Genome-Wide Association Study Based on Plant Height and Drought-Tolerance Indices Reveals Two Candidate Drought-Tolerance Genes in Sweet Sorghum
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Materials and Experimental Design
2.2. Measurements and Data Analysis
3. Results
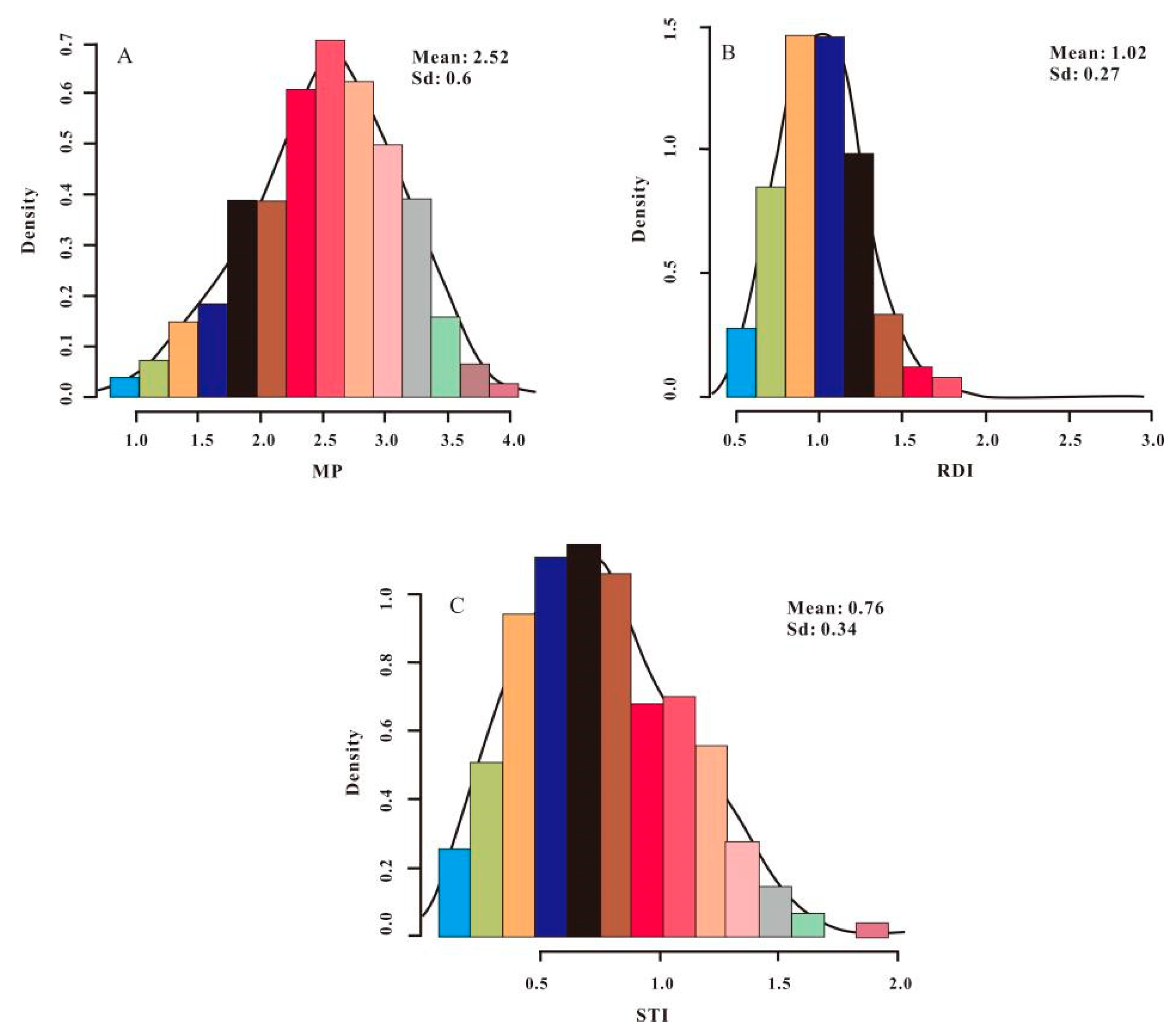
3.1. Distribution of PH and Drought-Tolerance Indices in Sweet Sorghum
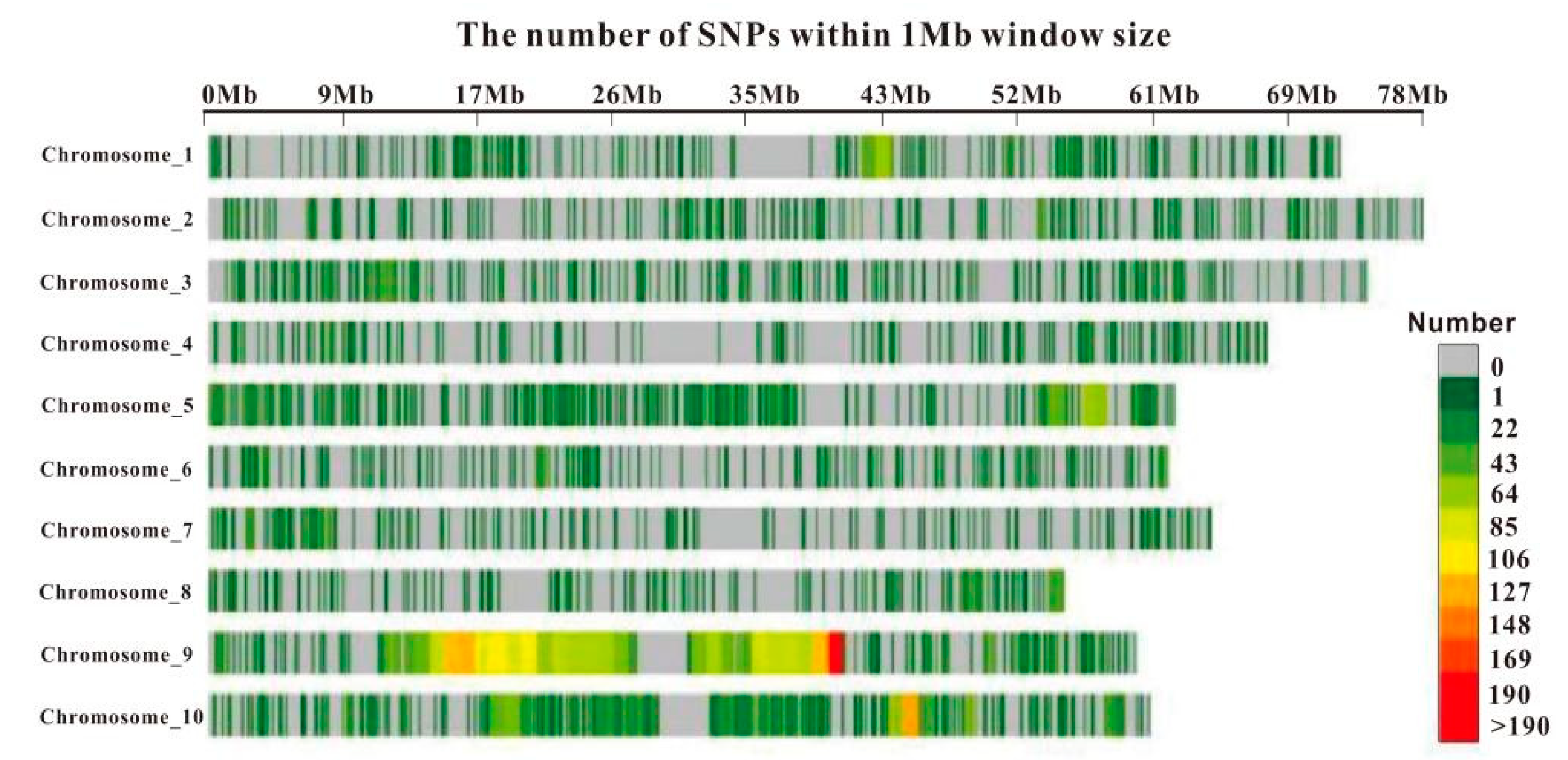
3.2. SNP Marker Density Distribution
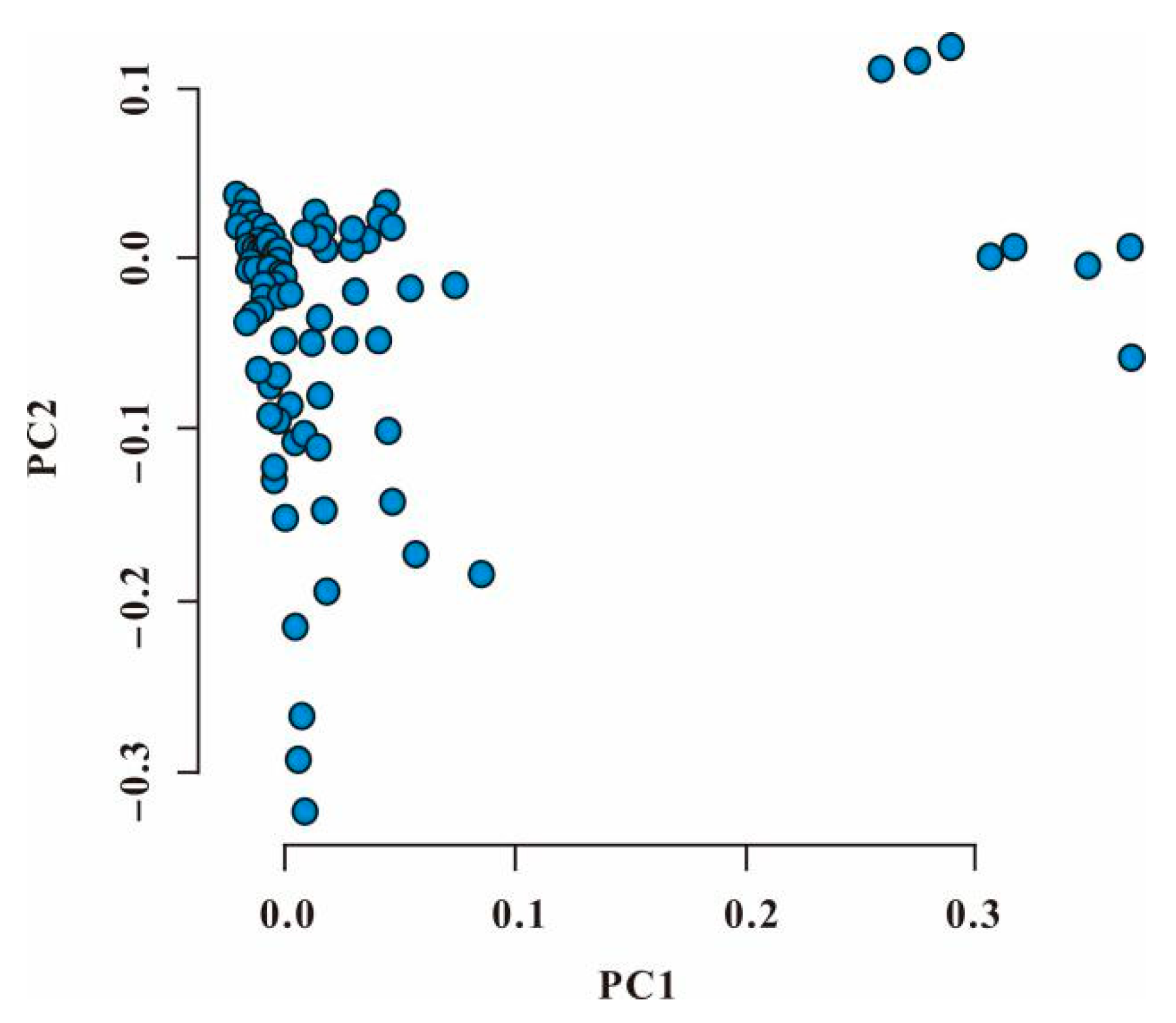
3.3. Principal Component Analysis of Genotype Data
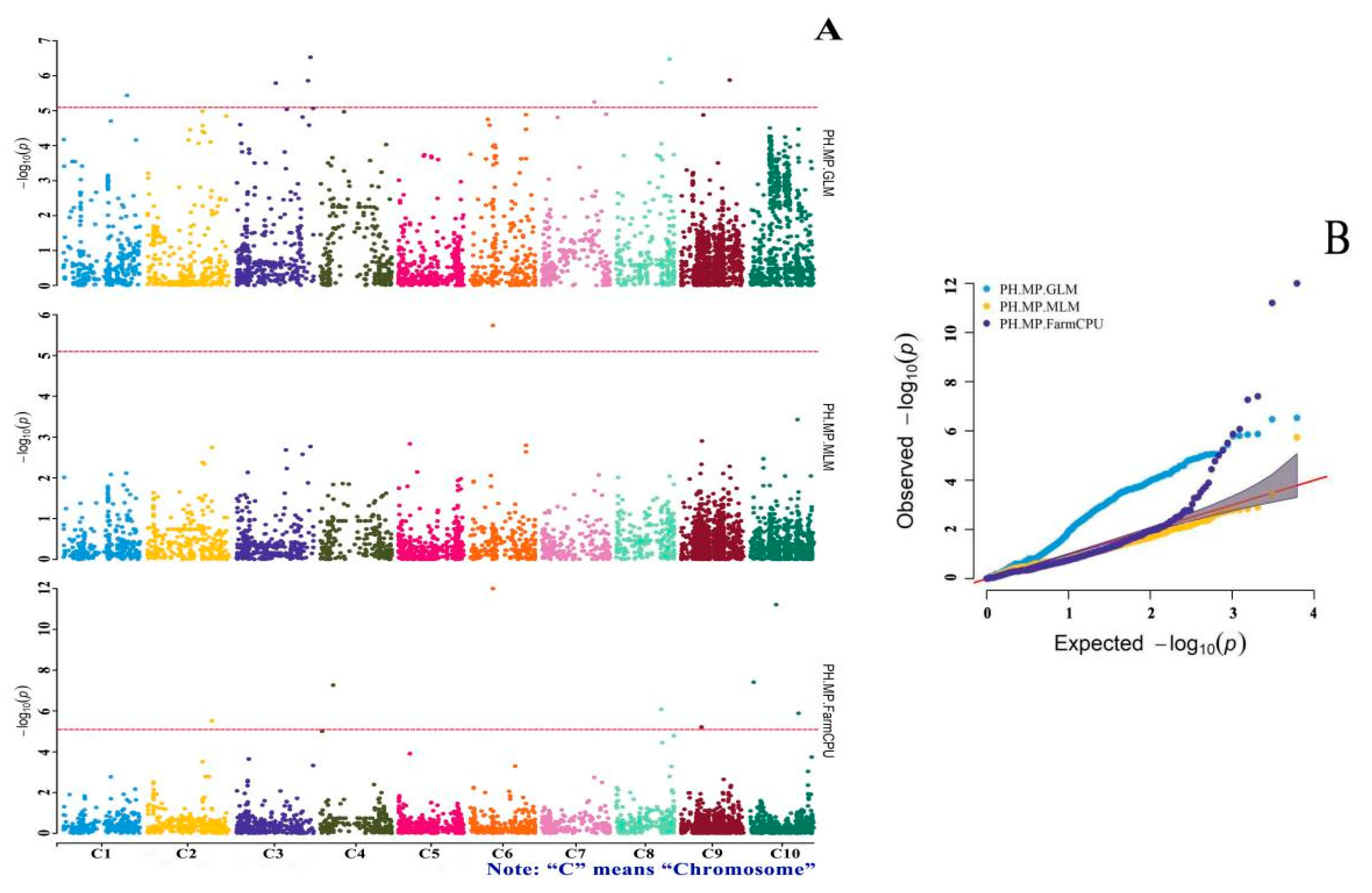
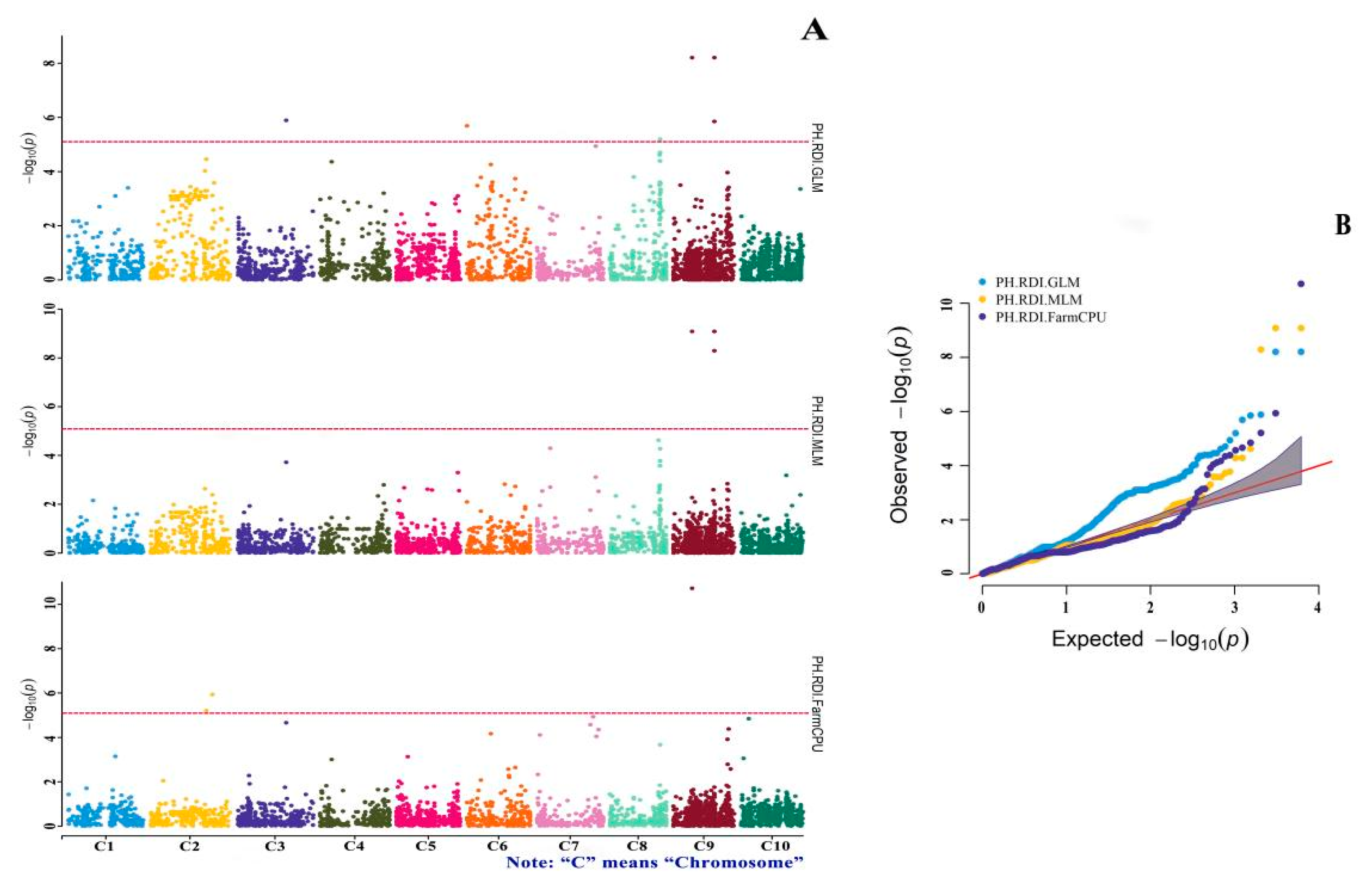
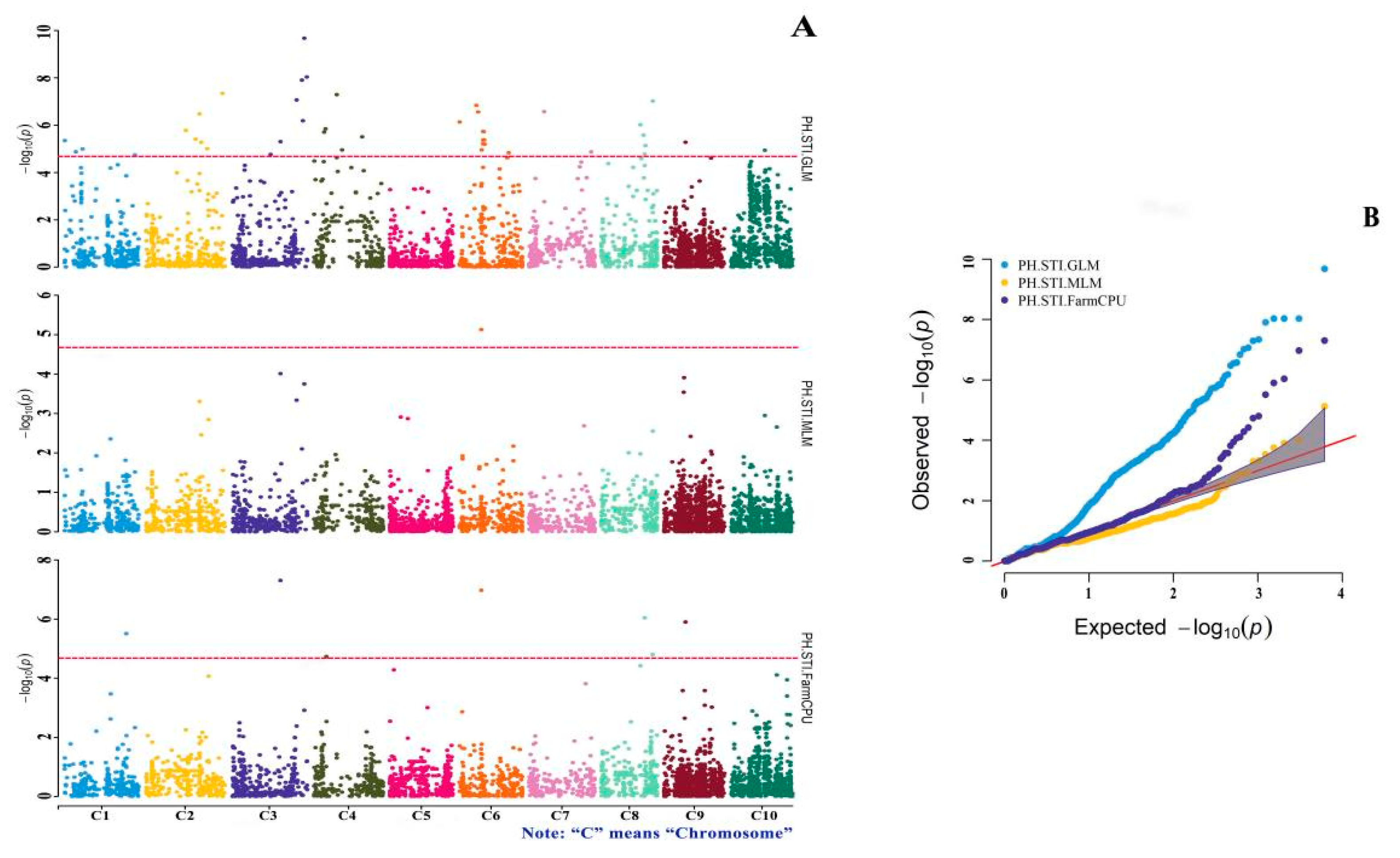
3.4. GWAS of Drought-Tolerance Indices
3.5. Gene Function Annotation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lei, Y.J.; Zhang, Y.F.; Zhang, M.H.; Liang, X.M.; Shao, X.J. Calculation and prediction of water resource ecological footprint in Xinjiang. Agric. Res. Arid. Areas 2017, 35, 142–150. [Google Scholar]
- Hu, M.F.; Tian, C.Y.; Zhao, Z.Y.; Wang, L.X. Salinization causes and research progress of technologies improving saline-alkali soil in Xinjiang. J. Northwest A&F Univ. 2012, 40, 111–117. [Google Scholar]
- Mohamed, M.F.M.; Emam, M.M.; Salama, K.H.A.; Morsy, A.A. Sorghum under saline conditions: Responses, tolerance mechanisms, and management strategies. Planta 2021, 254, 24. [Google Scholar]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef]
- Ma, X.; Feng, F.; Wei, H.; Mei, H.; Xu, K.; Chen, S.; Li, T.; Liang, X.; Liu, H.; Luo, L. Genome-Wide Association Study for Plant Height and Grain Yield in Rice under Contrasting Moisture Regimes. Front. Plant Sci. 2016, 7, 1801. [Google Scholar] [CrossRef]
- Guo, Z.; Yang, W.; Chang, Y.; Ma, X.; Tu, H.; Xiong, F.; Jiang, N.; Feng, H.; Huang, C.; Yang, P.; et al. Genome-Wide Association Studies of Image Traits Reveal Genetic Architecture of Drought Resistance in Rice. Mol. Plant. 2018, 11, 789–805. [Google Scholar] [CrossRef]
- Pantalião, G.F.; Narciso, M.; Guimarães, C.; Castro, A.; Colombari, J.M.; Breseghello, F.; Rodrigues, L.; Vianello, R.P.; Borba, T.O.; Brondani, C. Genome-wide association study (GWAS) for grain yield in rice cultivated under water deficit. Genetica 2016, 144, 651–664. [Google Scholar] [CrossRef]
