Influences of Naphthalene Concentration on Starch Anaerobic Digestion: Focusing on Digestion Performance, Extracellular Polymeric Substances and Function Microbial Community
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Procedures
2.2. Chemical Analysis
2.3. Data Analysis
2.4. EPS Extraction and Analysis
2.5. Molecular Docking
2.6. Sequencing and Function Prediction
3. Results and Discussion
3.1. Influences of Nap on Biogas
3.2. Influences of Nap on AD Performance
3.2.1. Changes of DOM Components
3.2.2. Variations of VFA
3.2.3. Changes of SCOD
3.3. Effects of Nap on Zeta Potential of Sludge Particles
3.4. Effects of Nap on EPS Production and Composition
3.5. Effects of Nap on the Activity of Key Enzymes
3.6. Microbial Community Variation Responsed to Nap Concentration
3.6.1. Genus Level
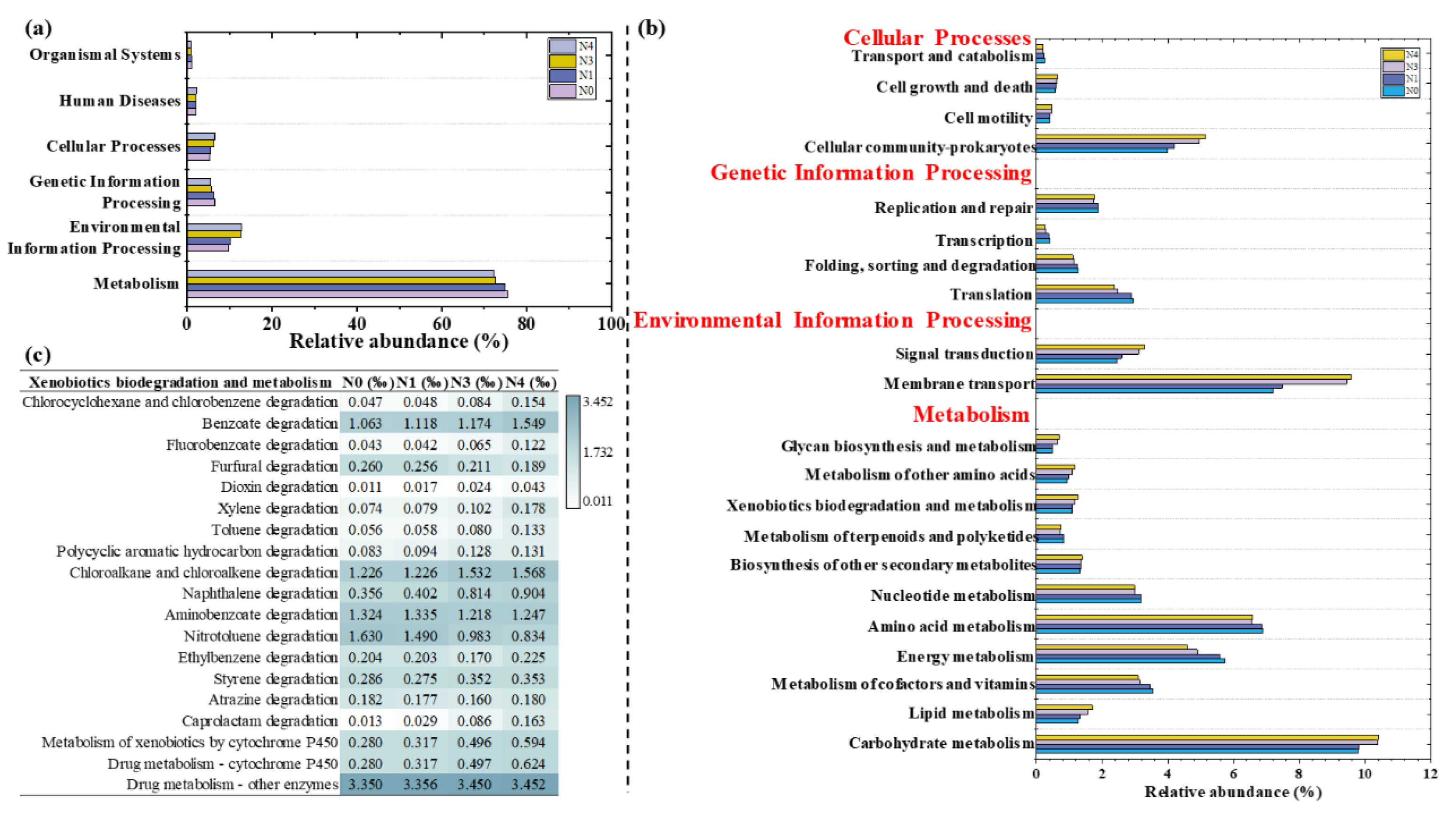
3.6.2. Potential Functions Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haritash, A.K.; Kaushik, C.P. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): A review. J. Hazard. Mater. 2009, 169, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zioła, N.; Słaby, K. The Content of Selected Heavy Metals and Polycyclic Aromatic Hydrocarbons (PAHs) in PM10 in Urban-Industrial Area. Sustainability 2020, 12, 5284. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, L.; Cai, Y.; Chen, Y. Distribution of polycyclic aromatic hydrocarbon (PAH) residues in several tissues of edible fishes from the largest freshwater lake in China, Poyang Lake, and associated human health risk assessment. Ecotoxicol. Environ. Saf. 2014, 104, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Hasinger, M.; Scherr, K.E.; Lundaa, T.; Bräuer, L.; Zach, C.; Loibner, A.P. Changes in iso-and n-alkane distribution during biodegradation of crude oil under nitrate and sulphate reducing conditions. J. Biotechnol. 2012, 157, 490–498. [Google Scholar] [CrossRef]
- Lv, W.; Schanbacher, F.L.; Yu, Z. Putting microbes to work in sequence: Recent advances in temperature-phased anaerobic digestion processes. Bioresour. Technol. 2010, 101, 9409–9414. [Google Scholar] [CrossRef]
