The Rediscovery of Traditional Maize Agrobiodiversity: A Study Case from Northern Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Germplasm
2.2. DNA Extraction and PCR Amplification
2.3. Statistical Analysis
3. Results and Discussion
3.1. Morphological Characterization
3.2. Genetic Characterization
3.3. PCoA Analysis
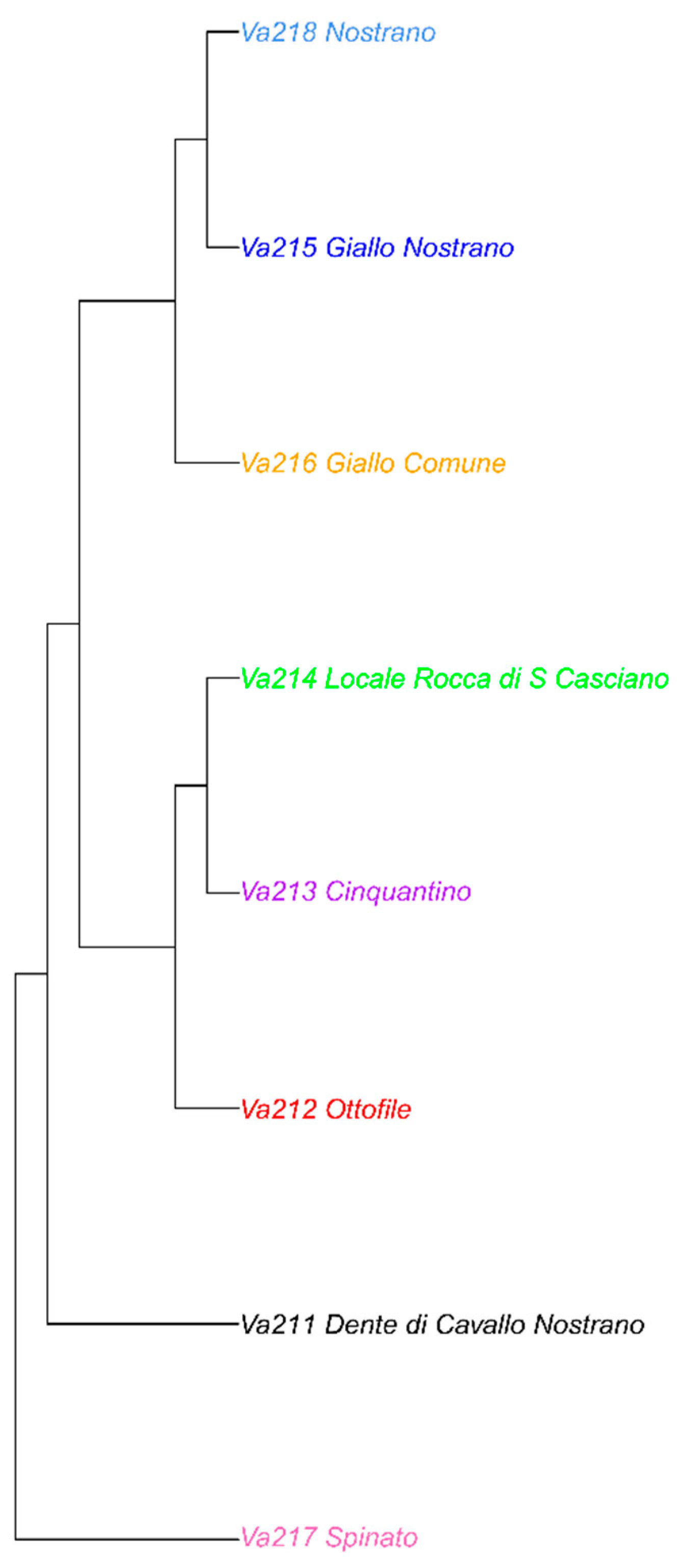
3.4. Phylogenetic Analysis
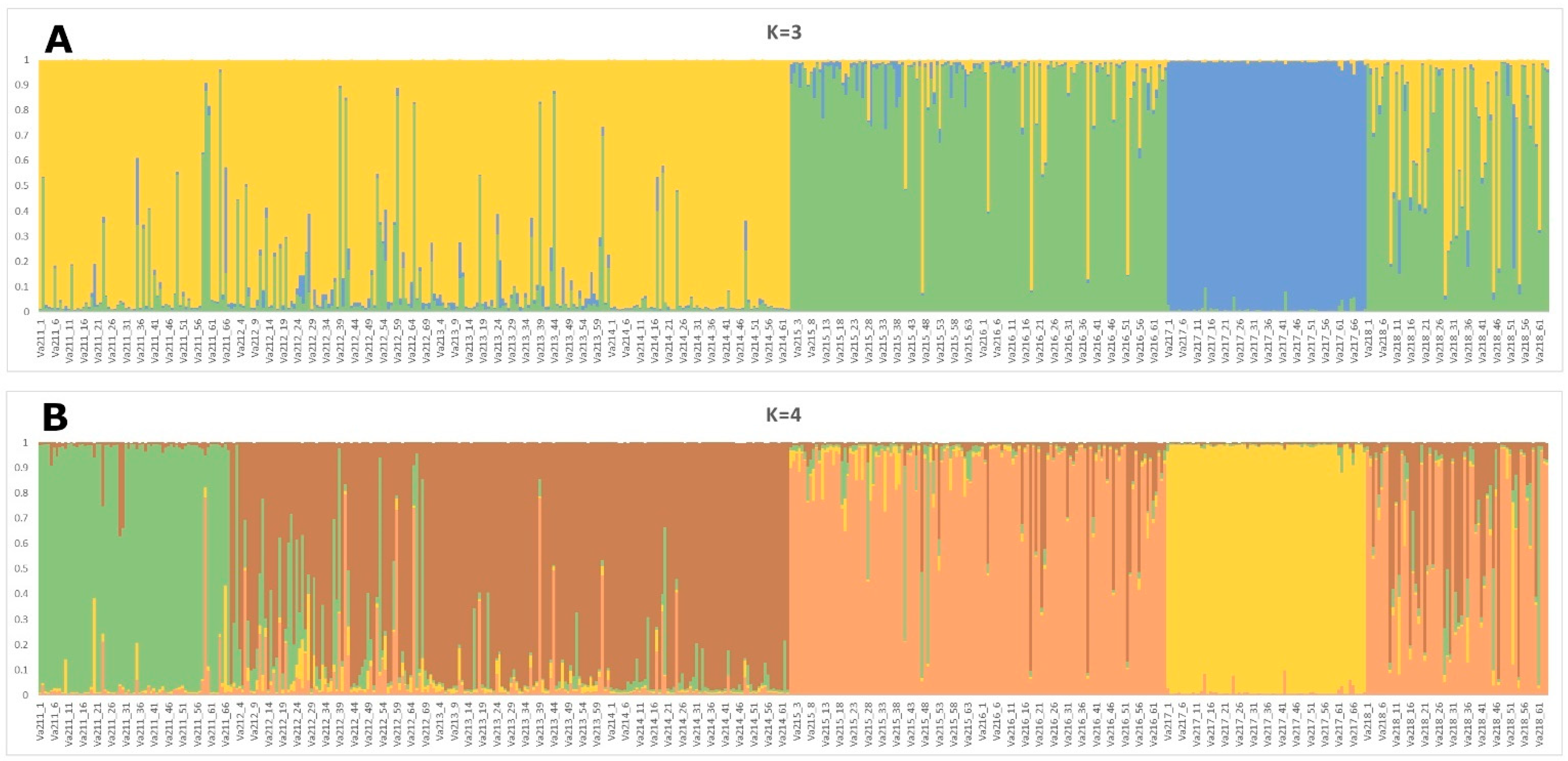
3.5. STRUCTURE Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Stitzer, M.C.; Ross-Ibarra, J. Maize domestication and gene interaction. New Phytol. 2018, 220, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Janick, J.; Caneva, G. The first images of maize in Europe. Maydica 2005, 50, 71–80. [Google Scholar]
- Brandolini, A.; Brandolini, A. Maize introduction, evolution and diffusion in Italy. Maydica 2009, 54, 233–242. [Google Scholar]
- Eschholz, T.W.; Stamp, P.; Peter, R.; Leipner, J.; Hund, A. Genetic structure and history of Swiss maize (Zea mays L. ssp. mays) landraces. Genet. Resour. Crop Evol. 2010, 57, 71–84. [Google Scholar] [CrossRef][Green Version]
- Oppong, A.; Bedoya, C.A.; Ewool, M.B.; Asante, M.D.; Thompson, R.; Adu-Dapaah, H.; Lamptey, J.N.L.; Ofori, K.; Offei, S.; Warburton, M. Bulk genetic characterization of Ghanaian maize landraces using microsatellite markers. Maydica 2014, 59, 1. [Google Scholar]
- Cömertpay, G.; Baloch, F.S.; Kilian, B.; Ülger, A.C.; Özkan, H. Diversity Assessment of Turkish Maize Landraces Based on Fluorescent Labelled SSR Markers. Plant. Mol. Biol. Rep. 2012, 30, 261–274. [Google Scholar] [CrossRef]
- Djemel, A.; Revilla, P.; Hanifi-Mekliche, L.; Malvar, R.A.; Álvarez, A.; Kelifi, L. Maize (Zea mays L.) from the Saharan oasis: Adaptation to temperate areas and agronomic performance. Genet. Resour. Crop Evol. 2012, 59, 1493–1504. [Google Scholar] [CrossRef]
- Qi-Lun, Y.; Ping, F.; Ke-Cheng, K.; Guang-Tang, P. Genetic diversity based on SSR markers in maize (Zea mays L.) landraces from Wuling mountain region in China. J. Genet. 2008, 87, 287–291. [Google Scholar] [CrossRef]
- Zapparoli, T.V. Il Granoturco; Biblioteca Agricola “Paravia”, G.B. Paravia & C.: Torino, Italy, 1930. [Google Scholar]
- Zapparoli, T.V. Derivati del granoturco Nostrano dell’Isola: Nostrano dell’Isola Finardi (S.M.), Isola Basso (S.M.) e Letizia (S.M.). Ital. Agric. 1939, 76, 317–326. [Google Scholar]