- Hoang, G.T.; Van Dinh, L.; Nguyen, T.T.; Ta, N.K.; Gathignol, F.; Mai, C.D.; Jouannic, S.; Tran, K.D.; Khuat, T.H.; Do, V.N.; et al. Genome-wide Association Study of a Panel of Vietnamese Rice Landraces Reveals New QTLs for Tolerance to Water Deficit During the Vegetative Phase. Rice 2019, 12, 4. [Google Scholar] [CrossRef]
- Sukumaran, S.; Dreisigacker, S.; Lopes, M.; Chavez, P.; Reynolds, M.P. Genome-wide association study for grain yield and related traits in an elite spring wheat population grown in temperate irrigated environments. Theor. Appl. Genet. 2015, 128, 353–363. [Google Scholar] [CrossRef]
- Mei, F.; Chen, B.; Du, L.; Li, S.; Zhu, D.; Chen, N.; Zhang, Y.; Li, F.; Wang, Z.; Cheng, X.; et al. A gain-of-function allele of a DREB transcription factor gene ameliorates drought tolerance in wheat. Plant Cell. 2022, 34, 4472–4494. [Google Scholar] [CrossRef]
- Zheng, X.; Qiao, L.; Liu, Y.; Wei, N.; Zhao, J.; Wu, B.; Yang, B.; Wang, J.; Zheng, J. Genome-Wide Association Study of Grain Number in Common Wheat from Shanxi Under Different Water Regimes. Front. Plant Sci. 2022, 12, 806295. [Google Scholar] [CrossRef] [PubMed]
- Mathew, I.; Shimelis, H.; Shayanowako, A.; Laing, M.; Chaplot, V. Genome-wide association study of drought tolerance and biomass allocation in wheat. PLoS ONE 2019, 14, e0225383. [Google Scholar] [CrossRef]
- Wang, N.; Wang, Z.P.; Liang, X.L.; Weng, J.F.; Lv, X.L.; Zhang, D.G.; Yang, J.; Yong, H.J.; Li, M.S.; Li, H.F.; et al. Identification of loci contributing to maize drought tolerance in a genome-wide association study. Euphytica 2016, 210, 165–179. [Google Scholar] [CrossRef]
- Wu, X.; Feng, H.; Wu, D.; Yan, S.; Zhang, P.; Wang, W.; Zhang, J.; Ye, J.; Dai, G.; Fan, Y.; et al. Using high-throughput multiple optical phenotyping to decipher the genetic architecture of maize drought tolerance. Genome Biol. 2021, 22, 185. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Liu, S.; Ferjani, A.; Li, J.; Yan, J.; Yang, X.; Qin, F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 2016, 48, 1233–1241. [Google Scholar] [CrossRef]
- Sun, M.; Li, Y.; Zheng, J.; Wu, D.; Li, C.; Li, Z.; Zang, Z.; Zhang, Y.; Fang, Q.; Li, W.; et al. A Nuclear Factor Y-B Transcription Factor, GmNFYB17, Regulates Resistance to Drought Stress in Soybean. Int. J. Mol. Sci. 2022, 23, 7242. [Google Scholar] [CrossRef]
