Use of Metagenomic Whole Genome Shotgun Sequencing Data in Taxonomic Assignment of Dipterygium glaucum Rhizosphere and Surrounding Bulk Soil Microbiomes, and Their Response to Watering
Abstract
1. Introduction
2. Materials and Methods
2.1. Watering Experiment and Soil Collection
2.2. DNA Extraction and Whole Genome Shotgun Sequencing
3. Results
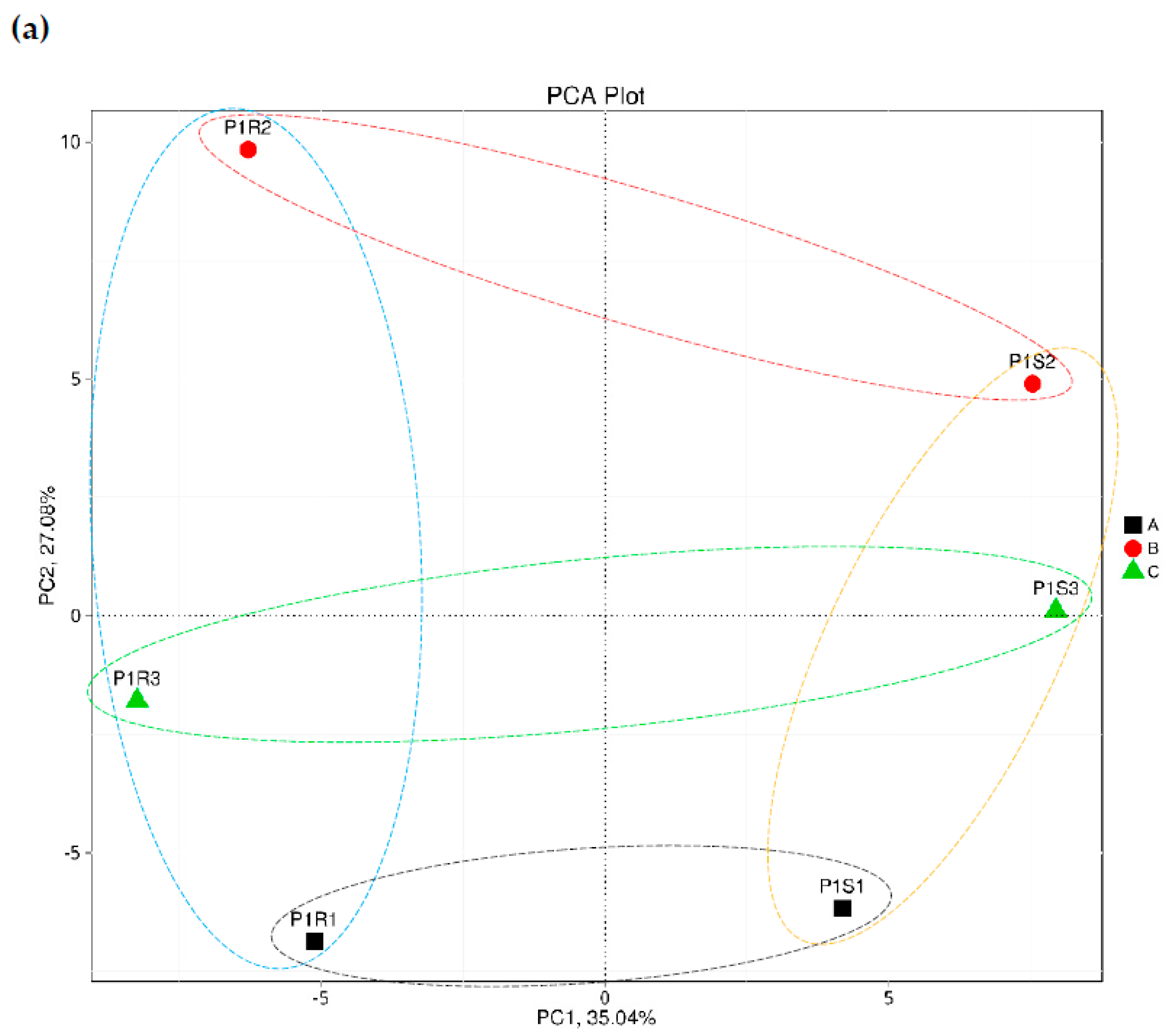
