Simple Sequence Repeats-Based Genetic Characterization and Varietal Identification of Potato Varieties Grown in Pakistan
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and SSR Analysis
2.3. Polymerase Chain Reaction (PCR)
2.4. Scoring and Data Analysis
3. Results
3.1. Genetic Diversity Parameters Revealed Based on SSR Markers
3.2. Cluster Analysis
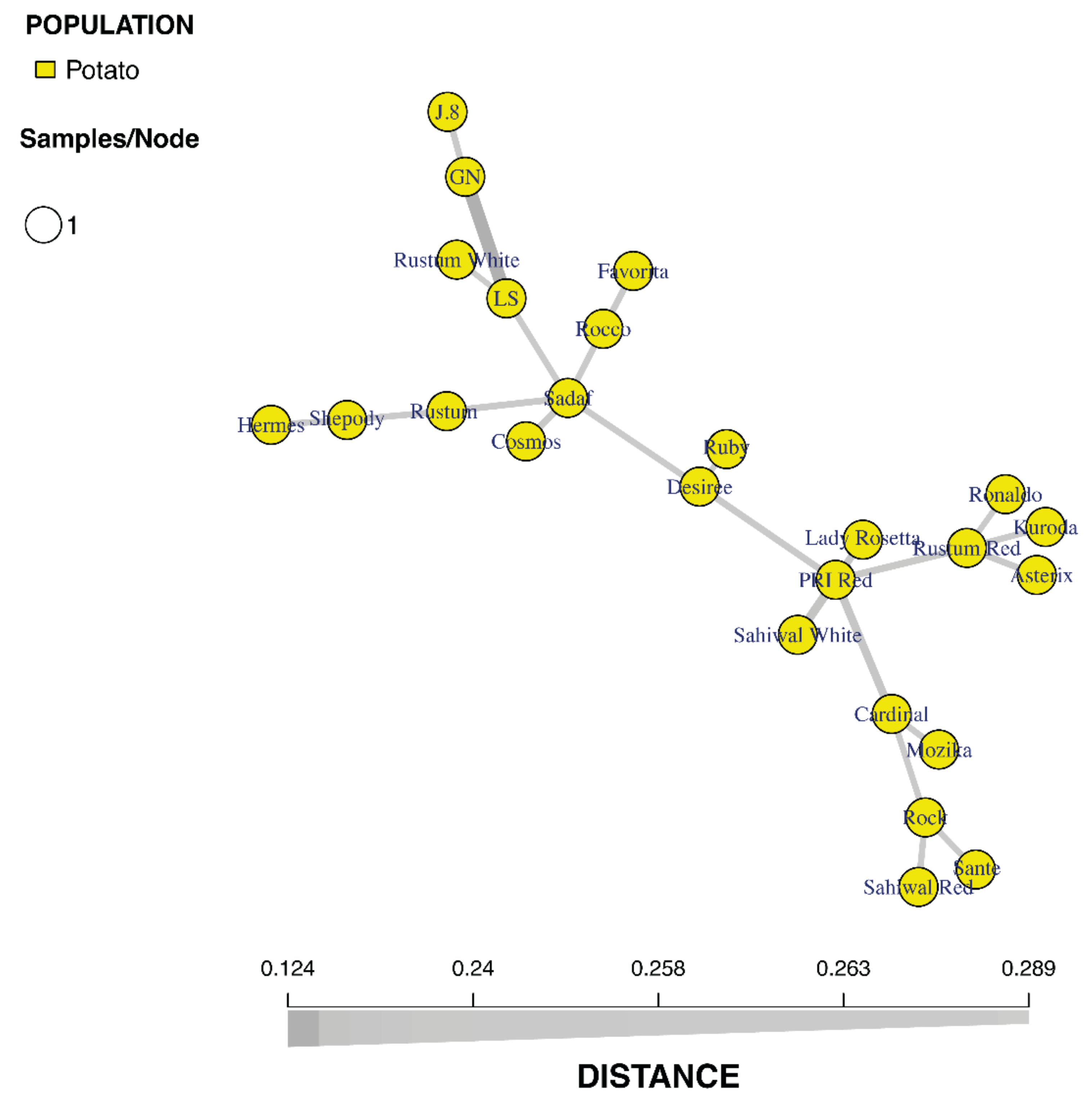
3.3. Minimum Spanning Network (MSN) Analysis
3.4. Varietal Identification Based on Unique Alleles Based on 35 SSR Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kafle, K.; Shriwastav, C.P.; Marasini, M. Influence of Integrated Nutrient Management Practices on Soil Properties and Yield of Potato (Solanum tuberosum L.) in an Inceptisol of Khajura, Banke. Int. J. Appl. Sci. Biotechnol. 2019, 7, 365–369. [Google Scholar] [CrossRef]
- Shahzad, R.; Jamil, S.; Ahmad, S.; Nisar, A.; Amina, Z.; Saleem, S.; Zaffar Iqbal, M.; Muhammad Atif, R.; Wang, X. Harnessing the Potential of Plant Transcription Factors in Developing Climate Resilient Crops to Improve Global Food Security: Current and Future Perspectives. Saudi J. Biol. Sci. 2021, 28, 2323–2341. [Google Scholar] [CrossRef] [PubMed]
- Economic Survey of Pakistan. 2021–2022. Available online: https://www.finance.gov.pk/survey_2022.html (accessed on 14 September 2022).