- Chen, L.; Fang, Y.; Li, X.; Zeng, K.; Chen, H.; Zhang, H.; Yang, H.; Cao, D.; Hao, Q.; Yuan, S.; et al. Identification of soybean drought-tolerant genotypes and loci correlated with agronomic traits contributes new candidate genes for breeding. Plant. Mol. Biol. 2020, 102, 109–122. [Google Scholar] [CrossRef]
- Spindel, J.E.; Dahlberg, J.; Colgan, M.; Hollingsworth, J.; Sievert, J.; Staggenborg, S.H.; Hutmacher, R.; Jansson, C.; Vogel, J.P. Association mapping by aerial drone reveals 213 genetic associations for Sorghum bicolor biomass traits under drought. BMC Genom. 2018, 19, 679. [Google Scholar] [CrossRef]
- Maina, F.; Harou, A.; Hamidou, F.; Morris, G.P. Genome-wide association studies identify putative pleiotropic locus mediating drought tolerance in sorghum. Plant Direct. 2022, 6, 413. [Google Scholar] [CrossRef]
- Hou, S.; Zhu, G.; Li, Y.; Li, W.; Fu, J.; Niu, E.; Li, L.; Zhang, D.; Guo, W. Genome-Wide Association Studies Reveal Genetic Variation and Candidate Genes of Drought Stress Related Traits in Cotton (Gossypium hirsutum L.). Front. Plant. Sci. 2018, 9, 1276. [Google Scholar] [CrossRef]
- Guan, S.Y.; Jiao, P.; Jiang, Z.Z.; Qi, Z.; Xia, H.F.; Qu, J.; Ma, Y.Y. Research Progress of MYB Transcription Factors in Plant Abiotic Stress. J. Jilin Agric. Univ. 2019, 41, 253–260. [Google Scholar]
- Xu, L.; Wang, Y.C.; He, X.L. Huang, Y.H.; Xu, Z.L.; Shao, H.B.; Zhang, D.Y. Isolation, Expression and Binding Function Analysis of the Transcription Factor GmMYB52 in Soybean. Acta Agron. Sin. 2017, 43, 1458–1467. [Google Scholar] [CrossRef]
- Wang, C.; Lu, G.; Hao, Y.; Guo, H.; Guo, Y.; Zhao, J.; Cheng, H. ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in transgenic cotton. Planta 2017, 246, 453–469. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Yang, J.; Ferreira, T.; Morris, A.P.; Medland, S.E.; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; DIAbetes Genetics Replication and Meta-Analysis (DIAGRAM) Consortium; Madden, P.A.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012, 44, 369–375. [Google Scholar] [CrossRef]
