Understanding the Implications of Predicted Function for Assessment of Rapid Bioremediation in a Farmland-Oilfield Mixed Area
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Field and Bioremediation
2.2. Sample Collection
2.3. Physicochemical Parameters
2.4. DNA Extraction and PCR Amplification
2.5. High-Throughput Sequencing and Bioinformatic Analysis
2.6. Quantification of Functional Genes
3. Results
3.1. The Profile of the Soil Microbial Community before and after Bioremediation
3.2. Functional Profiling of the Soil Microbiome before and after Bioremediation
3.3. Predicted Functional Changes before and after Bioremediation in Relation to PHs and PAHs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017, 223, 277–286. [Google Scholar] [CrossRef]
- Fitzgerald, T.; Kuwayama, Y.; Olmstead, S.; Thompson, A. Dynamic impacts of US energy development on agricultural land use. Energy Policy 2020, 137, 111163. [Google Scholar] [CrossRef]
- Kamalanathan, M.; Xu, C.; Schwehr, K.; Bretherton, L.; Beaver, M.; Doyle, S.M.; Genzer, J.; Hillhouse, J.; Sylvan, J.B.; Santschi, P.; et al. Extracellular Enzyme Activity Profile in a Chemically Enhanced Water Accommodated Fraction of Surrogate Oil: Toward Understanding Microbial Activities After the Deepwater Horizon Oil Spill. Front. Microbiol. 2018, 9, 798. [Google Scholar] [CrossRef]
- Sheng, Y.Z.; Liu, Y.; Yang, J.J.; Dong, H.L.; Liu, B.; Zhang, H.; Li, A.Y.; Wei, Y.Q.; Li, G.H.; Zhang, D.Y. History of petroleum disturbance triggering the depth-resolved assembly process of microbial communities in the vadose zone. J. Hazard. Mater. 2021, 402, 124060. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sha, C.Y.; Wu, J.; Su, J.H.; Wu, J.Q.; Wang, Q.; Tan, J.; Huang, S.F. Bacterial community response to petroleum contamination in brackish tidal marsh sediments in the Yangtze River Estuary, China. J. Environ. Sci. 2021, 99, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Wang, X.M.; Li, Y.H.; Ji, H.B. Remediation of Petroleum-Contaminated Soils with Microbial and Microbial Combined Methods: Advances, Mechanisms, and Challenges. Sustainability 2021, 13, 9267. [Google Scholar] [CrossRef]
- Wu, M.L.; Ma, C.; Wang, D.; Liu, H.; Zhu, C.C.; Xu, H.N. Nutrient drip irrigation for refractory hydrocarbon removal and microbial community shift in a historically petroleum-contaminated soil. Sci. Total Environ. 2020, 713, 136331. [Google Scholar] [CrossRef]