- Yildiz, M. Potato: From Incas to All over the World; BoD—Books on Demand: Norderstedt, Germany, 2018; ISBN 978-1-78923-254-7. [Google Scholar]
- Ramakrishnan, A.P.; Ritland, C.E.; Blas Sevillano, R.H.; Riseman, A. Review of Potato Molecular Markers to Enhance Trait Selection. Am. J. Potato Res. 2015, 92, 455–472. [Google Scholar] [CrossRef]
- Duan, Y.; Liu, J.; Xu, J.; Bian, C.; Duan, S.; Pang, W.; Hu, J.; Li, G.; Jin, L. DNA Fingerprinting and Genetic Diversity Analysis with Simple Sequence Repeat Markers of 217 Potato Cultivars (Solanum tuberosum L.) in China. Am. J. Potato Res. 2019, 96, 21–32. [Google Scholar] [CrossRef]
- Singh, A.; Singh, A.; Arya, M.; Sah, U.; Singh, A. Utility of Molecular Markers in Molecular Breeding for Integrated Crop Improvement. Bull. Environ. Pharmacol. Life Sci. 2019, 8, 1–8. [Google Scholar]
- Priyadarshini, L.; Samal, K.C.; Sahoo, J.P.; Mohapatra, U. Morphological, Biochemical and Molecular Characterization of Some Promising Potato (Solanum tuberosum L.) Cultivars of Odisha. J. Pharmacogn. Phytochem. 2020, 9, 1657–1664. [Google Scholar]
- Naznin, F.; Haque, M.A.; Rahman, M.H.S.; Rahman, M.H.A.; Mehedi, M.N.H. Molecular Diversity Analysis of Indigenous Potato Cultivars Using RAPD Markers. Asian J. Biochem. Genet. Mol. Biol. 2020, 6, 43–48. [Google Scholar] [CrossRef]
- Wang, F.; Li, F.; Wang, J. Genetic Diversity of Chinese and CIP Potato (Solanum tuberosum L.) Germplasm Assessed by Amplified Fragment Length Polymorphism (AFLP) Markers. Potato Res. 2013, 56, 167–178. [Google Scholar] [CrossRef]
- Berdugo-Cely, J.A.; Martínez-Moncayo, C.; Lagos-Burbano, T.C. Genetic Analysis of a Potato (Solanum tuberosum L.) Breeding Collection for Southern Colombia Using Single Nucleotide Polymorphism (SNP) Markers. PLoS ONE 2021, 16, e0248787. [Google Scholar] [CrossRef]
- Pandey, J.; Scheuring, D.C.; Koym, J.W.; Coombs, J.; Novy, R.G.; Thompson, A.L.; Holm, D.G.; Douches, D.S.; Miller, J.C.; Vales, M.I. Genetic Diversity and Population Structure of Advanced Clones Selected over Forty Years by a Potato Breeding Program in the USA. Sci. Rep. 2021, 11, 8344. [Google Scholar] [CrossRef]