- Valdman, E.; Battaglini, F.; Leite, S.; Valdman, B. Naphthalene detection by a bioluminescence sensor applied to wastewater samples. Sens. Actuators B Chem. 2004, 103, 7–12. [Google Scholar] [CrossRef]
- El-Naggar, M.; Hanafy, S.; Younis, A.M.; Ghandour, M.A.; El-Sayed, A.-A.Y. Seasonal and Temporal Influence on Polycyclic Aromatic Hydrocarbons in the Red Sea Coastal Water, Egypt. Sustainability 2021, 13, 11906. [Google Scholar] [CrossRef]
- Jin, B.; Niu, J.; Liu, Y.; Zhao, J.; Yin, Z. Effects of polycyclic aromatic hydrocarbons on sludge performance for denitrification and phosphorus removal. Chem. Eng. J. 2020, 397, 125552. [Google Scholar] [CrossRef]
- Verrhiest, G.J.; Clément, B.; Volat, B.; Montuelle, B.; Perrodin, Y. Interactions between a polycyclic aromatic hydrocarbon mixture and the microbial communities in a natural freshwater sediment. Chemosphere 2002, 46, 187–196. [Google Scholar] [CrossRef]
- Yu, H.; Xia, Q.; Yan, J.; Herreno-Saenz, D.; Wu, Y.-S.; Tang, I.-W.; Fu, P.P. Photoirradiation of polycyclic aromatic hydrocarbons with UVA light–a pathway leading to the generation of reactive oxygen species, lipid peroxidation, and DNA damage. Int. J. Env. Res. Public Health 2006, 3, 348–354. [Google Scholar] [CrossRef]
- Krishnamurthi, K.; Devi, S.S.; Chakrabarti, T. The genotoxicity of priority polycyclic aromatic hydrocarbons (PAHs) containing sludge samples. Toxicol. Mech. Methods 2007, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cebulska-Wasilewska, A.; Binkova, B.; Sram, R.J.; Kalina, I.; Popov, T.; Farmer, P.B. Repair competence assay in studies of the influence of environmental exposure to c-PAHs on individual susceptibility to induction of DNA damage. Mutat. Res.-Fundam. Mol. Mech. Mutag. 2007, 620, 155–164. [Google Scholar] [CrossRef] [PubMed]