- Delacuvellerie, A.; Cyriaque, V.; Gobert, S.; Benali, S.; Wattiez, R. The plastisphere in marine ecosystem hosts potential specific microbial degraders including Alcanivorax borkumensis as a key player for the low-density polyethylene degradation. J. Hazard. Mater. 2019, 380, 120899. [Google Scholar] [CrossRef]
- Zhou, H.H.; Huang, X.M.; Liang, Y.P.; Li, Y.H.; Xie, Q.L.; Zhang, C.F.; You, S.H. Enhanced bioremediation of hydraulic fracturing flowback and produced water using an indigenous biosurfactant-producing bacteria Acinetobacter sp. Y2. Chem. Eng. J. 2020, 397, 125348. [Google Scholar] [CrossRef]
- Ribicic, D.; Netzer, R.; Hazen, T.C.; Techtmann, S.M.; Drablos, F.; Brakstad, O.G. Microbial community and metagenome dynamics during biodegradation of dispersed oil reveals potential key-players in cold Norwegian seawater. Mar. Pollut. Bull. 2018, 129, 370–378. [Google Scholar] [CrossRef]
- Tuo, B.H.; Yan, J.B.; Fan, B.A.; Yang, Z.H.; Liu, J.Z. Biodegradation characteristics and bioaugmentation potential of a novel quinoline-degrading strain of Bacillus sp. isolated from petroleum-contaminated soil. Bioresour. Technol. 2012, 107, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Cerqueda-Garcia, D.; Garcia-Maldonado, J.Q.; Aguirre-Macedo, L.; Garcia-Cruz, U. A succession of marine bacterial communities in batch reactor experiments during the degradation of five different petroleum types. Mar. Pollut. Bull. 2020, 150, 110775. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.L.; Tang, J.; Liu, X.M.; Song, B.R.; Zhen, M.N.; Ashbolt, N.J. Vertical response of microbial community and degrading genes to petroleum hydrocarbon contamination in saline alkaline soil. J. Environ. Sci. 2019, 81, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Mangse, G.; Werner, D.; Meynet, P.; Ogbaga, C.C. Microbial community responses to different volatile petroleum hydrocarbon class mixtures in an aerobic sandy soil. Environ. Pollut. 2020, 264, 114738. [Google Scholar] [CrossRef]
- Olasanmi, I.O.; Thring, R.W. The Role of Biosurfactants in the Continued Drive for Environmental Sustainability. Sustainability 2018, 10, 4817. [Google Scholar] [CrossRef]