- Wang, M.; Wang, L.; Jiang, N.; Jia, T.; Luo, Z. A robust and efficient statistical method for genetic association studies using case and control samples from multiple cohorts. BMC Genom. 2013, 14, 88. [Google Scholar] [CrossRef]
- Zhang, Z.; Buckler, E.S.; Casstevens, T.M.; Bradbury, P.J. Software engineering the mixed model for genome-wide association studies on large samples. Brief Bioinform. 2009, 10, 664–675. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Korte, A.; Vilhjálmsson, B.J.; Segura, V.; Platt, A.; Long, Q.; Nordborg, M. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat. Genet. 2012, 44, 1066–1071. [Google Scholar] [CrossRef]
- Yang, J.; Zaitlen, N.A.; Goddard, M.E.; Visscher, P.M.; Price, A.L. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 2014, 46, 100–106. [Google Scholar] [CrossRef]
- Zhang, Z.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef]
- Li, M.; Liu, X.; Bradbury, P.; Yu, J.; Zhang, Y.M.; Todhunter, R.J.; Buckler, E.S.; Zhang, Z. Enrichment of statistical power for genome-wide association studies. BMC Biol. 2014, 12, 73. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Dai, Y.H.; Guan, Y.; Liu, M.Q.; Zhang, Q.K.; He, X.H. Dynamic Monitoring and Evaluation of Ecological Environment Quality in Alar Reclamation Area from 1990 to 2020. Bull. Soil Water Conserv. 2022, 42, 122–128. [Google Scholar]
- Amiri, R.; Bahraminejad, S.; Sasani, S.; Ghobadi, M. Genetic evaluation of 80 irrigated bread wheat genotypes for drought tolerance indices. Bulg. J. Agric. Sci. 2014, 20, 101–111. [Google Scholar]
- Li, C.; Sun, B.; Li, Y.; Liu, C.; Wu, X.; Zhang, D.; Shi, Y.; Song, Y.; Buckler, E.S.; Zhang, Z.; et al. Numerous genetic loci identified for drought tolerance in the maize nested association mapping populations. BMC Genom. 2016, 17, 894. [Google Scholar] [CrossRef]
- Khan, S.U.; Zheng, Y.; Chachar, Z.; Zhang, X.; Zhou, G.; Zong, N.; Leng, P.; Zhao, J. Dissection of Maize Drought Tolerance at the Flowering Stage Using Genome-Wide Association Studies. Genes 2022, 13, 564. [Google Scholar] [CrossRef]
- Khayatnezhad, M.; Zaefizadeh, M.; Gholamin, R.; Jamaati-E-Somarin, S. Study of Genetic Diversity and Path Analysis for Yield in Durum Wheat Genotypes under Water and Dry Conditions. World Appl. Sci. J. 2010, 9, 655–665. [Google Scholar]