- State Environmental Protection Administration. The Water and Wastewater Monitoring Analysis Method Editorial Board. Water and Wastewater Monitoring Analysis Method, 4th ed.; China Environmental Science Press: Beijing, China, 2002; pp. 38–47. (In Chinese)
- Song, L.; Li, D.; Cao, X.; Tang, Y.; Liu, R.; Niu, Q.; Li, Y.Y. Optimizing biomethane production of mesophilic chicken manure and sheep manure digestion: Mono-digestion and co-digestion kinetic investigation, autofluorescence analysis and microbial community assessment. J. Environ. Manag. 2019, 237, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Jing, M.; Han, G.; Wan, J.; Zhang, S.; Liu, R. Catalase and superoxide dismutase response and the underlying molecular mechanism for naphthalene. Sci. Total Environ. 2020, 736, 139567. [Google Scholar] [CrossRef] [PubMed]
- Wemheuer, F.; Taylor, J.; Daniel, R.; Johnston, E.; Meinicke, P.; Thomas, T.; Wemheuer, B. Tax4Fun2: Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome 2020, 15, 11. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Gan, L.; Chen, Z.L. Biodegradation of naphthalene by strain Bacillus fusiformis (BFN). J. Hazard. Mater. 2010, 182, 771–777. [Google Scholar] [CrossRef]
- Morasch, B.; Annweiler, E.; Warthmann, R.J.; Meckenstock, R.U. The use of a solid adsorber resin for enrichment of bacteria with toxic substrates and to identify metabolites: Degradation of naphthalene, o -, and m -xylene by sulfate-reducing bacteria. J. Microbiol. Methods 2001, 44, 183–191. [Google Scholar] [CrossRef]
- Chang, S.; Liu, X.; Ren, B.; Liu, B. Effects of LB Broth, Naphthalene Concentration, and Acetone on the Naphthalene Degradation Activities by Pseudomonas putida G7. Water Environ. Res. 2015, 87, 61–67. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, Z.; Tang, S. Characterization of dissolved organic matter in a submerged membrane bioreactor by using three-dimensional excitation and emission matrix fluorescence spectroscopy. Water Res. 2009, 43, 1533–1540. [Google Scholar] [CrossRef]
