.29 | 0.03 | <0.0001 | 0.94 | 0.935 |
| Available P | POPS1 | 0.83 | 0.07 | <0.0001 | 0.94 | 0.9432 |
| Available K | BIK3 | 0.08 | 0.03 | 0.1465 | 0.32 | 0.9712 |
| Consortium | 0.58 | 0.02 | <0.0001 | 0.65 | ||
| Urease | TAIK1 | 0.28 | 0.05 | <0.0001 | 0.94 | 0.9367 |
| Acid Phosphatase | POPS1 | 0.66 | 0.01 | <0.0001 | 0.96 | 0.9731 |
| Consortium | 0.36 | 0.02 | 0.0929 | 0.01 | ||
| Alkaline Phosphatase | POPS1 | 0.12 | 0.04 | 0.1 | 0.07 | 0.9657 |
| BIK3 | 0.67 | 0.06 | 0.0005 | 0.8 | ||
| TAIK1 | 0.07 | 0.01 | 0.0057 | 0.1 | ||
| Dehydrogenase | PIK1 | 0.61 | 0.07 | <0.0001 | 0.96 | 0.9641 |
| Plant N | BIK3 | 0.76 | 0.03 | 0.0206 | 0.51 | 0.738 |
| Consortium | 0.8 | 0.06 | 0.0424 | 0.23 | ||
| Plant P | BIK3 | 0.25 | 0.03 | <0.0001 | 0.93 | 0.9349 |
| Plant K | PIK1 | 0.45 | 0.05 | 0.0438 | 0.06 | 0.9822 |
| POPS1 | 0.72 | 0.07 | 0.0343 | 0.04 | ||
| BIK3 | 0.67 | 0.05 | 0.1244 | 0.01 | ||
| Consortium | 0.72 | 0.1 | <0.0001 | 0.88 |
| Soil and Plant Characteristics | Parameters | Estimates | Standard Error | Probability | Partial R-Square | Model R-Square |
|---|---|---|---|---|---|---|
| pH | POPS1 | 0.07 | 0.09 | 0.0877 | 0.9 | 0.93 |
| BIK3 | 0.05 | 0.07 | <0.0001 | 0.04 | ||
| EC | BIK3 | 0.06 | 0.06 | <0.0001 | 0.95 | 0.9624 |
| Consortium | 0.06 | 0.02 | 0.1458 | 0.01 | ||
| OC | PIK1 | 0.67 | 0.07 | <0.0001 | 0.94 | 0.9381 |
| Available N | TAIK1 | 0.10 | 0.01 | <0.0001 | 0.94 | 0.9432 |
| Available P | PIK1 | 0.07 | 0.01 | <0.0001 | 0.95 | 0.9483 |
| Available K | BIK3 | 0.07 | 0.04 | 0.0965 | 0.58 | 0.9398 |
| Consortium | 0.18 | 0.03 | 0.0113 | 0.36 | ||
| Urease | Consortium | 0.14 | 0.07 | <0.0001 | 0.95 | 0.9494 |
| Acid Phosphatase | POPS1 | 0.13 | 0.09 | <0.0001 | 0.97 | 0.9827 |
| Consortium | 0.81 | 0.05 | 0.0726 | 0.01 | ||
| Alkaline Phosphatase | POPS1 | 0.68 | 0.01 | 0.0983 | 0.06 | 0.9716 |
| BIK3 | 0.45 | 0.08 | 0.0003 | 0.82 | ||
| TAIK1 | 0.52 | 0.07 | 0.0044 | 0.09 | ||
| Dehydrogenase | PIK1 | 0.25 | 0.02 | <0.0001 | 0.97 | 0.9699 |
| Plant N | PIK1 | 0.40 | 0.06 | 0.0472 | 0.21 | 0.7497 |
| BIK3 | 0.70 | 0.10 | 0.015 | 0.54 | ||
| Plant P | POPS1 | 0.51 | 0.03 | 0.0928 | 0.02 | 0.9673 |
| BIK3 | 0.59 | 0.01 | <0.0001 | 0.95 | ||
| Plant K | PIK1 | 0.65 | 0.07 | 0.0944 | 0.04 | 0.9807 |
| POPS1 | 0.86 | 0.01 | 0.0702 | 0.03 | ||
| BIK3 | 0.73 | 0.05 | 0.0665 | 0.02 | ||
| Consortium | 0.96 | 0.01 | <0.0001 | 0.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanama, S.; Pesari, M.; Rajendran, G.; Gali, U.D.; Rathod, S.; Panuganti, R.; Chilukuri, S.; Chinnaswami, K.; Kumar, S.; Minkina, T.; et al. Correlation of the Effect of Native Bioagents on Soil Properties and Their Influence on Stem Rot Disease of Rice. Sustainability 2023, 15, 11768. https://doi.org/10.3390/su151511768
Vanama S, Pesari M, Rajendran G, Gali UD, Rathod S, Panuganti R, Chilukuri S, Chinnaswami K, Kumar S, Minkina T, et al. Correlation of the Effect of Native Bioagents on Soil Properties and Their Influence on Stem Rot Disease of Rice. Sustainability. 2023; 15(15):11768. https://doi.org/10.3390/su151511768
Chicago/Turabian StyleVanama, Sowmya, Maruthi Pesari, Gobinath Rajendran, Uma Devi Gali, Santosha Rathod, Rajanikanth Panuganti, Srivalli Chilukuri, Kannan Chinnaswami, Sumit Kumar, Tatiana Minkina, and et al. 2023. "Correlation of the Effect of Native Bioagents on Soil Properties and Their Influence on Stem Rot Disease of Rice" Sustainability 15, no. 15: 11768. https://doi.org/10.3390/su151511768
APA StyleVanama, S., Pesari, M., Rajendran, G., Gali, U. D., Rathod, S., Panuganti, R., Chilukuri, S., Chinnaswami, K., Kumar, S., Minkina, T., Sansinenea, E., & Keswani, C. (2023). Correlation of the Effect of Native Bioagents on Soil Properties and Their Influence on Stem Rot Disease of Rice. Sustainability, 15(15), 11768. https://doi.org/10.3390/su151511768