- Khudur, L.S.; Shahsavari, E.; Miranda, A.F.; Morrison, P.D.; Nugegoda, D.; Ball, A.S. Evaluating the efficacy of bioremediating a diesel-contaminated soil using ecotoxicological and bacterial community indices. Environ. Sci. Pollut. Res. 2015, 22, 14809–14819. [Google Scholar] [CrossRef]
- Barak, H.; Brenner, A.; Sivan, A.; Kushmaro, A. Temporal distribution of microbial community in an industrial wastewater treatment system following crash and during recovery periods. Chemosphere 2020, 258, 127271. [Google Scholar] [CrossRef]
- Bae, H.S.; Huang, L.B.; White, J.R.; Wang, J.; DeLaune, R.D.; Ogram, A. Response of microbial populations regulating nutrient biogeochemical cycles to oiling of coastal saltmarshes from the Deepwater Horizon oil spill. Environ. Pollut. 2018, 241, 136–147. [Google Scholar] [CrossRef]
- Boccadoro, C.; Krolicka, A.; Receveur, J.; Aeppli, C.; Le Floch, S. Microbial community response and migration of petroleum compounds during a sea-ice oil spill experiment in Svalbard. Mar. Environ. Res. 2018, 142, 214–233. [Google Scholar] [CrossRef]
- dos Santos, E.D.; Silva, I.S.; Simoes, T.H.N.; Simioni, K.C.M.; Oliveira, V.M.; Grossman, M.J.; Durrant, L.R. Correlation of soil microbial community responses to contamination with crude oil with and without chromium and copper. Int. Biodeterior. Biodegrad. 2012, 70, 104–110. [Google Scholar] [CrossRef]
- Gofstein, T.R.; Perkins, M.; Field, J.; Leigh, M.B. The Interactive Effects of Crude Oil and Corexit 9500 on Their Biodegradation in Arctic Seawater. Appl. Environ. Microb. 2020, 86, e01194-20. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, H.Z.; Salam, D.A. Response of sediment microbial communities to crude oil contamination in marine sediment microbial fuel cells under ferric iron stimulation. Environ. Pollut. 2020, 263, 114658. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kim, M.S.; Kim, J.G.; Kim, S.O. Use of Soil Enzymes as Indicators for Contaminated Soil Monitoring and Sustainable Management. Sustainability 2020, 12, 8209. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.H.; Niu, J.J.; Huang, W.K.; Peng, D.L.; Xiao, Y.H.; Zhang, X.; Liang, Y.L.; Liu, X.D.; Yin, H.Q. Comparison of microbial taxonomic and functional shift pattern along contamination gradient. BMC Microbiol. 2016, 16, 110. [Google Scholar] [CrossRef][Green Version]