- Cao, S.; Sun, F.; Lu, D.; Zhou, Y. Characterization of the refractory dissolved organic matters (rDOM) in sludge alkaline fermentation liquid driven denitrification: Effect of HRT on their fate and transformation. Water Res. 2019, 159, 135–144. [Google Scholar] [CrossRef]
- Kun, L.; Yinguang, C.; Naidong, X.; Xiong, Z.; Mu, L. Effect of humic acids with different characteristics on fermentative short-chain fatty acids production from waste activated sludge. Environ. Sci. Technol. 2015, 49, 4929–4936. [Google Scholar]
- Karthikeyan, O.P.; Selvam, A.; Wong, J. Hydrolysis-acidogenesis of food waste in solid-liquid-separating continuous stirred tank reactor (SLS-CSTR) for volatile organic acid production. Bioresour. Technol. 2016, 200, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Leng, L.; Yang, P.; Singh, S.; Zhuang, H.; Xu, L.; Chen, W.-H.; Dolfing, J.; Li, D.; Zhang, Y.; Zeng, H.; et al. A review on the bioenergetics of anaerobic microbial metabolism close to the thermodynamic limits and its implications for digestion applications. Bioresour. Technol. 2018, 247, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Yue, L.; Cheng, J.; Zhang, H.; Yuan, L.; Hua, J.; Dong, H.; Li, Y.-Y.; Zhou, J. Inhibition of N-Vanillylnonanamide in anaerobic digestion of lipids in food waste: Microorganisms damage and blocked electron transfer. J. Hazard. Mater. 2020, 399, 123098. [Google Scholar] [CrossRef] [PubMed]
- Lunardi, C.; Gomes, A.; Rocha, F.; Tommaso, J.; Patience, G. Experimental methods in chemical engineering: Zeta potential. Can. J. Chem. Eng. 2021, 99, 627–639. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, Y.; Gao, H.; Fan, X.; Guo, Y.; Wang, H.; Zheng, H. Variations in macro and micro physicochemical properties of activated sludge under a moderate oxidation- in situ coagulation conditioning: Relationship between molecular structure and dewaterability. Water Res. 2019, 155, 245–254. [Google Scholar] [CrossRef]