- Saeidi, M.; Abdoli, M.; Azhand, M.; Khas-amiri, M. Evaluation of drought resistance of barley (Hordeum vulgare L.) cultivars using agronomic characteristics and drought tolerance indices. Albanian J. Agric. Sci. 2013, 12, 545–554. [Google Scholar]
- Aditya, J.; Bhartiya, A.; Pal, R.S.; Kant, L.; Pattanayak, A. Identification of drought tolerant, high yielding rice genotypes for rainfed upland ecosystem of uttarakhand hills through different drought tolerance indices. J. Environ. Biol. 2022, 43, 306–316. [Google Scholar] [CrossRef]
- Mueen, A.K.; Hafiz Muhammad, F.U.; Iqbal, M.; Rehman, A.; Chattha, W.S. Evaluation of high-yielding wheat (Triticum aestivum L.) varieties under water limitation. Plant. Genet. Resour. 2021, 19, 245–251. [Google Scholar]
- Ballesta, P.; Mora, F.; Del, P.A. Association mapping of drought tolerance indices in wheat: QTL-rich regions on chromosome 4A. Sci. Agric. 2020, 77, 153. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh, B.I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.P.; Chen, P.; Hong, W.J.; Zhao, X.Y.; Liu, X.M. Research Progress of MYB Transcription Factor Family in Arabidopsis thaliana. Life Sci. Res. 2016, 20, 555–560. [Google Scholar]
- Xiong, H.; Li, J.; Liu, P.; Duan, J.; Zhao, Y.; Guo, X.; Li, Y.; Zhang, H.; Ali, J.; Li, Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 2014, 9, e92913. [Google Scholar] [CrossRef]
- Ma, H.; Liu, C.; Li, Z.; Ran, Q.; Xie, G.; Wang, B.; Fang, S.; Chu, J.; Zhang, J. ZmbZIP4 Contributes to Stress Resistance in Maize by Regulating ABA Synthesis and Root Development. Plant Physiol. 2018, 178, 753–770. [Google Scholar] [CrossRef]
| Chromosome | Length (bp) | No. of SNPs | Average Density (SNPs/Mb) |
|---|---|---|---|
| 1 | 72,621,628 | 392 | 5.4 |
| 2 | 77,923,599 | 340 | 4.4 |
| 3 | 74,347,826 | 379 | 5.1 |
| 4 | 67,928,809 | 243 | 3.6 |
| 5 | 61,993,318 | 615 | 10.0 |
| 6 | 61,563,909 | 284 | 4.6 |
| 7 | 64,298,007 | 266 | 4.1 |
| 8 | 54,875,046 | 271 | 4.9 |
| 9 | 59,493,343 | 2401 | 40.4 |
| 10 | 60,355,397 | 995 | 16.5 |
| Marker | Variant | Effect | p-Value | Methods |
|---|---|---|---|---|
| SNP-9-46359555 | T/C | 0.259 | 1.34 × 10−6 | MP.GLM |
| SNP-9-32010157 | T/C | −0.800 | 5.43 × 10−6 | MP.FarmCPU |