- Mendes, L.W.; Tsai, S.M. Distinct taxonomic and functional composition of soil microbiomes along the gradient forest-restinga-mangrove in southeastern Brazil. Antonie Leeuwenhoek Int. J. Gen. 2018, 111, 101–114. [Google Scholar] [CrossRef]
- Manzano-Marin, A.; Oceguera-Figueroa, A.; Latorre, A.; Jimenez-Garcia, L.F.; Moya, A. Solving a Bloody Mess: B-Vitamin Independent-Metabolic Convergence among Gammaproteobacterial Obligate Endosymbionts from Blood-Feeding Arthropods and the Leech Haementeria officinalis. Genome Biol. Evol. 2015, 7, 2871–2884. [Google Scholar] [CrossRef]
- Sun, S.; Jones, R.B.; Fodor, A.A. Inference-based accuracy of metagenome prediction tools varies across sample types and functional categories. Microbiome 2020, 8, 46. [Google Scholar] [CrossRef]
- Sun, B.; Bai, Z.H.; Bao, L.J.; Xue, L.X.; Zhang, S.W.; Wei, Y.X.; Zhang, Z.Y.; Zhuang, G.Q.; Zhuang, X.L. Bacillus subtilis biofertilizer mitigating agricultural ammonia emission and shifting soil nitrogen cycling microbiomes. Environ. Int. 2020, 144, 105989. [Google Scholar] [CrossRef]
- Fan, R.J.; Ma, W.P.; Zhang, H.L. Microbial community responses to soil parameters and their effects on petroleum degradation during bio-electrokinetic remediation. Sci. Total Environ. 2020, 748, 142463. [Google Scholar] [CrossRef]
- Silva, D.P.; Duarte, G.; Villela, H.D.M.; Santos, H.F.; Rosado, P.M.; Rosado, J.G.; Rosado, A.S.; Ferreira, E.M.; Soriano, A.U.; Peixoto, R.S. Adaptable mesocosm facility to study oil spill impacts on corals. Ecol. Evol. 2019, 9, 5172–5185. [Google Scholar] [CrossRef] [PubMed]
- Chaineau, C.H.; Rougeux, G.; Yepremian, C.; Oudot, J. Effects of nutrient concentration on the biodegradation of crude oil and associated microbial populations in the soil. Soil Biol. Biochem. 2005, 37, 1490–1497. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Bacosa, H.P.; Erdner, D.L.; Rosenheim, B.E.; Shetty, P.; Seitz, K.W.; Baker, B.J.; Liu, Z.F. Hydrocarbon degradation and response of seafloor sediment bacterial community in the northern Gulf of Mexico to light Louisiana sweet crude oil. ISME J. 2018, 12, 2532–2543. [Google Scholar] [CrossRef]
- Niepceron, M.; Martin-Laurent, F.; Crampon, M.; Portet-Koltalo, F.; Akpa-Vinceslas, M.; Legras, M.; Bru, D.; Bureau, F.; Bodilis, J. GammaProteobacteria as a potential bioindicator of a multiple contamination by polycyclic aromatic hydrocarbons (PAHs) in agricultural soils. Environ. Pollut. 2013, 180, 199–205. [Google Scholar] [CrossRef]