- Zhang, Z.; Guo, L.; Wang, Y.; Li, F.; Zhao, Y.; Gao, M.; She, Z. Degradation and transformation of extracellular polymeric substances (EPS) and dissolved organic matters (DOM) during two-stage anaerobic digestion with waste sludge. Int. J. Hydrogen Energy 2017, 42, 9619–9629. [Google Scholar] [CrossRef]
- Tian, X.; Shen, Z.; Han, Z.; Zhou, Y. The effect of extracellular polymeric substances on exogenous highly toxic compounds in biological wastewater treatment: An overview. Bioresour. Technol. Rep. 2019, 5, 28–42. [Google Scholar] [CrossRef]
- Hermann, J.H.; Frans, J.W.; Sikkema, J.; Keweloh, H.; de Bont, J.A. Mechanisms of resistance of whole cells to toxic organic solvents. Elsevier Current Trends 1994, 12, 409–415. [Google Scholar]
- Tan, Y.; Zheng, C.; Cai, T.; Niu, C.; Wang, S.; Pan, Y.; Lu, X.; Zhen, G.; Qian, G.; Zhao, Y. Anaerobic bioconversion of petrochemical wastewater to biomethane in a semi-continuous bioreactor: Biodegradability, mineralization behaviors and methane productivity. Bioresour. Technol. 2020, 304, 123005. [Google Scholar] [CrossRef]
- Li, S.; Cao, Y.; Zhao, Z.; Zhang, Y. Regulating Secretion of Extracellular Polymeric Substances through Dosing Magnetite and Zerovalent Iron Nanoparticles to Affect Anaerobic Digestion Mode. ACS Sustain. Chem. Eng. 2019, 7, 9655–9662. [Google Scholar] [CrossRef]
- Tay, J.-H.; Liu, Q.-S.; Liu, Y. The role of cellular polysaccharides in the formation and stability of aerobic granules. Lett. Appl. Microbiol. 2001, 33, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, J.; Deng, D.; Li, R.; Guo, C.; Ma, J.; Chen, M. Investigation of extracellular polymeric substances (EPS) in four types of sludge: Factors influencing EPS properties and sludge granulation. J. Water Process Eng. 2021, 40, 101924. [Google Scholar] [CrossRef]
- Park, M.; Snyder, S.A. Sample handling and data processing for fluorescent excitation-emission matrix (EEM) of dissolved organic matter (DOM). Chemosphere 2018, 193, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chen, H. Correlation of porous structure, mass transfer and enzymatic hydrolysis of steam exploded corn stover. Chem. Eng. Sci. 2013, 104, 1036–1044. [Google Scholar] [CrossRef]