- Schumacher, C.; Thümecke, S.; Schilling, F.; Köhl, K.; Kopka, J.; Sprenger, H.; Hincha, D.K.; Walther, D.; Seddig, S.; Peters, R.; et al. Genome-Wide Approach to Identify Quantitative Trait Loci for Drought Tolerance in Tetraploid Potato (Solanum tuberosum L.). Int. J. Mol. Sci. 2021, 22, 6123. [Google Scholar] [CrossRef]
- Kumar, K.P.S. Traditional and Medicinal Uses of Banana. J. Pharmacogn. Phytochem. 2012, 1, 13. [Google Scholar]
- Bali, S.; Sathuvalli, V.; Brown, C.; Novy, R.; Ewing, L.; Debons, J.; Douches, D.; Coombs, J.; Navarre, D.; Whitworth, J. Genetic Fingerprinting of Potato Varieties from the Northwest Potato Variety Development Program. Am. J. Potato Res. 2017, 94, 54–63. [Google Scholar] [CrossRef]
- Spanoghe, M.; Marique, T.; Rivière, J.; Lanterbecq, D.; Gadenne, M. Investigation and Development of Potato Parentage Analysis Methods Using Multiplexed SSR Fingerprinting. Potato Res. 2015, 58, 43–65. [Google Scholar] [CrossRef]
- Lee, K.-J.; Sebastin, R.; Cho, G.-T.; Yoon, M.; Lee, G.-A.; Hyun, D.-Y. Genetic Diversity and Population Structure of Potato Germplasm in RDA-Genebank: Utilization for Breeding and Conservation. Plants 2021, 10, 752. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a Genetic Linkage Map in Man Using Restriction Fragment Length Polymorphisms. Am. J. Hum. Genet. 1980, 32, 314. [Google Scholar]
- Dice, L.R. Measures of the Amount of Ecologic Association between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R Package for Genetic Analysis of Populations with Clonal or Partially Clonal Reproduction. PeerJ 2013. PrePrints. [Google Scholar] [CrossRef]
- Li, X.Y.; Xu, H.X.; Chen, J.W. Rapid Identification of Red-Flesh Loquat Cultivars Using EST-SSR Markers Based on Manual Cultivar Identification Diagram Strategy. Genet. Mol. Res. 2014, 13, 3384–3394. [Google Scholar] [CrossRef]
- Jian, Y.; Yan, W.; Xu, J.; Duan, S.; Li, G.; Jin, L. Genome-Wide Simple Sequence Repeat Markers in Potato: Abundance, Distribution, Composition, and Polymorphism. DNA Res. 2021, 28, dsab020. [Google Scholar] [CrossRef]
- Fulladolsa, A.C.; Navarro, F.M.; Kota, R.; Severson, K.; Palta, J.P.; Charkowski, A.O. Application of Marker Assisted Selection for Potato Virus Y Resistance in the University of Wisconsin Potato Breeding Program. Am. J. Potato Res. 2015, 92, 444–450. [Google Scholar] [CrossRef]