- Cui, Z.S.; Xu, G.S.; Gao, W.; Li, Q.; Yang, B.J.; Yang, G.P.; Zheng, L. Isolation and characterization of Cycloclasticus strains from Yellow Sea sediments and biodegradation of pyrene and fluoranthene by their syntrophic association with Marinobacter strains. Int. Biodeterior. Biodegrad. 2014, 91, 45–51. [Google Scholar] [CrossRef]
- Liu, R.Y.; Gao, Y.X.; Ji, Y.F.; Zhang, Y.; Yang, M. Characteristics of hydrocarbon hydroxylase genes in a thermophilic aerobic biological system treating oily produced wastewater. Water Sci. Technol. 2015, 71, 123–130. [Google Scholar] [CrossRef]
- Moser, R.; Stahl, U. Insights into the genetic diversity of initial dioxygenases from PAH-degrading bacteria. Appl. Microbiol. Biotechnol. 2001, 55, 609–618. [Google Scholar] [CrossRef]
- Leys, N.M.E.J.; Ryngaert, A.; Bastiaens, L.; Verstraete, W.; Top, E.M.; Springael, D. Occurrence and phylogenetic diversity of Sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons. Appl. Environ. Microb. 2004, 70, 1944–1955. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Huang, Y.; Zhang, Z.T.; Hao, H.; Wang, H. Absence of the nahG-like gene caused the syntrophic interaction between Marinobacter and other microbes in PAH-degrading process. J. Hazard. Mater. 2020, 384, 121387. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.J.; Penttinen, P.; Mikkonen, A.; Lindstrom, K. Bacterial community changes in response to oil contamination and perennial crop cultivation. Environ. Sci. Pollut. Res. 2018, 25, 14575–14584. [Google Scholar] [CrossRef]
- Ribicic, D.; McFarlin, K.M.; Netzer, R.; Brakstad, O.G.; Winkler, A.; Throne-Holst, M.; Storseth, T.R. Oil type and temperature dependent biodegradation dynamics—Combining chemical and microbial community data through multivariate analysis. BMC Microbiol. 2018, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Galazka, A.; Grzadziel, J.; Galazka, R.; Ukalska-Jaruga, A.; Strzelecka, J.; Smreczak, B. Genetic and Functional Diversity of Bacterial Microbiome in Soils with Long Term Impacts of Petroleum Hydrocarbons. Front. Microbiol. 2018, 9, 1923. [Google Scholar] [CrossRef]
- Robichaud, K.; Stewart, K.; Labrecque, M.; Hijri, M.; Cherewyk, J.; Amyot, M. An ecological microsystem to treat waste oil contaminated soil: Using phytoremediation assisted by fungi and local compost, on a mixed-contaminant site, in a cold climate. Sci. Total Environ. 2019, 672, 732–742. [Google Scholar] [CrossRef]