- Zapparoli, T.V. Due granoturchi selezionati: Rostrato Cajo Duilio (S.M.), Giallo tondo S. Pancrazio (S.M.). Ital. Agric. 1939, 76, 609–614. [Google Scholar]
- Zapparoli, T.V. Il granoturco Marano. Ital. Agric. 1939, 76, 155–159. [Google Scholar]
- Zapparoli, T.V. Il granoturco Scagliolo 23A. Ital. Agric. 1939, 76, 239–245. [Google Scholar]
- Bertolini, M.; Verderio, A.; Motto, M.; Berardo, N.; Brugna, E.; Balduini, C. Mais in Lombardia: Varietà Tradizionali; Regione Lombardia: Milano, Italy, 2002. [Google Scholar]
- Mariani, G. Possibilità di costituzione di mais ibridi destinati ad ambienti difficili con linee tratte da popolazioni locali. Maydica 1964, 9, 41–59. [Google Scholar]
- Ottaviano, E. Primi risultati di ricerche per l’ottenimento di mais ibridi da foraggio. Maydica 1964, 9, 99–109. [Google Scholar]
- Ottaviano, E. Selezione ricorrente per l’ottenimento di nuovi ibridi da foraggio. Maydica 1967, 12, 47–49. [Google Scholar]
- Brandolini, A.; Brandolini, A. Il Mais in Italia. Storia Naturale e Agricola, 2nd ed.; CRF Press: Bergamo, Italy, 2006. [Google Scholar]
- Bianchi, A.; Ghatnekar, M.V.; Ghidoni, A. Knobs in Italian maize. Chromosoma 1963, 14, 601–617. [Google Scholar] [CrossRef]
- Ghatnekar, M.V. Heterochromatic knobs in Itaian maize population and the evolution of maize in Italy. Cytologia 1965, 30, 402–425. [Google Scholar] [CrossRef]
- Ghatnekar, M.V. Spontaneous chromosome aberrations and abnormal behaviours in Italian maize populations. Cytologia 1965, 30, 426–436. [Google Scholar] [CrossRef]
- Bianchi, A.; Borghi, B.; Lorenzoni, C.; Pozzi, M.; Salamini, F. Mendelian characters in Italian maize. Maize Genet. Coop. News Lett. 1964, 38, 89–91. [Google Scholar]
- Lorenzoni, C.; Pozzi, M.; Salamini, F. Frequenze di mutanti in popolazioni di mais italiani. Genet. Agric. 1965, 18, 146–158. [Google Scholar]
- Salamini, F. Variabilità mendeliana nelle varietà italiane di mais. Maydica 1968, 13, 1–45. [Google Scholar]
- Ardenghi, N.M.G.; Rossi, G.; Guzzon, F. Back to beaked: Zea mays subsp. Mays Rostrata Group in northern Italy, refugia and revival of open-pollinated maize landraces in an intensive cropping system. PeerJ 2018, 6, e5123. [Google Scholar]