| SNP-8-54701961 | C/T | −0.280 | 5.27 × 10−6 | MP.FarmCPU |
| SNP-8-50726311 | C/G | −0.180 | 3.37 × 10−7 | MP.GLM |
| SNP-8-42803746 | G/A | −0.100 | 1.22 × 10−7 | MP.FarmCPU |
| SNP-8-42803746 | G/A | −0.140 | 1.57 × 10−6 | MP.GLM |
| SNP-7-49761327 | G/C | −0.503 | 5.68 × 10−6 | MP.GLM |
| SNP-6-21397577 | T/C | −0.776 | 1.73 × 10−12 | MP.FarmCPU |
| SNP-6-21397577 | T/C | −0.745 | 1.84 × 10−6 | MP.MLM |
| SNP-4-1724051 | C/T | −0.099 | 3.23 × 10−6 | MP.FarmCPU |
| SNP-4-12346536 | A/T | −0.293 | 2.50 × 10−9 | MP.FarmCPU |
| SNP-3-71564436 | A/C | −0.336 | 2.98 × 10−7 | MP.GLM |
| SNP-3-69226243 | A/G | −0.287 | 1.40 × 10−6 | MP.GLM |
| SNP-3-38408072 | C/A | −0.140 | 1.63 × 10−6 | MP.GLM |
| SNP-2-62168227 | A/C | 0.137 | 1.72 × 10−6 | MP.FarmCPU |
| SNP-1-60603003 | T/C | 0.316 | 3.69 × 10−6 | MP.GLM |
| SNP-10-45922678 | T/C | −0.208 | 1.57 × 10−7 | MP.FarmCPU |
| SNP-10-3166515 | C/T | 0.349 | 1.21 × 10−9 | MP.FarmCPU |
| SNP-10-24652464 | T/C | 0.561 | 5.82 × 10−12 | MP.FarmCPU |
| Marker | Variant | Effect | p-Value | Methods |
|---|---|---|---|---|
| SNP-9-40211466 | A/T | −0.920 | 1.42 × 10−6 | RDI.GLM |
| SNP-9-40211466 | A/T | −1.479 | 5.15 × 10−9 | RDI.MLM |
| SNP-9-40211299 | T/A | −1.059 | 6.28 × 10−9 | RDI.GLM |
| SNP-9-40211299 | T/A | −1.504 | 8.32 × 10−10 | RDI.MLM |
| SNP-9-18625001 | G/A | −0.991 | 3.42 × 10−18 | RDI.FarmCPU |
| SNP-9-18625001 | G/A | −1.059 | 6.28 × 10−9 | RDI.GLM |
| SNP-9-18625001 | G/A | −1.504 | 8.32 × 10−10 | RDI.MLM |
| SNP-8-48976460 | T/C | −0.237 | 6.46 × 10−6 | RDI.GLM |
| SNP-7-59879988 | T/A | 0.098 | 8.80 × 10−8 | RDI.FarmCPU |
| SNP-7-57772159 | T/A | −0.059 | 3.74 × 10−7 | RDI.FarmCPU |
| SNP-7-51913862 | T/C | 0.069 | 5.20 × 10−8 | RDI.FarmCPU |
| SNP-7-2876065 | A/G | −0.102 | 5.05 × 10−7 | RDI.FarmCPU |
| SNP-6-23330464 | C/G | −0.064 | 5.51 × 10−7 | RDI.FarmCPU |
| SNP-6-153673 | A/G | −0.091 | 2.07 × 10−6 | RDI.GLM |
| SNP-3-48129228 | T/G | 0.183 | 3.09 × 10−8 | RDI.FarmCPU |
| SNP-3-48129228 | T/G | 0.270 | 1.30 × 10−6 | RDI.GLM |
| SNP-2-60765047 | T/C | −0.064 | 5.09 × 10−10 | RDI.FarmCPU |
| SNP-2-54990996 | T/A | −0.076 | 3.20 × 10−9 | RDI.FarmCPU |
| SNP-10-7591972 | C/T | −0.125 | 1.82 × 10−7 | RDI.FarmCPU |
| Marker | Variant | Effect | p-Value | Methods |