- Feng, X.; Liu, Z.; Jia, X.; Lu, W. Distribution of Bacterial Communities in Petroleum-Contaminated Soils from the Dagang Oilfield, China. Trans. Tianjin Univ. 2020, 26, 22–32. [Google Scholar] [CrossRef]
- Harmsen, J.; Rietra, R.P.J.J. 25 years monitoring of PAHs and petroleum hydrocarbons biodegradation in soil. Chemosphere 2018, 207, 229–238. [Google Scholar] [CrossRef]
- Lu, C.; Hong, Y.; Liu, J.; Gao, Y.; Ma, Z.; Yang, B.; Ling, W.; Waigi, M.G. A PAH-degrading bacterial community enriched with contaminated agricultural soil and its utility for microbial bioremediation. Environ. Pollut. 2019, 251, 773–782. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Murarka, P.; Srivastava, P. Biodesulfurization of petroleum wastes. In Biovalorisation of Wastes to Renewable Chemicals and Biofuels; Elsevier: Amsterdam, The Netherlands, 2020; pp. 165–187. [Google Scholar]
- Govil, T. Taxonomical Diversity of Extremophiles in the Deep Biosphere. In Microbial Diversity in the Genomic Era; Academic Press: Cambridge, MA, USA, 2019; pp. 631–656. [Google Scholar]
- Nie, M.Q.; Nie, H.Y.; He, M.L.; Lin, Y.Y.; Wang, L.; Jin, P.K.; Zhang, S.Y. Immobilization of biofilms of Pseudomonas aeruginosa NY3 and their application in the removal of hydrocarbons from highly concentrated oil-containing wastewater on the laboratory scale. J. Environ. Manag. 2016, 173, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.J.; Liu, W.M.; Tian, S.H.; Wang, W.; Qi, Q.G.; Jiang, P.; Gao, X.M.; Li, F.J.; Li, H.Y.; Yu, H.W. Petroleum Hydrocarbon-Degrading Bacteria for the Remediation of Oil Pollution Under Aerobic Conditions: A Perspective Analysis. Front. Microbiol. 2018, 9, 2885. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.C.; Chang, T.C.; Liao, C.S.; Chang, Y.T. A Comparison of the Microbial Community and Functional Genes Present in Free-Living and Soil Particle-Attached Bacteria from an Aerobic Bioslurry Reactor Treating High-Molecular-Weight PAHs. Sustainability 2019, 11, 1088. [Google Scholar] [CrossRef]
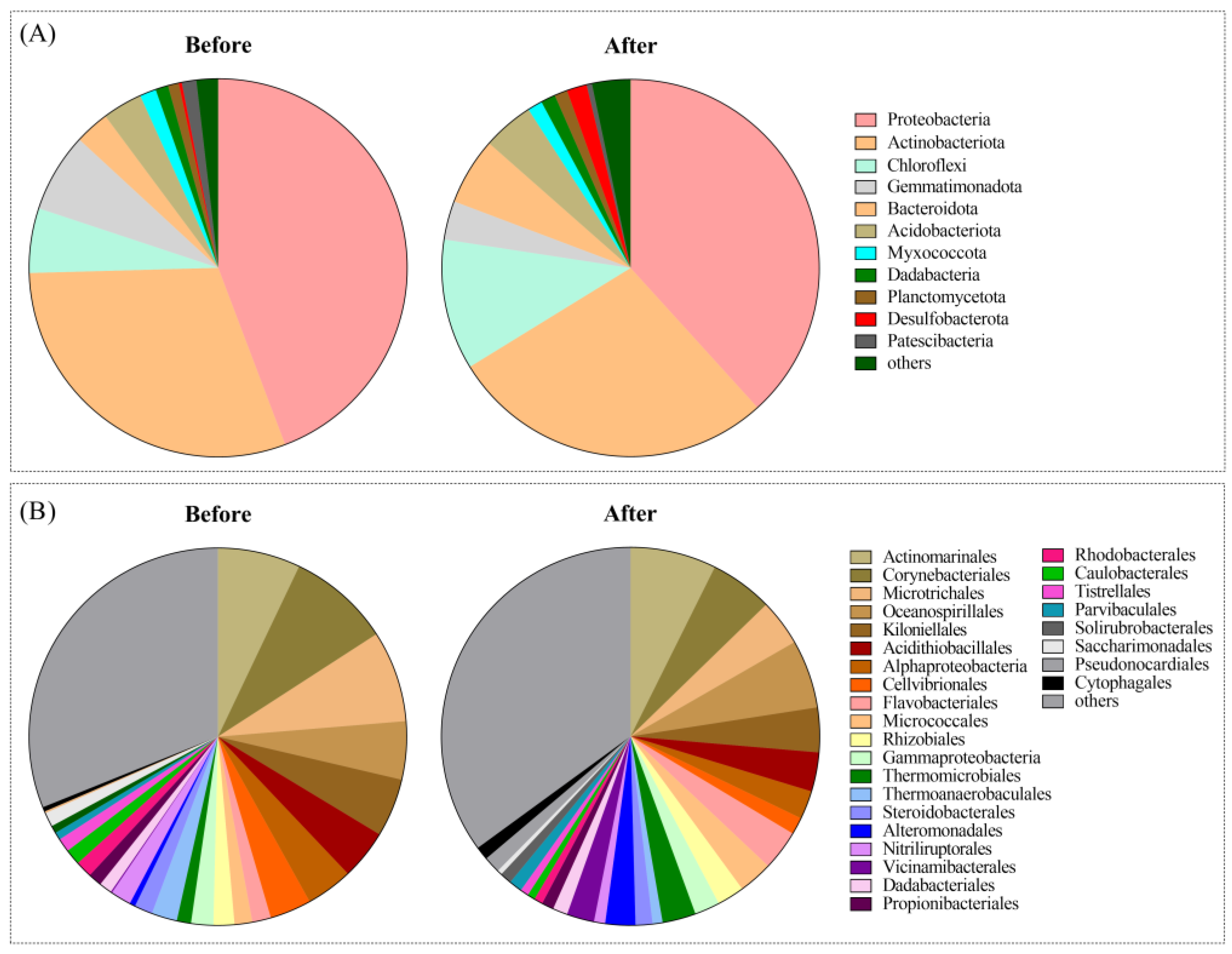


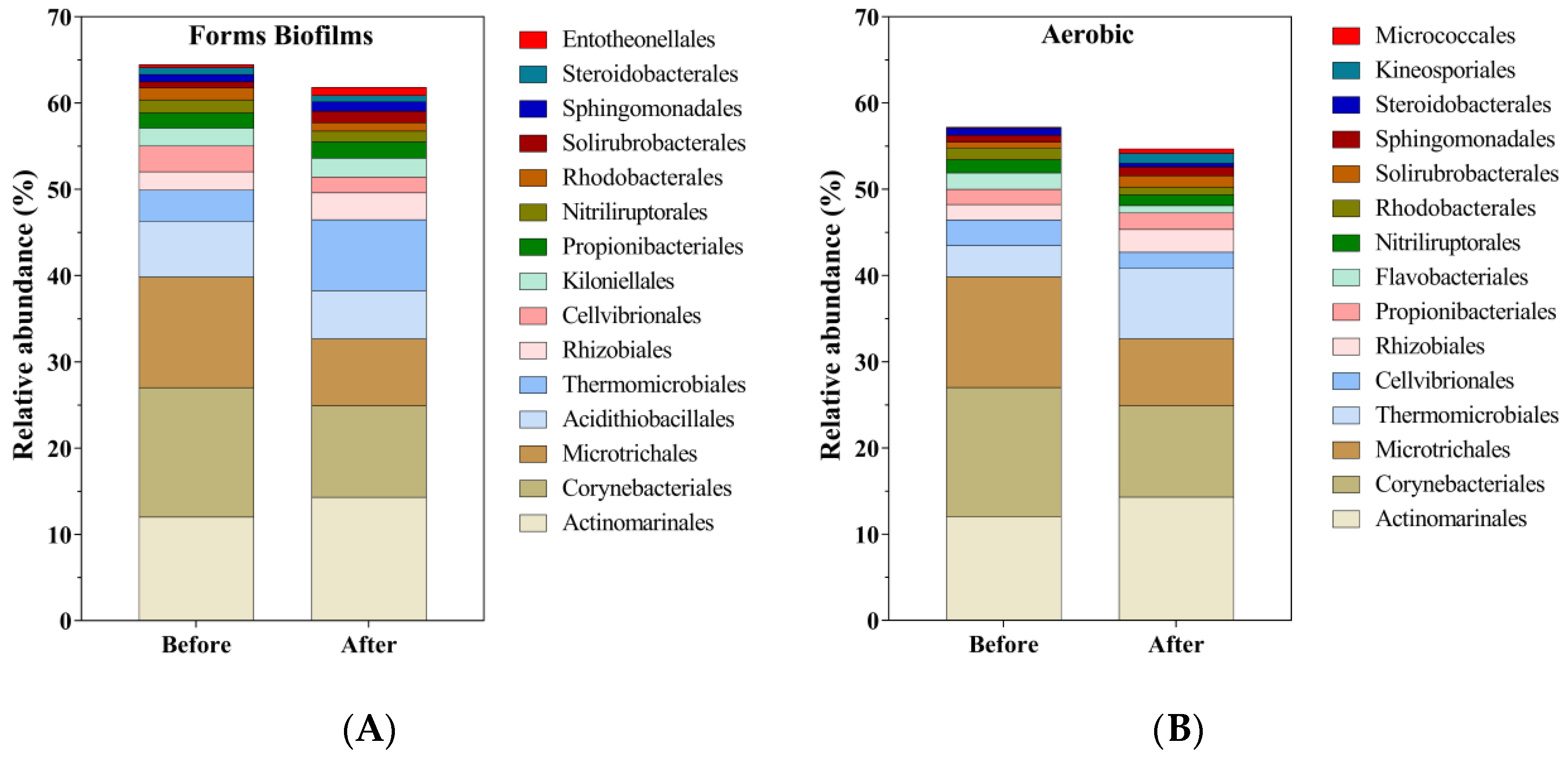
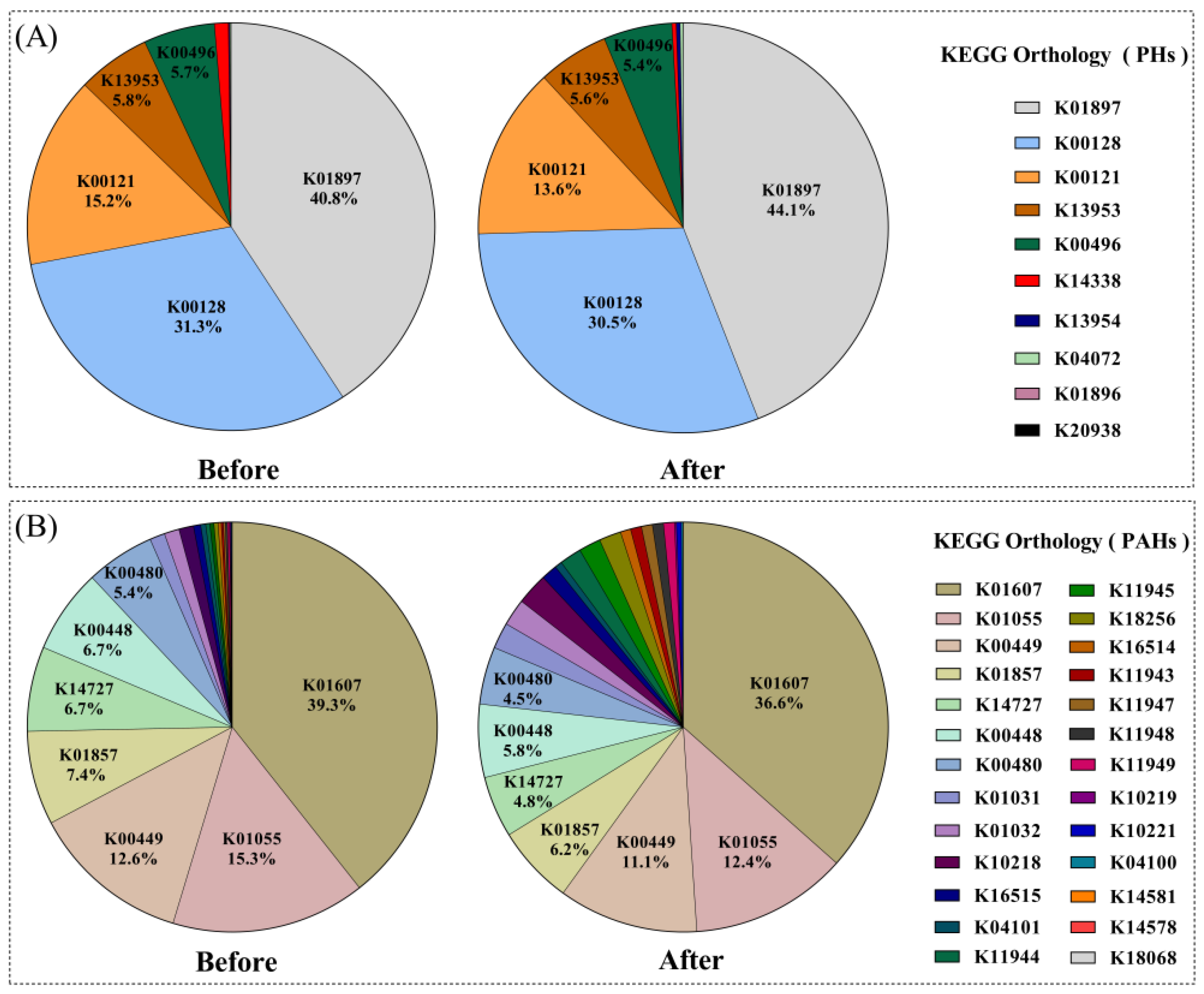

| Phylum | Sub-Taxa a | Before (%) | After (%) | Change Rate b (%) | Targets | Reference |
|---|---|---|---|---|---|---|