- Cassani, E.; Puglisi, D.; Cantaluppi, E.; Landoni, M.; Giupponi, L.; Giorgi, A.; Pilu, R. Genetic studies regarding the control of seed pigmentation of an ancient European pointed maize (Zea mays L.) rich in phlobaphenes: The “Nero Spinoso” from the Camonica valley. Genet. Resour. Crop Evol. 2017, 64, 761–773. [Google Scholar] [CrossRef]
- Barcaccia, G.; Lucchin, M.; Parrini, P. Characterization of a flint maize (Zea mays var. indurata) Italian landrace, II. Genetic diversity and relatedness assessed by SSR and Inter-SSR molecular markers. Genet. Resour. Crop Evol. 2003, 50, 253–271. [Google Scholar]
- Bitocchi, E.; Nanni, L.; Rossi, M.; Rau, D.; Bellucci, E.; Giardini, A.; Buonamici, A.; Vendramin, G.G.; Papa, R. Introgression from modern hybrid varieties into landrace populations of maize (Zea mays ssp. mays L.) in central Italy. Mol. Ecol. 2009, 18, 603–621. [Google Scholar]
- Palumbo, F.; Galla, G.; Martínez-Bello, L.; Barcaccia, G. Venetian Local Corn (Zea mays L.) Germplasm: Disclosing the Genetic Anatomy of Old Landraces Suited for Typical Cornmeal Mush Production. Diversity 2017, 9, 32. [Google Scholar] [CrossRef]
- Giupponi, L.; Leoni, V.; Colombo, F.; Cassani, E.; Hejna, M.; Rossi, L.; Pilu, R. Characterization of “Mais delle Fiorine” (Zea mays L.) and nutritional, morphometric and genetic comparison with other maize landraces of Lombardy region (Northern Italy). Genet. Resour. Crop Evol. 2021, 68, 2075–2091. [Google Scholar] [CrossRef]
- Stagnati, L.; Martino, M.; Soffritti, G.; Lanubile, A.; Ravasio, A.; Marocco, A.; Rossi, G.; Busconi, M. Microsatellite and morphological characterization of three Rostrato di Val Chiavenna (Sondrio, Italy) maize (Zea mays L.) accessions. Genet. Resour. Crop Evol. 2021, 68, 3025–3038. [Google Scholar] [CrossRef]
- Stagnati, L.; Soffritti, G.; Martino, M.; Lanubile, A.; Desiderio, F.; Ravasio, A.; Marocco, A.; Rossi, G.; Busconi, M. Morphological and Genetic Characterization of Local Maize Accessions from Emilia Romagna Region, Italy. Sustainability 2022, 14, 91. [Google Scholar] [CrossRef]
- Hartings, H.; Berardo, N.; Mazzinelli, G.F.; Valoti, P.; Verderio, A.; Motto, M. Assessment of genetic diversity and relationships among maize (Zea mays L.) Italian landraces by morphological traits and AFLP profiling. Theor. Appl. Genet. 2008, 117, 831–842. [Google Scholar] [CrossRef]