- Klein, M.I.; Duarte, S.; Xiao, J.; Mitra, S.; Foster, T.H.; Koo, H. Structural and molecular basis of the role of starch and sucrose in streptococcus mutans biofilm development. Appl. Environ. Microbiol. 2009, 75, 837–841. [Google Scholar] [CrossRef]
- Gunn, J.S.; Bakaletz, L.O.; Wozniak, D.J. What’s on the Outside Matters: The Role of the Extracellular Polymeric Substance of Gram-negative Biofilms in Evading Host Immunity and as a Target for Therapeutic Intervention. J. Biol. Chem. 2016, 291, 12538–12546. [Google Scholar] [CrossRef]
- Fan, W.; Yuan, G.; Li, Q.; Wei, L. Antibacterial mechanisms of methyl gallate against Ralstonia solanacearum. Australas. Plant Path. 2014, 43, 1–7. [Google Scholar] [CrossRef]
- Zhu, J.-K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Nekoei, A.R.; Vatanparast, M. π-Hydrogen bonding and aromaticity: A systematic interplay study. Phys. Chem. Chem. Phys. 2019, 21, 623–630. [Google Scholar] [CrossRef]
- Li, R.; Duan, N.; Zhang, Y.; Liu, Z.; Li, B.; Zhang, D.; Dong, T. Anaerobic co-digestion of chicken manure and microalgae Chlorella sp.: Methane potential, microbial diversity and synergistic impact evaluation. Waste Manag. 2017, 68, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Yong, N.; Zhao, J.; Tang, Y.; Guo, P.; Yang, Y.; Wu, X.; Zhao, F. Species Divergence vs. Functional Convergence Characterizes Crude Oil Microbial Community Assembly. Front. Microbiol. 2016, 7, 1254. [Google Scholar]
- Conrad, R.; Klose, M.; Claus, P. Phosphate inhibits acetotrophic methanogenesis on rice roots. Appl. Environ. Microbiol. 2000, 66, 828–831. [Google Scholar] [CrossRef] [PubMed]
- Sonthiphand, P.; Hall, M.W.; Neufeld, J.D. Biogeography of anaerobic ammonia-oxidizing (anammox) bacteria. Front. Microbiol. 2014, 5, 399. [Google Scholar] [CrossRef]