| Proteobacteria | Alcanivorax | 4.74 | 5.58 | 17.72 | PHs | [8] |
| Porticoccaceae | 3.19 | 1.31 | −58.93 | PHs | [10] | |
| Rhodobacteraceae | 2.28 | 0.44 | −80.70 | PHs | [12] | |
| Woeseia | 1.39 | 1.17 | −15.83 | PHs | [36] | |
| Alteromonadaceae | 0.04 | 0 | −100.00 | PHs | [3] | |
| Gammaproteobacteria | 1.86 | 2.09 | 12.37 | PAHs | [37] | |
| Marinobacter | 0.44 | 2.15 | 388.64 | PAHs | [38] | |
| Burkholderiales | 0.24 | 1.23 | 412.50 | PAHs | [39] | |
| Pseudomonas | 0.19 | 0.04 | −78.95 | PAHs | [40] | |
| Sphingomonas | 0.19 | 0.33 | 73.68 | PAHs | [41] | |
| Halomonas | 0.05 | 0.08 | 60.00 | PAHs | [42] | |
| Acinetobacter | 0.01 | 0.02 | 100.00 | PHs, PAHs | [9] | |
| Actinobacterota | Mycobacterium | 8.16 | 4.66 | −42.89 | PHs, PAHs | [7] |
| Microbacteriaceae | 1.36 | 0.54 | −60.29 | PHs | [43] | |
| Rhodococcus | 0.19 | 0.03 | −84.21 | PAHs | [40] | |
| Bacteroidetes | Flavobacteriaceae | 1.16 | 1.86 | 60.34 | PAHs | [44] |
| Firmicutes | Bacillus | 0.04 | 0.28 | 600.00 | PAHs | [11] |
| Total | 25.53 | 21.81 | −14.57 |
| Target | KO IDs | Descriptions [Enzyme Codes] |
|---|---|---|
| PHs | K00496 | alkane 1-monooxygenase [EC:1.14.15.3] |
| K20938 | long-chain alkane monooxygenase [EC:1.14.14.28] | |
| K13953 | alcohol dehydrogenase, propanol-preferring [EC:1.1.1.1] | |
| K13954 | alcohol dehydrogenase [EC:1.1.1.1] | |
| K00121 | S-(hydroxymethyl) glutathione dehydrogenase/alcohol dehydrogenase [EC: 1.1.1.284 1.1.1.1] | |
| K04072 | acetaldehyde dehydrogenase/alcohol dehydrogenase [EC:1.2.1.10 1.1.1.1] | |
| K00128 | aldehyde dehydrogenase (NAD+) [EC:1.2.1.3] | |
| K14338 | cytochrome P450/NADPH-cytochrome P450 reductase [EC:1.14.14.1 1.6.2.4] | |
| K01897 | long-chain acyl-CoA synthetase [EC:6.2.1.3] | |
| K01896 | medium-chain acyl-CoA synthetase [EC:6.2.1.2] | |
| PAHs | K11943 | PAH dioxygenase large subunit [EC:1.13.11.-] |
| K11944 | PAH dioxygenase small subunit [EC:1.13.11.-] | |
| K11945 | extradiol dioxygenase [EC:1.13.11.-] | |
| K11947 | aldehyde dehydrogenase [EC:1.2.1.-] | |
| K11948 | 1-hydroxy-2-naphthoate dioxygenase [EC:1.13.11.38] | |
| K11949 | 4-(2-carboxyphenyl)-2-oxobut-3-enoate aldolase [EC:4.1.2.34] | |
| K00480 | salicylate hydroxylase [EC:1.14.13.1] | |
| K18068 | phthalate 4,5-dioxygenase [EC:1.14.12.7] | |
| K00448 | protocatechuate 3,4-dioxygenase, alpha subunit [EC:1.13.11.3] | |