- Ignjatović-Micić, D.; Drinić, S.M.; Nikolić, A.; Lazić-Jancić, V. SSR analysis for genetic structure and diversity determination of maize local populations from former Yugoslavia territories. Genetika 2008, 44, 1517–1524. [Google Scholar] [CrossRef] [PubMed]
- Lucchin, M.; Barcaccia, G.; Parrini, P. Characterization of a flint maize (Zea mays L. convar. mays) Italian landrace: I. Morpho-phenological and agronomic traits. Genet. Resour. Crop Evol. 2003, 50, 315–327. [Google Scholar]
- Canella, M.; Ardenghi, N.M.G.; Müller, J.V.; Rossi, G.; Guzzon, F. An updated checklist of plant agrobiodiversity of northern Italy. Genet. Resour. Crop Evol. 2022, 69, 2159–2178. [Google Scholar] [CrossRef]
- CRA. Schede Varietà Emilia-Romagna; CRA: Rome, Italy, 2005. [Google Scholar]
- Troyer, F. Temperate Corn—Background, Behavior, and Breeding. In Specialty Corns, 2nd ed.; Hallauer, A.R., Ed.; CRC Press: Boca Raton, FL, USA, 2000; p. 74. ISBN 9780429127458. [Google Scholar]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar]
- Liu, K.; Muse, S.V. PowerMarker: Integrated analysis environment for genetic marker data. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.; Jasieniuk, M. Polysat: An R package for polyploid microsatellite analysis. Mol. Ecol. Resour. 2011, 11, 562–566. [Google Scholar] [CrossRef] [PubMed]
- Schliep, K.P. phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2018, 35, 526–528. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 8, 2611–2620. [Google Scholar] [CrossRef]
- Bonciarelli, F. Studio agronomico comparato delle popolazioni umbre di mais. Maydica 1961, 6, 35–61. [Google Scholar]
- Gage, J.L.; White, M.R.; Edwards, J.W.; Kaeppler, S.; de Leon, N. Selection Signatures Underlying Dramatic Male Inflorescence Transformation During Modern Hybrid Maize Breeding. Genetics 2018, 210, 1125–1138. [Google Scholar] [CrossRef] [PubMed]
- Saltini, A. Messi e armenti di Romagna nei versi dell’ultimo emulo di Virgilio. Romagna Arte Stor. 2000, 2000, 59. [Google Scholar]
- Senior, M.L.; Murphy, J.P.; Goodman, M.M.; Stuber, C.W. Utility of SSRs for Determining Genetic Similarities an Relationships in Maize Using an Agarose Gel System. Crop. Sci. 1998, 38, 1088–1098. [Google Scholar] [CrossRef]