|---|---|---|---|---|
| SNP-1-60603003 | T/C | 0.122 | 3.11 × 10−6 | STI.FarmCPU |
| SNP-9-21304761 | G/C | 0.169 | 1.26 × 10−6 | STI.FarmCPU |
| SNP-9-21304761 | G/C | 0.210 | 5.34 × 10−6 | STI.GLM |
| SNP-8-50726311 | C/G | −0.112 | 9.58 × 10−8 | STI.GLM |
| SNP-8-43496503 | A/G | −0.136 | 7.33 × 10−6 | STI.GLM |
| SNP-8-42803746 | G/A | −0.068 | 9.16 × 10−7 | STI.FarmCPU |
| SNP-8-41546525 | T/C | −0.161 | 2.65 × 10−6 | STI.GLM |
| SNP-8-38378608 | G/T | −0.157 | 9.75 × 10−7 | STI.GLM |
| SNP-7-14894170 | T/C | −0.198 | 2.70 × 10−7 | STI.GLM |
| SNP-6-24638786 | T/C | −0.116 | 6.40 × 10−6 | STI.GLM |
| SNP-6-24155091 | C/G | −0.119 | 4.23 × 10−6 | STI.GLM |
| SNP-6-23766693 | G/C | −0.120 | 1.90 × 10−6 | STI.GLM |
| SNP-6-23020118 | T/C | −0.114 | 6.21 × 10−6 | STI.GLM |
| SNP-6-22967016 | C/T | −0.114 | 4.32 × 10−6 | STI.GLM |
| SNP-6-22966971 | A/G | −0.120 | 1.86 × 10−6 | STI.GLM |
| SNP-6-21397577 | T/C | −0.398 | 1.07 × 10−7 | STI.FarmCPU |
| SNP-6-21397577 | T/C | −0.426 | 7.49 × 10−6 | STI.MLM |
| SNP-6-18184340 | T/C | −0.138 | 2.78 × 10−7 | STI.GLM |
| SNP-6-16486758 | T/C | −0.147 | 1.48 × 10−7 | STI.GLM |
| SNP-6-153673 | A/G | −0.108 | 7.35 × 10−7 | STI.GLM |
| SNP-4-47497728 | T/C | −0.178 | 3.13 × 10−6 | STI.GLM |
| SNP-4-22602002 | A/G | −0.249 | 5.12 × 10−8 | STI.GLM |
| SNP-4-11561108 | T/G | −0.164 | 1.44 × 10−6 | STI.GLM |
| SNP-4-11561107 | G/C | −0.164 | 1.44 × 10−6 | STI.GLM |
| SNP-4-10100565 | G/C | −0.206 | 1.98 × 10−6 | STI.GLM |
| SNP-3-73977961 | C/A | −0.145 | 9.30 × 10−9 | STI.GLM |
| SNP-3-73977959 | T/A | −0.145 | 9.30 × 10−9 | STI.GLM |
| SNP-3-73977947 | T/A | −0.145 | 9.30 × 10−9 | STI.GLM |
| SNP-3-71564436 | A/C | −0.246 | 2.12 × 10−10 | STI.GLM |
| SNP-3-70162204 | A/G | −0.165 | 6.51 × 10−7 | STI.GLM |
| SNP-3-69226243 | A/G | −0.201 | 1.24 × 10−8 | STI.GLM |
| SNP-3-63949295 | G/C | −0.213 | 8.63 × 10−8 | STI.GLM |
| SNP-3-48129228 | T/G | 0.240 | 4.95 × 10−8 | STI.FarmCPU |
| SNP-3-48129228 | T/G | 0.293 | 4.99 × 10−6 | STI.GLM |
| SNP-2-75743471 | G/T | −0.159 | 4.57 × 10−8 | STI.GLM |
| SNP-2-54990998 | T/C | −0.120 | 5.33 × 10−6 | STI.GLM |
| SNP-2-54990996 | T/A | −0.120 | 5.33 × 10−6 | STI.GLM |
| SNP-2-53243721 | C/T | 0.486 | 3.35 × 10−7 | STI.GLM |