- Rivière, D.; Desvignes, V.; Pelletier, E.; Chaussonnerie, S.; Guermazi, S.; Weissenbach, J.; Li, T.; Camacho, P.; Sghir, A. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 2009, 3, 700–714. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Shin, S.G.; Han, G.; Koo, T.; Hwang, S. Bacteria and archaea communities in full-scale thermophilic and mesophilic anaerobic digesters treating food wastewater: Key process parameters and microbial indicators of process instability. Bioresour. Technol. 2017, 245, 689–697. [Google Scholar] [CrossRef]
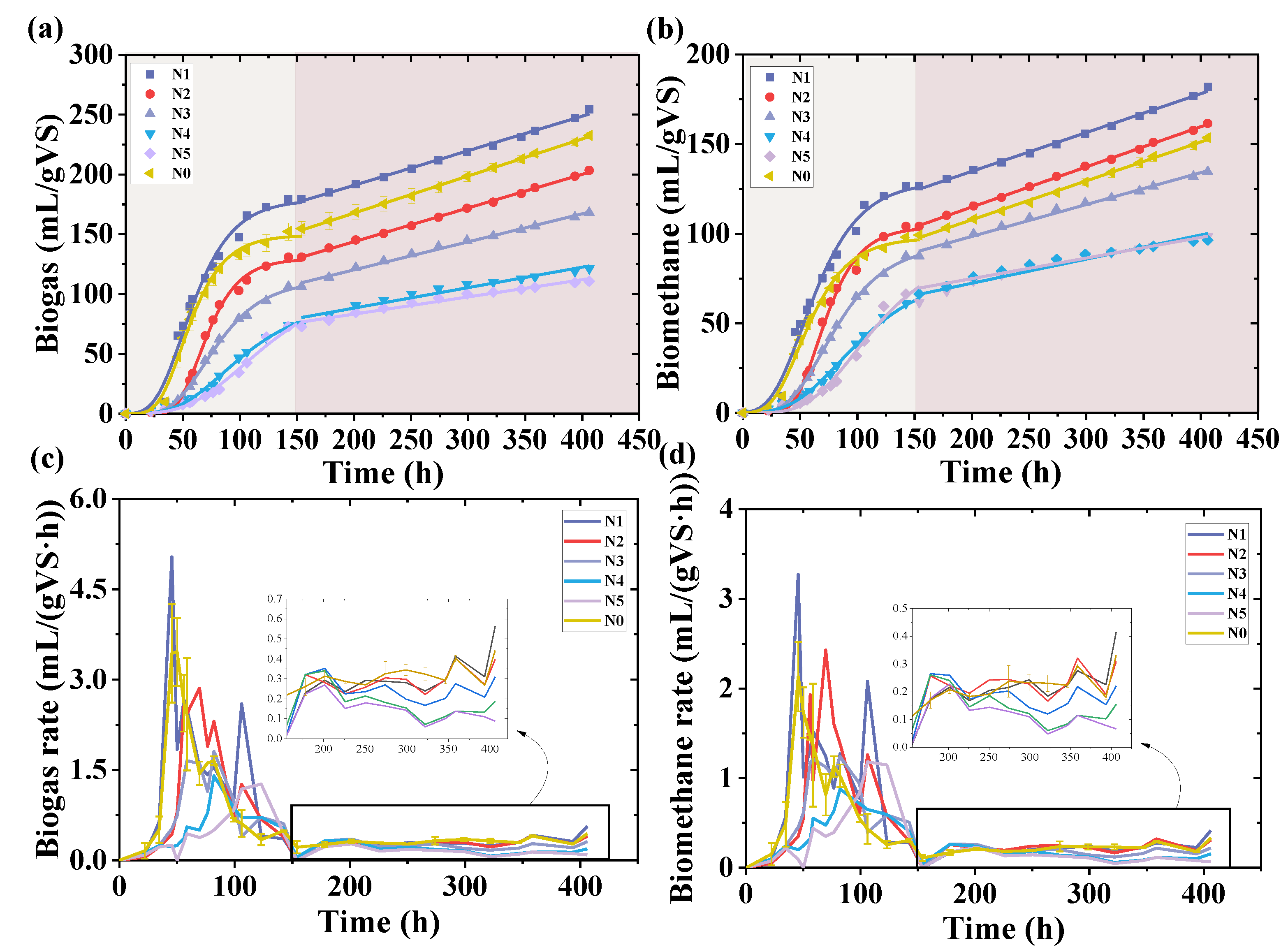
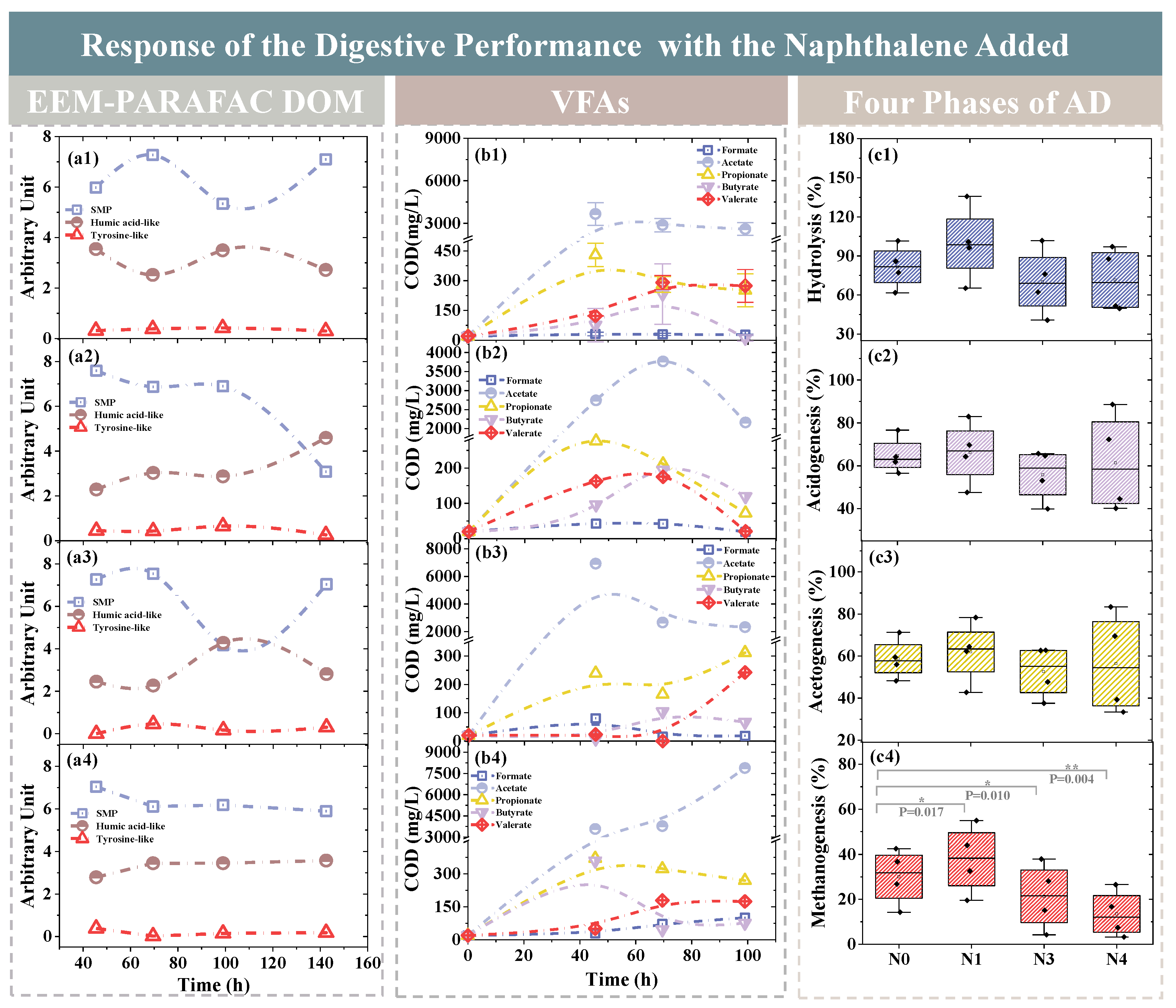
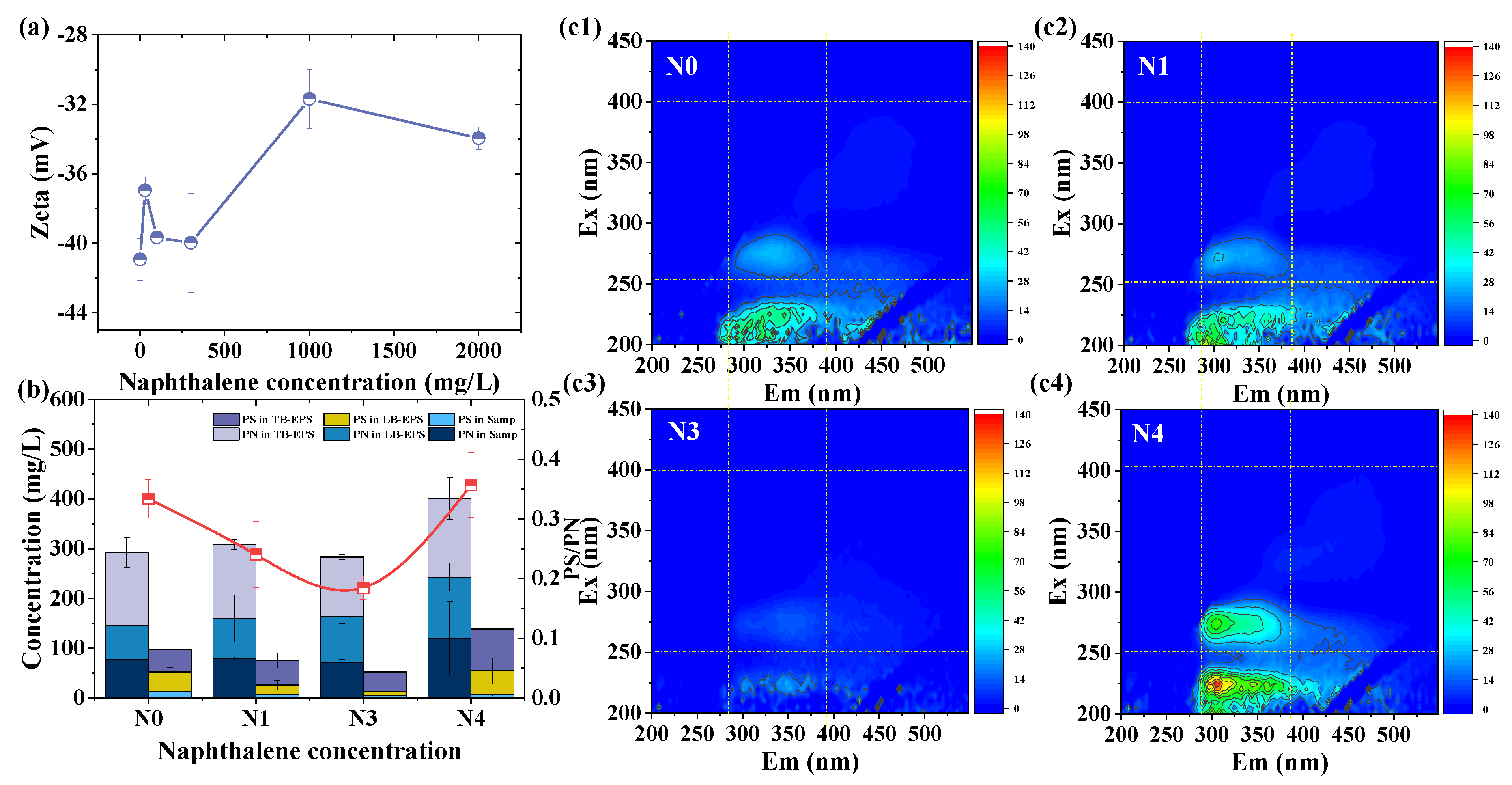

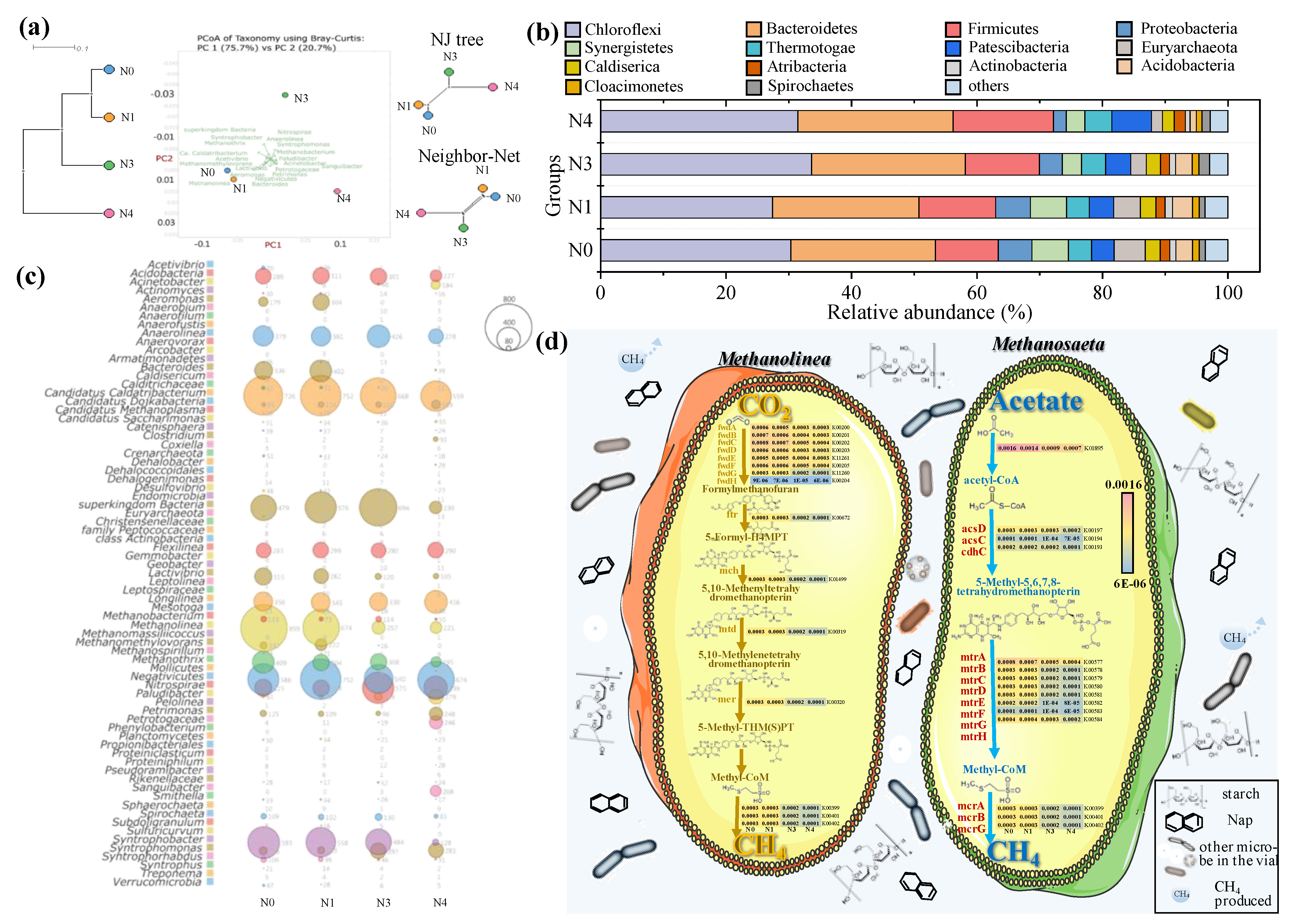
| Group | Sludge (g) | Basal Medium (mL) | Starch (g) | Nap (mg/L) | Methanol (mL) | Methane (mL) |
|---|---|---|---|---|---|---|
| N0 | 15 | 90 | 0.5 | 0 | 0.5 | 415.25 |
| N1 | 15 | 90 | 0.5 | 30 | 0.5 | 415.25 |
| N2 | 15 | 90 | 0.5 | 100 | 0.5 | 415.25 |
| N3 | 15 | 90 | 0.5 | 300 | 0.5 | 415.25 |
| N4 | 15 | 90 | 0.5 | 1000 | 0.5 | 415.25 |
| N5 | 15 | 90 | 0.5 | 2000 | 0.5 | 415.25 |
| Model | Parameters | N0 | N1 | N2 | N3 | N4 | N5 |
|---|---|---|---|---|---|---|---|
| Gompertz (Biogas) | P0 (mL/gVS) | 149.32 | 178.90 | 129.52 | 111.49 | 92.77 | 109.97 |
| Standard error | 3.02 | 5.70 | 2.37 | 1.97 | 4.75 | 16.26 | |
| K mL/(gVS·h) | 2.66 | 2.72 | 2.47 | 1.44 | 0.84 | 0.82 | |
| Standard error | 0.18 | 0.25 | 0.13 | 0.05 | 0.03 | 0.04 | |
| A (h) | 28.10 | 25.35 | 44.87 | 40.15 | 44.75 | 53.90 | |
| Standard error | 1.91 | 2.93 | 1.21 | 1.00 | 1.56 | 2.83 | |
| Reduced chi-sqr | 24.86 | 66.93 | 12.15 | 3.34 | 3.68 | 11.04 | |
| Adj. R-Square | 0.99 | 0.99 | 1.00 | 1.00 | 1.00 | 0.99 | |
| Residual Sum of Squares | 298.33 | 803.13 | 145.82 | 40.09 | 44.15 | 132.47 | |
| Root-MSE (SD) | 4.99 | 8.18 | 3.49 | 1.83 | 1.92 | 3.32 | |
| NRMSE | 0.03 | 0.05 | 0.03 | 0.02 | 0.03 | 0.05 | |
| AIC | 50.85 | 65.71 | 40.11 | 20.75 | 22.19 | 38.67 | |
| Gompertz (Biomethane) | P0 (mL/gVS) | 97.73 | 129.14 | 103.77 | 93.68 | 77.59 | 90.86 |
| Standard error | 1.97 | 4.47 | 2.14 | 1.30 | 3.52 | 10.69 | |