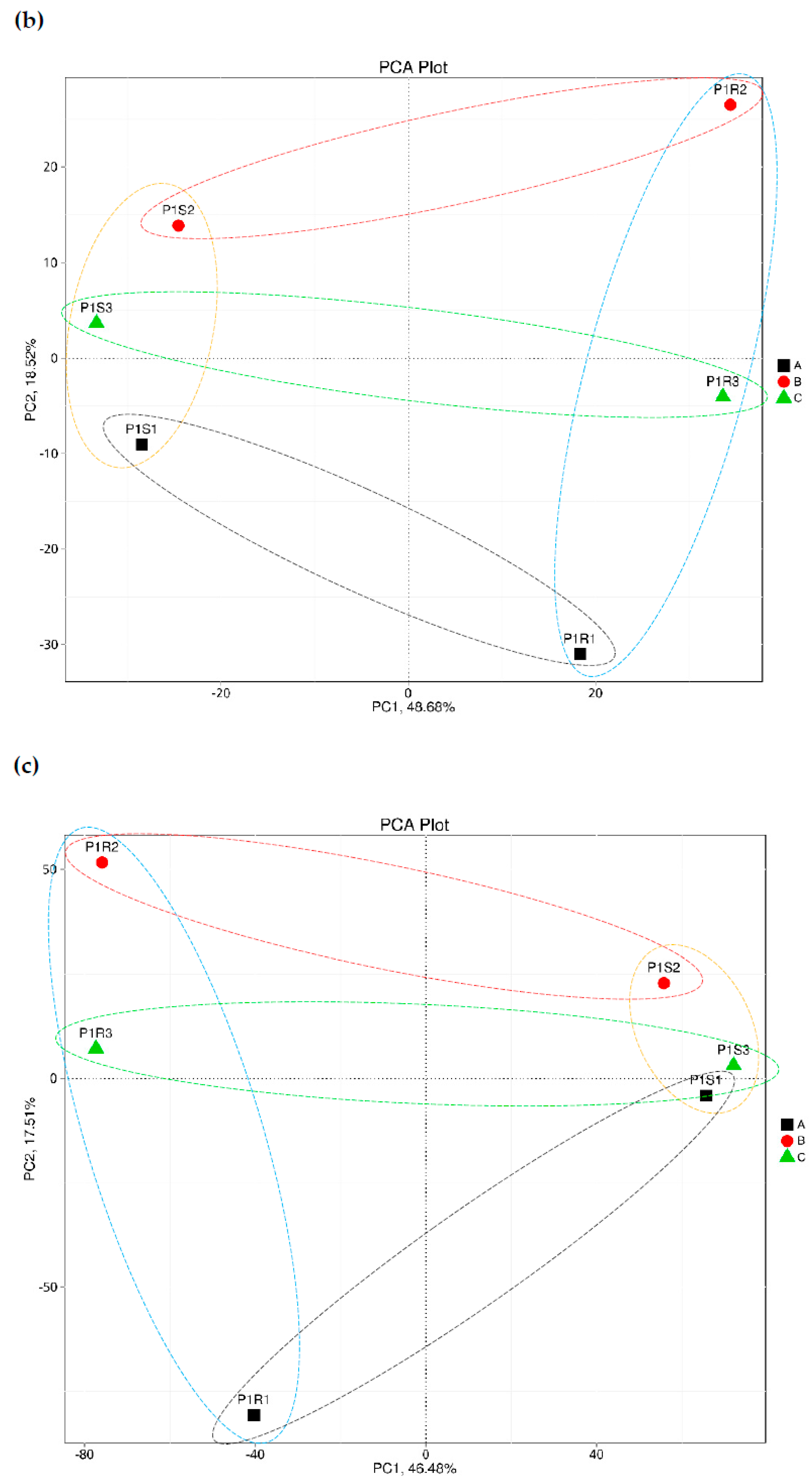
3.1. Correlation Coefficient and Principal Component Analyses