| SNP-2-49409474 | C/G | −0.176 | 3.93 × 10−6 | STI.GLM |
| SNP-2-39852357 | A/G | −0.205 | 1.67 × 10−6 | STI.GLM |
| SNP-1-159169 | G/T | −0.182 | 4.54 × 10−6 | STI.GLM |
| SNP | Genes | Gene Function Annotation |
|---|---|---|
| SNP-1-159169 | Sb01g000300.1 | transcriptional corepressor Leunig-homolog-like [Sorghum bicolor], |
| Zea mays LOC100285229 (pco116270) | ||
| SNP-1-60603003 | Sb01g037050.1 | TF TGA2.2 [Sorghum bicolor], |
| Zea mays putative bZIP TF | ||
| (LOC100274089) | ||
| SNP-1-60603003 | Sb01g037050.2 | TF TGA2.2 [Sorghum bicolor], |
| Zea mays putative bZIP TF | ||
| (LOC100274089) | ||
| SNP-1-60603003 | Sb01g037050.3 | TF TGA2.2 [Sorghum bicolor], |
| Setaria italica TF HBP-1b(c1)-like (LOC101767047), | ||
| transcript variant X1, mRNA | ||
| SNP-1-60603003 | Sb01g037050.4 | TF TGA2.2 [Sorghum bicolor], |
| Zea mays putative bZIP TF | ||
| (LOC100274089) | ||
| SNP-8-50726311 | Sb08g019720.1 | TF LUX-like [Oryza brachyantha], |
| Sorghum bicolor hypothetical protein, mRNA, | ||
| MYB family TF EFM, Arabidopsis thaliana |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xin, Y.; Gao, L.; Hu, W.; Gao, Q.; Yang, B.; Zhou, J.; Xu, C. Genome-Wide Association Study Based on Plant Height and Drought-Tolerance Indices Reveals Two Candidate Drought-Tolerance Genes in Sweet Sorghum. Sustainability 2022, 14, 14339. https://doi.org/10.3390/su142114339
Xin Y, Gao L, Hu W, Gao Q, Yang B, Zhou J, Xu C. Genome-Wide Association Study Based on Plant Height and Drought-Tolerance Indices Reveals Two Candidate Drought-Tolerance Genes in Sweet Sorghum. Sustainability. 2022; 14(21):14339. https://doi.org/10.3390/su142114339
Chicago/Turabian StyleXin, Yue, Lina Gao, Wenming Hu, Qi Gao, Bin Yang, Jianguo Zhou, and Cuilian Xu. 2022. "Genome-Wide Association Study Based on Plant Height and Drought-Tolerance Indices Reveals Two Candidate Drought-Tolerance Genes in Sweet Sorghum" Sustainability 14, no. 21: 14339. https://doi.org/10.3390/su142114339
APA StyleXin, Y., Gao, L., Hu, W., Gao, Q., Yang, B., Zhou, J., & Xu, C. (2022). Genome-Wide Association Study Based on Plant Height and Drought-Tolerance Indices Reveals Two Candidate Drought-Tolerance Genes in Sweet Sorghum. Sustainability, 14(21), 14339. https://doi.org/10.3390/su142114339