- Anoumaa, M.; Yao, N.K.; Kouam, E.B.; Kanmegne, G.; Machuka, E.; Osama, S.K.; Nzuki, I.; Kamga, Y.B.; Fonkou, T.; Omokolo, D.N. Genetic Diversity and Core Collection for Potato (Solanum tuberosum L.) Cultivars from Cameroon as Revealed by SSR Markers. Am. J. Potato Res. 2017, 94, 449–463. [Google Scholar] [CrossRef]
- Favoretto, P.; Veasey, E.A.; de Melo, P.C.T. Molecular Characterization of Potato Cultivars Using SSR Markers. Hortic. Bras. 2011, 29, 542–547. [Google Scholar] [CrossRef]
- Braun, A.; Wenzel, G. Molecular Analysis of Genetic Variation in Potato (Solarium tuberosum L.). I. German Cultivars and Advanced Clones. Potato Res. 2004, 47, 81–92. [Google Scholar] [CrossRef]
- Milbourne, D.; Meyer, R.C.; Collins, A.J.; Ramsay, L.D.; Gebhardt, C.; Waugh, R. Isolation, Characterisation and Mapping of Simple Sequence Repeat Loci in Potato. Mol. Gen. Genet. 1998, 259, 233–245. [Google Scholar] [CrossRef]
- Liao, H.; Guo, H. Using SSR to Evaluate the Genetic Diversity of Potato Cultivars from Yunnan Province (SW China). Acta Biol. Crac. Ser. Bot. 2014, 56, 16–27. [Google Scholar] [CrossRef]
- Elibariki, G.; Njahira, M.; Wanjala, B.; Hosea, K.; Ndunguru, J. Original Article Genetic Diversity and Identification of Duplicates in Selected Tanzanian Farmer- Preferred Cassava Landraces Using Simple Sequence Repeat (SSR) Markers. Int. J. Res. Plant Sci. 2013, 3, 81–87. [Google Scholar]
- Kishine, M.; Tsutsumi, K.; Kitta, K. A Set of Tetra-Nucleotide Core Motif SSR Markers for Efficient Identification of Potato (Solanum tuberosum) Cultivars. Breed. Sci. 2017, 67, 544–547. [Google Scholar] [CrossRef]
- Geleta, L.F.; Labuschagne, M.T.; Viljoen, C.D. Genetic Variability in Pepper (Capsicum annuum L.) Estimated by Morphological Data and Amplified Fragment Length Polymorphism Markers. Biodivers. Conserv. 2005, 14, 2361–2375. [Google Scholar] [CrossRef]


| Primer | Annealing Temp (°C) | Total No. of Scorable Bands | Total no. of Alleles N | No. of Monomorphic Alleles | No. of Poly-Morphic Alleles = K | Ratio of Polymorphic Loci P | Size bp | Unique Bands | PIC |
|---|---|---|---|---|---|---|---|---|---|
| S122 | 52 | 62 | 4 | 2 | 2 | 50 | 130–190 | 0 | 0.65 |
| S168 | 52 | 124 | 8 | 2 | 6 | 75 | 50–400 | 0 | 0.83 |
| S151 | 52 | 102 | 6 | 1 | 5 | 83.33 | 50–300 | 0 | 0.80 |
| S184 | 52 | 98 | 8 | 0 | 8 | 100 | 170–550 | 1 | 0.81 |