- Sharma, L.; Prasanna, B.M.; Ramesh, B. Analysis of phenotypic and microsatellite-based diversity of maize landraces in India, especially from the North East Himalayan region. Genetica 2010, 138, 619–631. [Google Scholar] [CrossRef]


| Accession | Accession Name | Sampling Location | Racial Complex | Local Race | Agro-Ecotype | Plant Height (cm) | Ear Height (cm) | Tasseling (GDD) | Physiological Maturity (GDD) |
|---|---|---|---|---|---|---|---|---|---|
| Va211 | Dente di Cavallo Nostrano | San Lorenzo-Riccione (FC) | Conici Vitrei e derivati (Conical Flints and Derived Races) | Poliranghi (Multi Rowed Flints) | Spadone | 202 | 105 | 646 | 1430 |
| Va212 | Ottofile | Ricò-Meldola (FC) | Ottofile Vitrei e Derivati (Eight-Rowed Flints and Derived Races) | Derivati 10–12 file (10–12 Rowed Derived Flints) | Derivati 10–12 file (10–12 Rowed Derived Flints) | 230 | 100 | 646 | 1446 |
| Va213 | Cinquantino | Ravaldino in Monte (FC) | Conici Vitrei e derivati (Conical Flints and Derived Races) | Barbina | Barbina | 220 | 95 | 646 | 1430 |
| Va214 | Locale Rocca di San Casciano | Rocca San Casciano (FC) | Conici Vitrei e derivati (Conical Flints and Derived Races | Barbina | Maggese | 210 | 85 | 632 | 1366 |
| Va215 | Giallo Nostrano- Giallo Predappio | Predappio (FC) | Conici Vitrei e derivati (Conical Flints and Derived Races) | Poliranghi (Multi Rowed Flints) | Spadone | 220 | 122 | 646 | 1446 |
| Va216 | Giallo Comune | Santa Sofia (FC) | Conici Vitrei e derivati (Conical Flints and Derived Races) | Poliranghi (Multi Rowed Flints) | Culaccione | 220 | 90 | 632 | 1398 |
| Va217 | Spinato | Cesena (FC) | Dentati Bianchi e Gialli (Dent corn) | Dentati Bianchi Moderni (Modern White dent) | Dentato Giallo Moderno (Modern Yellow Dent) | 265 | 122 | 799 | 1476 |
| Va218 | Nostrano | Sogliano sul Rubicone (FC) | Ottofile Vitrei e derivati (Eight-Rowed Flints and Derived Races) | Cannellino | Cannellino | 245 | 102 | 662 | 1446 |
| Marker Name | Locus | Forward Primer 5′-3′ | Reverse Primer 5′-3′ | LG | Ta (°C) | Size (bp) | Reference |
|---|---|---|---|---|---|---|---|
| M302 | phi127 | ATATGCATTGCCTGGAACTGGAAGGA | [VIC]AATTCAAACACGCCTCCCGAGTGT | 2 | 58 | 100–120 | [29] |
| M304 | phi076 | TTCTTCCGCGGCTTCAATTTGACC | [6FAM]GCATCAGGACCCGCAGAGTC | 4 | 58 | 150–200 | [29] |
| M306 | phi031 | GCAACAGGTTACATGAGCTGACGA | [PET]CCAGCGTGCTGTTCCAGTAGTT | 6 | 58 | 180–220 | [29] |