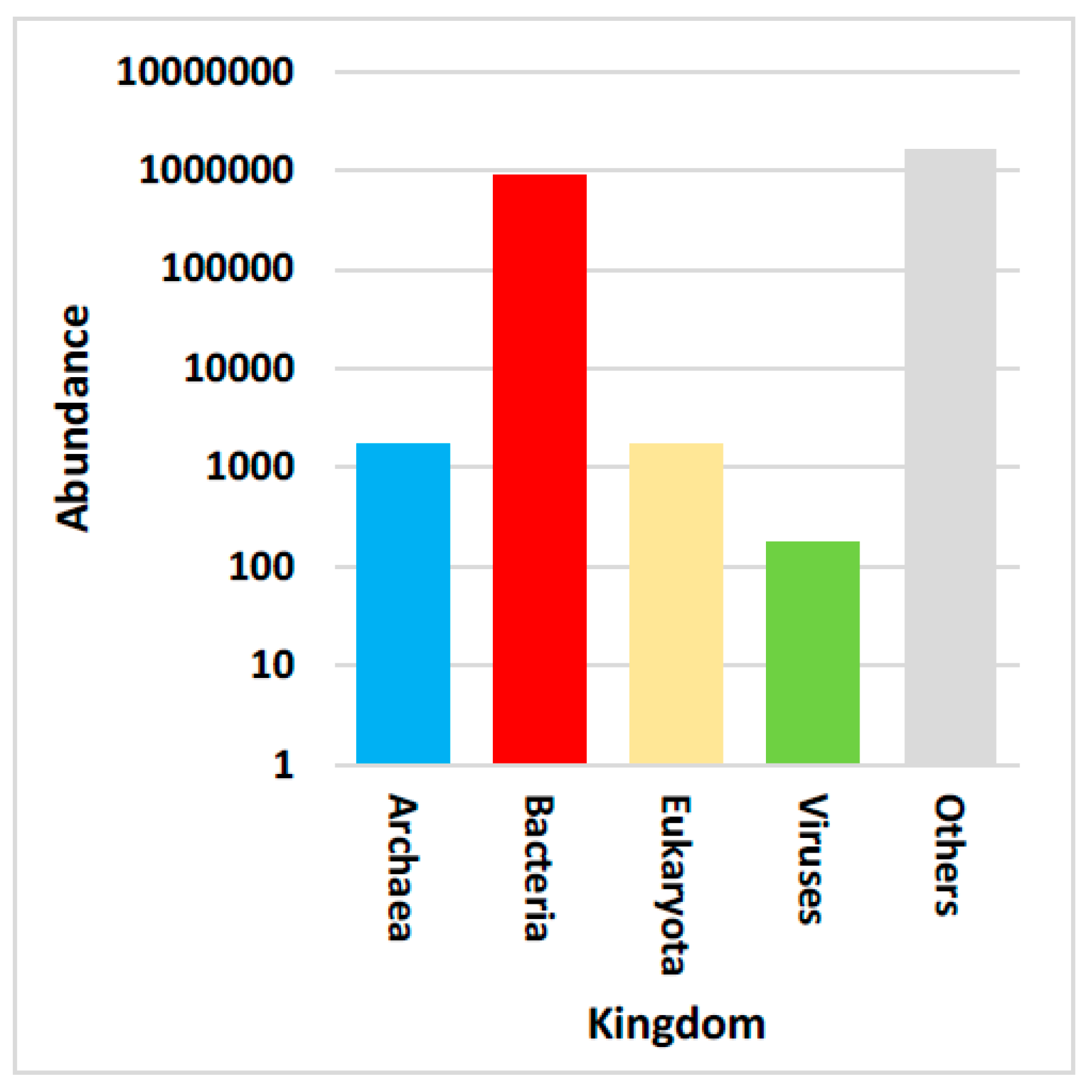
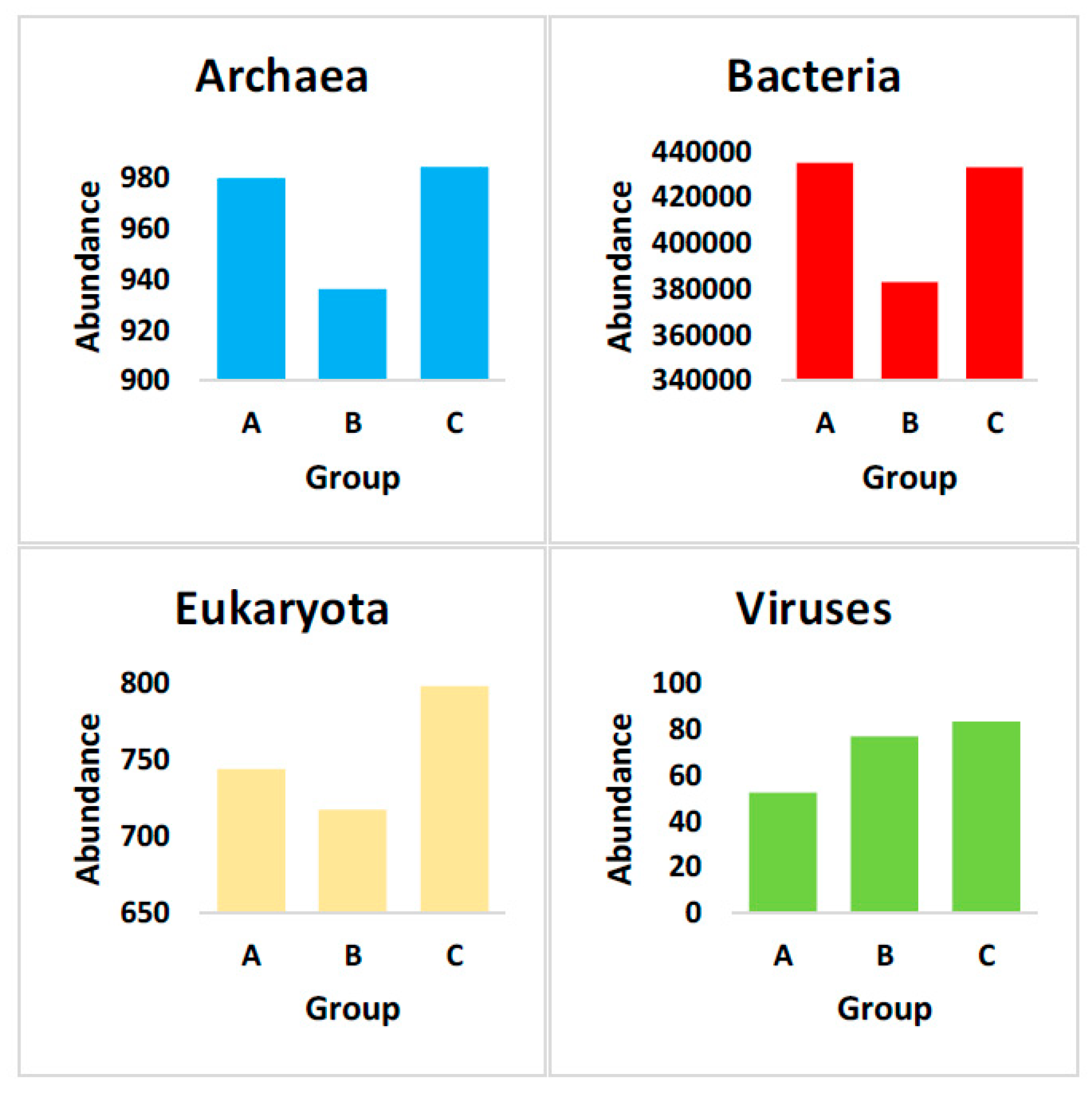
3.2. Taxonomic Assignment Based on Gene Abundance
4. Discussion