| S118 | 52 | 115 | 5 | 2 | 3 | 60 | 40–260 | 0 | 0.79 |
| S192 | 52 | 91 | 8 | 0 | 8 | 100 | 180–310 | 1 | 0.83 |
| S174 | 56 | 103 | 14 | 1 | 13 | 92.85 | 120–310 | 2 | 0.86 |
| S25 | 56 | 127 | 20 | 1 | 19 | 95 | 160–800 | 8 | 0.87 |
| S182 | 52 | 123 | 11 | 0 | 11 | 100 | 130–370 | 0 | 0.87 |
| S187 | 52 | 116 | 8 | 2 | 6 | 75 | 40–350 | 1 | 0.83 |
| S162 | 52 | 44 | 3 | 0 | 3 | 100 | 160–390 | 0 | 0.65 |
| S148 | 52 | 69 | 9 | 0 | 9 | 100 | 180–490 | 1 | 0.86 |
| S188 | 52 | 100 | 11 | 0 | 11 | 100 | 90–400 | 2 | 0.86 |
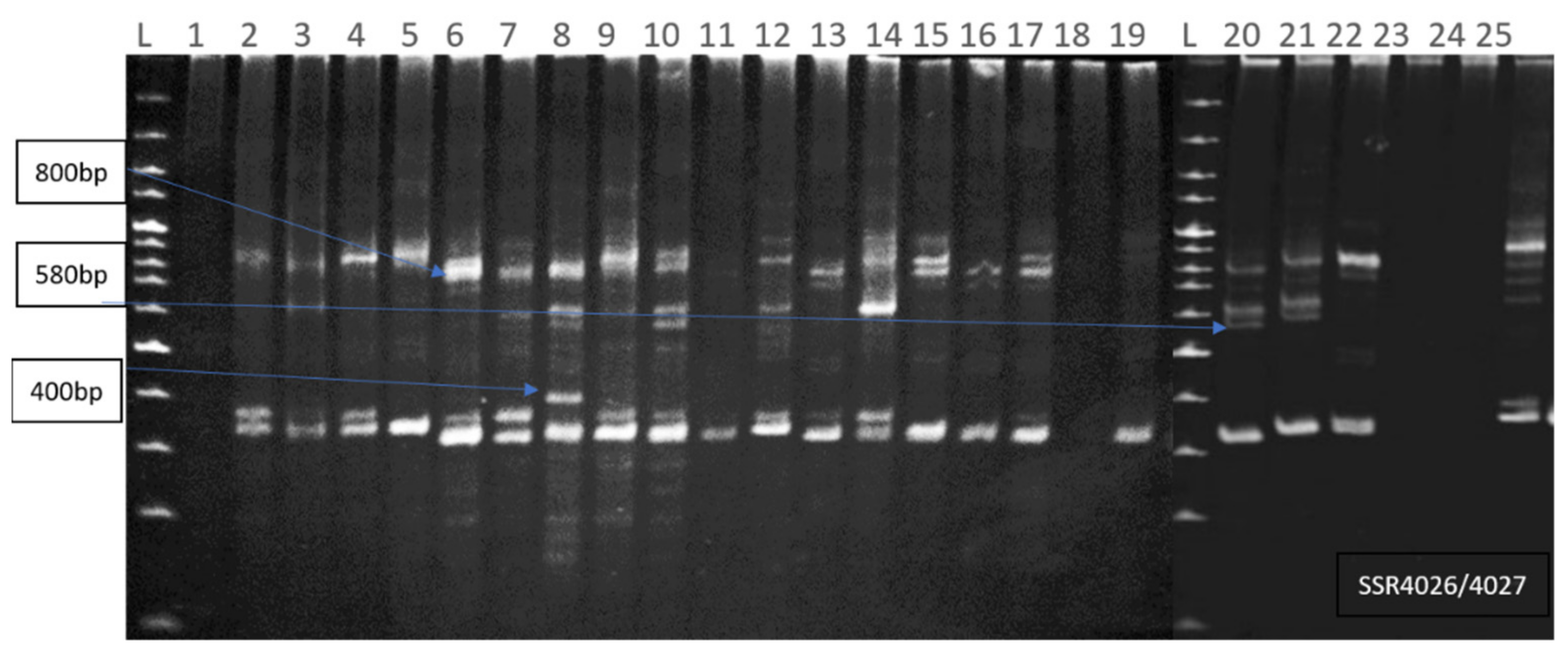
| S4026/4027 | 52 | 80 | 12 | 0 | 12 | 100 | 340–1000 | 3 | 0.87 |
| S8242 | 52 | 71 | 9 | 0 | 9 | 100 | 200–490 | 3 | 0.76 |
| S12002 | 56 | 68 | 4 | 1 | 3 | 75 | 40–245 | 0 | 0.68 |
| 31924 | 56 | 67 | 5 | 0 | 5 | 100 | 150–260 | 0 | 0.74 |
| 43016 | 54 | 49 | 5 | 0 | 5 | 100 | 50–210 | 1 | 0.73 |
| 46514 | 54 | 65 | 4 | 0 | 4 | 100 | 130–220 | 0 | 0.68 |
| STM0037 | 52 | 36 | 2 | 0 | 2 | 100 | 70–90 | 0 | 0.47 |
| STM0031 | 52 | 71 | 8 | 0 | 8 | 100 | 50–300 | 2 | 0.79 |
| ST10032 | 52 | 55 | 4 | 0 | 4 | 100 | 50–140 | 1 | 0.65 |
| S170 | 52 | 68 | 5 | 0 | 5 | 100 | 70–150 | 1 | 0.74 |
| S189 | 56 | 50 | 4 | 0 | 4 | 100 | 190–220 | 1 | 0.66 |
| S183 | 50 | 48 | 3 | 0 | 3 | 100 | 170–190 | 0 | 0.62 |
| S120 | 52 | 55 | 3 | 2 | 1 | 33.33 | 50–200 | 0 | 0.59 |
| S153 | 52 | 43 | 3 | 0 | 3 | 100 | 150–190 | 0 | 0.62 |
| 164010 | 56 | 49 | 3 | 2 | 1 | 33.33 | 30–250 | 0 | 0.57 |
| STM1106 | 52 | 82 | 6 | 1 | 5 | 83.33 | 40–200 | 0 | 0.78 |
| STM5114 | 52 | 67 | 3 | 1 | 2 | 66.66 | 40–310 | 0 | 0.66 |
| S7 | 52 | 71 | 6 | 0 | 6 | 100 | 50–190 | 0 | 0.79 |
| S180 | 54 | 22 | 1 | 0 | 1 | 100 | 190 | 0 | 0 |
| STM1104 | 54 | 21 | 2 | 0 | 2 | 100 | 180–190 | 0 | 0.36 |
| STM1052 | 52 | 28 | 4 | 0 | 4 | 100 | 210–250 | 1 | 0.36 |
| ST10004 | 52 | 33 | 3 | 0 | 3 | 100 | 90–110 | 0 | 0.64 |
| Total | 2473 | 214 | 18 | 196 | 3122.83 | 24.4 | |||
| Average | 70.65714 | 6.114286 | 0.514286 | 5.6 | 89.2237 | 0.69 |
| S. No. | Variety Name | SSR Marker |
|---|---|---|
| 1 | Mozika | S174 (310bp), S25 (430bp) |
| 2 | Cardinal | Did not produce a unique allele |
| 3 | Sahiwal White | Did not produce a unique allele |
| 4 | PRI Red | Did not produce a unique allele |
| 5 | Lady Rosetta | Did not produce a unique allele |