| K mL/(gVS·h) | 1.50 | 1.70 | 1.88 | 1.13 | 0.67 | 0.81 | |
| Standard error | 0.09 | 0.14 | 0.10 | 0.03 | 0.02 | 0.05 | |
| A (h) | 24.99 | 24.00 | 44.71 | 39.28 | 42.17 | 56.37 | |
| Standard error | 1.88 | 2.94 | 1.34 | 0.73 | 1.36 | 2.70 | |
| Reduced chi-sqr | 8.23 | 29.14 | 8.93 | 1.15 | 1.83 | 9.66 | |
| Adj. R-Square | 0.98 | 0.99 | 1.00 | 0.99 | 0.98 | 0.99 | |
| Residual Sum of Squares | 98.70 | 349.68 | 107.17 | 13.81 | 21.91 | 115.87 | |
| Root MSE (SD) | 2.87 | 5.40 | 2.99 | 1.07 | 1.35 | 3.11 | |
| NRMSE | 0.02 | 0.05 | 0.03 | 0.01 | 0.02 | 0.03 | |
| AIC | 34.26 | 53.23 | 35.50 | 4.76 | 11.68 | 36.67 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, Y.; Li, J.; Xue, H.; Liu, Y.; Qiao, J.; Tang, J.; Liu, R.; Niu, Q. Influences of Naphthalene Concentration on Starch Anaerobic Digestion: Focusing on Digestion Performance, Extracellular Polymeric Substances and Function Microbial Community. Sustainability 2022, 14, 16377. https://doi.org/10.3390/su142416377
Yao Y, Li J, Xue H, Liu Y, Qiao J, Tang J, Liu R, Niu Q. Influences of Naphthalene Concentration on Starch Anaerobic Digestion: Focusing on Digestion Performance, Extracellular Polymeric Substances and Function Microbial Community. Sustainability. 2022; 14(24):16377. https://doi.org/10.3390/su142416377
Chicago/Turabian StyleYao, Yilin, Jingyi Li, Hanhan Xue, Yutong Liu, Junpeng Qiao, Jingchun Tang, Rutao Liu, and Qigui Niu. 2022. "Influences of Naphthalene Concentration on Starch Anaerobic Digestion: Focusing on Digestion Performance, Extracellular Polymeric Substances and Function Microbial Community" Sustainability 14, no. 24: 16377. https://doi.org/10.3390/su142416377
APA StyleYao, Y., Li, J., Xue, H., Liu, Y., Qiao, J., Tang, J., Liu, R., & Niu, Q. (2022). Influences of Naphthalene Concentration on Starch Anaerobic Digestion: Focusing on Digestion Performance, Extracellular Polymeric Substances and Function Microbial Community. Sustainability, 14(24), 16377. https://doi.org/10.3390/su142416377