| K00449 | protocatechuate 3,4-dioxygenase, beta subunit [EC:1.13.11.3] | |
| K01857 | 3-carboxy-cis, cis-muconate cycloisomerase [EC:5.5.1.2] | |
| K01607 | 4-carboxymuconolactone decarboxylase [EC:4.1.1.44] | |
| K14727 | 3-oxoadipate enol-lactonase/4-carboxymuconolactone decarboxylase [EC:3.1.1.24 4.1.1.44] | |
| K01055 | 3-oxoadipate enol-lactonase [EC:3.1.1.24] | |
| K01031 | 3-oxoadipate CoA-transferase, alpha subunit [EC:2.8.3.6] | |
| K01032 | 3-oxoadipate CoA-transferase, beta subunit [EC:2.8.3.6] | |
| K18256 | 3,4-dihydroxyphthalate decarboxylase [EC:4.1.1.69] | |
| K04100 | protocatechuate 4,5-dioxygenase, alpha chain [EC:1.13.11.8] | |
| K04101 | protocatechuate 4,5-dioxygenase, beta chain [EC:1.13.11.8] | |
| K10219 | 2-hydroxy-4-carboxymuconate semialdehyde hemiacetal dehydrogenase [EC:1.1.1.312] | |
| K10221 | 2-pyrone-4,6-dicarboxylate lactonase [EC:3.1.1.57] | |
| K16514 | 4-oxalomesaconate tautomerase [EC:5.3.2.8] | |
| K16515 | 4-oxalomesaconate hydratase [EC:4.2.1.83] | |
| K10218 | 4-hydroxy-4-methyl-2-oxoglutarate aldolase [EC:4.1.3.17] | |
| K14578 | naphthalene 1,2-dioxygenase ferredoxin component | |
| K14581 | naphthalene 1,2-dioxygenase ferredoxin reductase component [EC:1.18.1.7] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Wu, S.; Zhang, Y.; Bohu, T.; Bai, Z.; Zhuang, X. Understanding the Implications of Predicted Function for Assessment of Rapid Bioremediation in a Farmland-Oilfield Mixed Area. Sustainability 2022, 14, 2248. https://doi.org/10.3390/su14042248
Wang H, Wu S, Zhang Y, Bohu T, Bai Z, Zhuang X. Understanding the Implications of Predicted Function for Assessment of Rapid Bioremediation in a Farmland-Oilfield Mixed Area. Sustainability. 2022; 14(4):2248. https://doi.org/10.3390/su14042248
Chicago/Turabian StyleWang, Haoyu, Shanghua Wu, Yuxiu Zhang, Tsing Bohu, Zhihui Bai, and Xuliang Zhuang. 2022. "Understanding the Implications of Predicted Function for Assessment of Rapid Bioremediation in a Farmland-Oilfield Mixed Area" Sustainability 14, no. 4: 2248. https://doi.org/10.3390/su14042248
APA StyleWang, H., Wu, S., Zhang, Y., Bohu, T., Bai, Z., & Zhuang, X. (2022). Understanding the Implications of Predicted Function for Assessment of Rapid Bioremediation in a Farmland-Oilfield Mixed Area. Sustainability, 14(4), 2248. https://doi.org/10.3390/su14042248