| 6 | Kuroda | 4026/4027 (800bp), S148 (480bp) |
| 7 | Rustum Red | S25 (185bp) |
| 8 | Ronaldo | S25 (510bp), S188 (400bp), 4026/4027 (400bp) |
| 9 | Desiree | S192 (185bp) |
| 10 | Rock | S189 (220bp) |
| 11 | Asterix | S174 (230), STM0031 (150bp, 170bp, 250bp) |
| 12 | Ruby | Did not produce a unique allele |
| 13 | Hermes | 8242 (480bp) |
| 14 | Sahiwal Red | Did not produce a unique allele |
| 15 | Sante | Did not produce a unique allele |
| 16 | Shepody | S170 (90bp) |
| 17 | Rustum | S187 (200bp) |
| 18 | Sadaf | 8242 (410bp) |
| 19 | Cosmos | Did not produce a unique allele |
| 20 | Rustum white | 4026/4027 (580bp), ST10032 (130bp) |
| 21 | LS | Did not produce a unique allele |
| 22 | GN | S25 (395bp) |
| 23 | J.8 | Did not produce a unique allele |
| 24 | Favorita | S25 (480bp, 600bp), S148 (550bp) |
| 25 | Rocco | STM1052 (250bp), 43016 (130bp), STM0031 (300bp), 8242 (280bp), S25 (700bp), S188 (190bp) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muhammad, A.; Noor, S.; Hussain, I.; Ali, K.; Shahzad, A.; Numan, M.; Zeshan, M.; Ijaz ul Hassan, S.; Ali, G.M. Simple Sequence Repeats-Based Genetic Characterization and Varietal Identification of Potato Varieties Grown in Pakistan. Sustainability 2022, 14, 11561. https://doi.org/10.3390/su141811561
Muhammad A, Noor S, Hussain I, Ali K, Shahzad A, Numan M, Zeshan M, Ijaz ul Hassan S, Ali GM. Simple Sequence Repeats-Based Genetic Characterization and Varietal Identification of Potato Varieties Grown in Pakistan. Sustainability. 2022; 14(18):11561. https://doi.org/10.3390/su141811561
Chicago/Turabian StyleMuhammad, Aish, Saima Noor, Iqbal Hussain, Kazim Ali, Armaghan Shahzad, Mian Numan, Muhammad Zeshan, Syed Ijaz ul Hassan, and Ghulam Muhammad Ali. 2022. "Simple Sequence Repeats-Based Genetic Characterization and Varietal Identification of Potato Varieties Grown in Pakistan" Sustainability 14, no. 18: 11561. https://doi.org/10.3390/su141811561
APA StyleMuhammad, A., Noor, S., Hussain, I., Ali, K., Shahzad, A., Numan, M., Zeshan, M., Ijaz ul Hassan, S., & Ali, G. M. (2022). Simple Sequence Repeats-Based Genetic Characterization and Varietal Identification of Potato Varieties Grown in Pakistan. Sustainability, 14(18), 11561. https://doi.org/10.3390/su141811561