| M308 | umc1075 | GAGAGATGACAGACACATCCTTGG | [6FAM]ACATTTATGATACCGGGAGTTGGA | 8 | 58 | 130–150 | [29] |
| M310 | phi084 | AGAAGGAATCCGATCCATCCAAGC | [PET]CACCCGTACTTGAGGAAAACCC | 10 | 58 | 140–170 | [29] |
| M24 | umc1327 | AGGGTTTTGCTCTTGGAATCTCTC | [NED]GAGGAAGGAGGAGGTCGTATCGT | 8 | 64 | 100–120 | MaizeGDB |
| M33 | p-bnlg176 | AGTTCACGTCCAGCTGAATGACAG | [6FAM]CGCGCATCGCATGCTTATCCTA | 1 | 62 | 140–170 | MaizeGDB |
| M78 | umc1941 | ACGACGAGACTCTGTTCTGGTTCT | [NED]AGGAGGATTACGTCAATCTGTTCG | 5 | 64 | 110–130 | MaizeGDB |
| M90 | umc1401 | CTCTGGTCCATCCTCATCGACT | [PET]TCTCTTGATCACATATCGATCCCA | 7 | 62 | 180–200 | MaizeGDB |
| M193 | umc1786 | ACCGTGACTTCCTCCTCATAACTG | [VIC]CATTTTTCGCATTTAGGAAATCCA | 8 | 60 | 180–220 | MaizeGDB |
| Locus | N | Na | Ne | I | Ho | uHe | PIC | F | FIS | FIT | FST | Nm |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| phi127 | 6.00 | 3.25 | 2.07 | 0.79 | 0.50 | 0.49 | 0.54 | −0.01 | −0.02 | 0.18 | 0.20 | 0.99 |
| phi076 | 5.00 | 3.75 | 2.19 | 0.88 | 0.52 | 0.54 | 0.55 | 0.03 | 0.03 | 0.17 | 0.14 | 1.48 |
| phi031 | 11.00 | 5.00 | 2.73 | 1.14 | 0.57 | 0.60 | 0.74 | 0.05 | 0.04 | 0.26 | 0.23 | 0.85 |
| umc1075 | 10.00 | 4.75 | 2.84 | 1.16 | 0.54 | 0.63 | 0.73 | 0.13 | 0.14 | 0.29 | 0.18 | 1.11 |
| phi084 | 4.00 | 2.63 | 1.79 | 0.65 | 0.43 | 0.43 | 0.38 | −0.03 | −0.02 | 0.11 | 0.13 | 1.69 |
| umc1327 | 7.00 | 3.50 | 2.06 | 0.82 | 0.52 | 0.49 | 0.48 | −0.09 | −0.07 | 0.06 | 0.13 | 1.74 |
| umc1786 | 9.00 | 5.38 | 2.67 | 1.13 | 0.42 | 0.59 | 0.64 | 0.25 | 0.27 | 0.37 | 0.13 | 1.69 |
| p-bnlg176 | 6.00 | 3.38 | 2.13 | 0.83 | 0.39 | 0.48 | 0.56 | 0.22 | 0.19 | 0.39 | 0.25 | 0.77 |
| umc1941 | 6.00 | 3.75 | 2.18 | 0.82 | 0.51 | 0.47 | 0.58 | −0.09 | −0.10 | 0.22 | 0.28 | 0.63 |
| umc1401 | 4.00 | 3.13 | 2.62 | 1.00 | 0.50 | 0.58 | 0.62 | 0.16 | 0.14 | 0.27 | 0.15 | 1.36 |
| Mean | 6.80 | 3.85 | 2.33 | 0.92 | 0.49 | 0.53 | 0.58 | 0.06 | 0.06 | 0.23 | 0.18 | 1.23 |
| SE | 2.44 | 0.17 | 0.08 | 0.04 | 0.02 | 0.02 | 0.03 | 0.04 | 0.03 | 0.02 | 0.13 | |
| Va211 | 50.00 | 5.00 | 2.38 | 0.98 | 0.41 | 0.52 | # | 0.19 | 0.22 | 0.37 | 0.19 | 1.04 |
| Va212 | 47.00 | 4.70 | 2.53 | 1.06 | 0.54 | 0.60 | # | 0.10 | 0.10 | 0.16 | 0.07 | 3.36 |
| Va213 | 34.00 | 3.40 | 2.28 | 0.90 | 0.50 | 0.54 | # | 0.09 | 0.08 | 0.23 | 0.16 | 1.29 |
| Va214 | 35.00 | 3.50 | 2.02 | 0.83 | 0.50 | 0.49 | # | −0.02 | −0.01 | 0.23 | 0.24 | 0.79 |
| Va215 | 39.00 | 3.90 | 2.80 | 1.08 | 0.53 | 0.61 | # | 0.13 | 0.14 | 0.18 | 0.05 | 4.65 |
| Va216 | 42.00 | 4.20 | 2.55 | 1.01 | 0.53 | 0.55 | # | 0.02 | 0.05 | 0.18 | 0.14 | 1.51 |
| Va217 | 27.00 | 2.70 | 1.71 | 0.60 | 0.40 | 0.36 | # | −0.10 | −0.11 | 0.37 | 0.44 | 0.32 |
| Va218 | 34.00 | 3.40 | 2.34 | 0.92 | 0.52 | 0.56 | # | 0.05 | 0.08 | 0.20 | 0.14 | 1.60 |
| Mean | 38.50 | 3.85 | 2.33 | 0.92 | 0.49 | 0.53 | 0.06 | 0.07 | 0.24 | 0.18 | 1.82 | |
| SE | 7.58 | 0.17 | 0.08 | 0.04 | 0.50 | 0.02 | 0.03 | 0.10 | 0.08 | 0.12 | 1.45 |
| Landrace | Locus | Allele | Allele Frequency |
|---|---|---|---|
| Va211 Dente di Cavallo Nostrano | phi031 | 212 | 0.017 |
| 220 | 0.017 | ||
| umc1075 | 122 | 0.023 | |
| 124 | 0.031 | ||
| 134 | 0.289 | ||
| 144 | 0.039 | ||
| umc1327 | 62 | 0.015 | |
| umc1796 | 126 | 0.024 | |
| Va212 Ottofile | phi127 | 102 | 0.015 |
| 111 | 0.015 | ||
| umc1075 | 120 | 0.016 | |
| phi084 | 152 | 0.007 | |
| p-bnlg176 | 132 | 0.015 | |
| umc1941 | 92 | 0.016 | |
| Va213 Cinquantino | phi031 | 196 | 0.032 |
| Va216_Giallo_Comune | phi031 | 184 | 0.008 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stagnati, L.; Soffritti, G.; Desiderio, F.; Lanubile, A.; Zambianchi, S.; Marocco, A.; Rossi, G.; Busconi, M. The Rediscovery of Traditional Maize Agrobiodiversity: A Study Case from Northern Italy. Sustainability 2022, 14, 12110. https://doi.org/10.3390/su141912110
Stagnati L, Soffritti G, Desiderio F, Lanubile A, Zambianchi S, Marocco A, Rossi G, Busconi M. The Rediscovery of Traditional Maize Agrobiodiversity: A Study Case from Northern Italy. Sustainability. 2022; 14(19):12110. https://doi.org/10.3390/su141912110
Chicago/Turabian StyleStagnati, Lorenzo, Giovanna Soffritti, Francesca Desiderio, Alessandra Lanubile, Sara Zambianchi, Adriano Marocco, Graziano Rossi, and Matteo Busconi. 2022. "The Rediscovery of Traditional Maize Agrobiodiversity: A Study Case from Northern Italy" Sustainability 14, no. 19: 12110. https://doi.org/10.3390/su141912110
APA StyleStagnati, L., Soffritti, G., Desiderio, F., Lanubile, A., Zambianchi, S., Marocco, A., Rossi, G., & Busconi, M. (2022). The Rediscovery of Traditional Maize Agrobiodiversity: A Study Case from Northern Italy. Sustainability, 14(19), 12110. https://doi.org/10.3390/su141912110