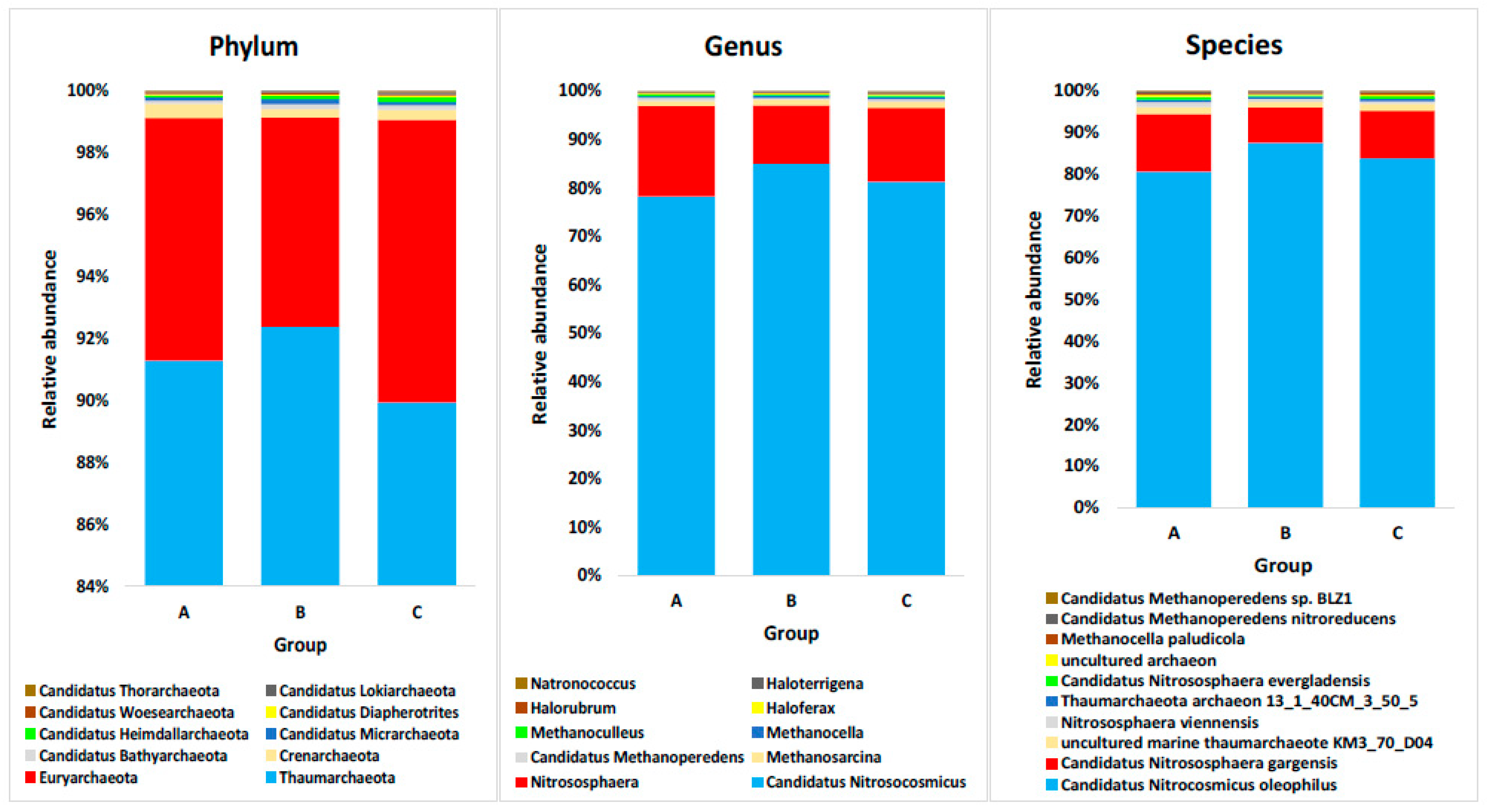
4.1. Domain Archaea
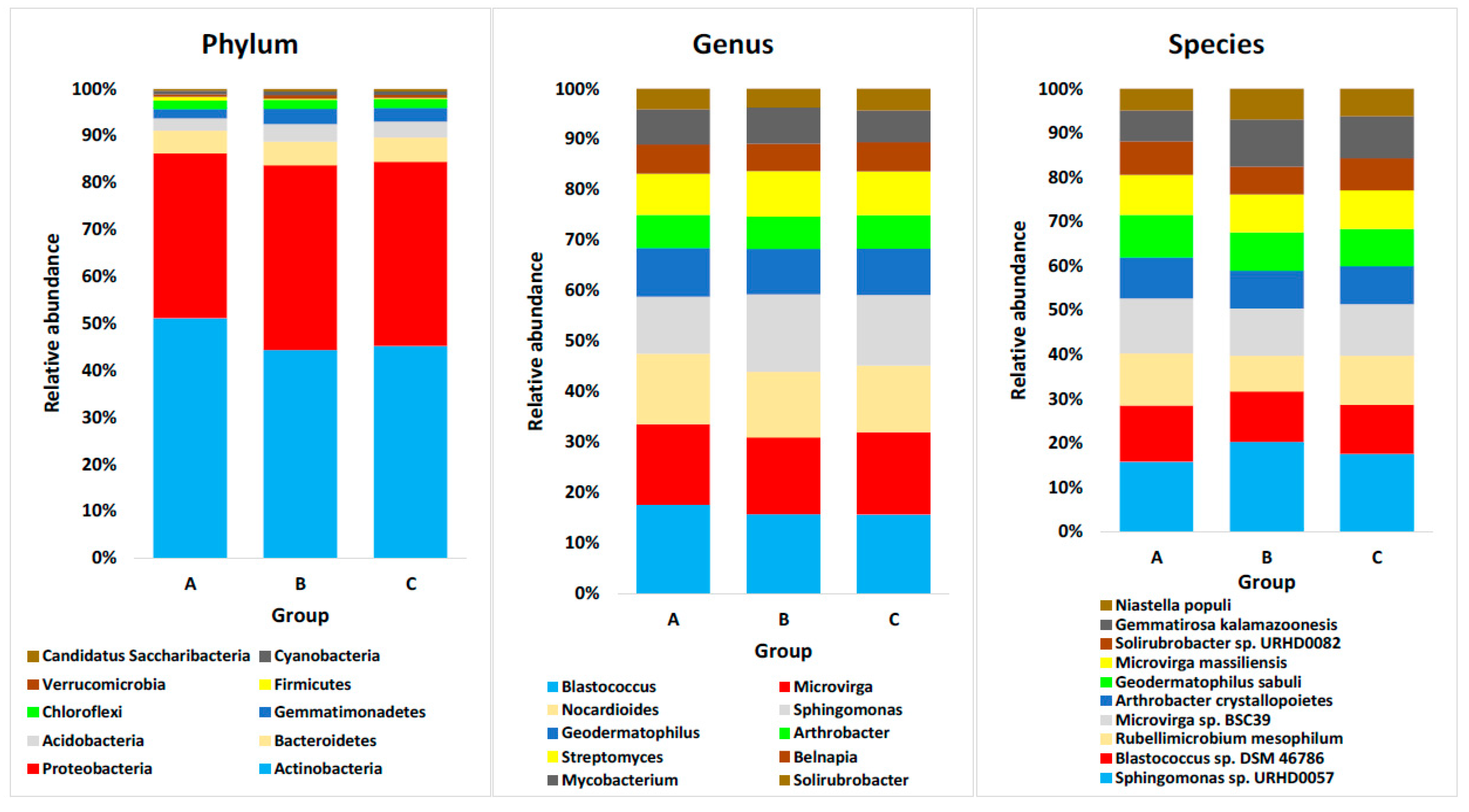
4.2. Domain Bacteria
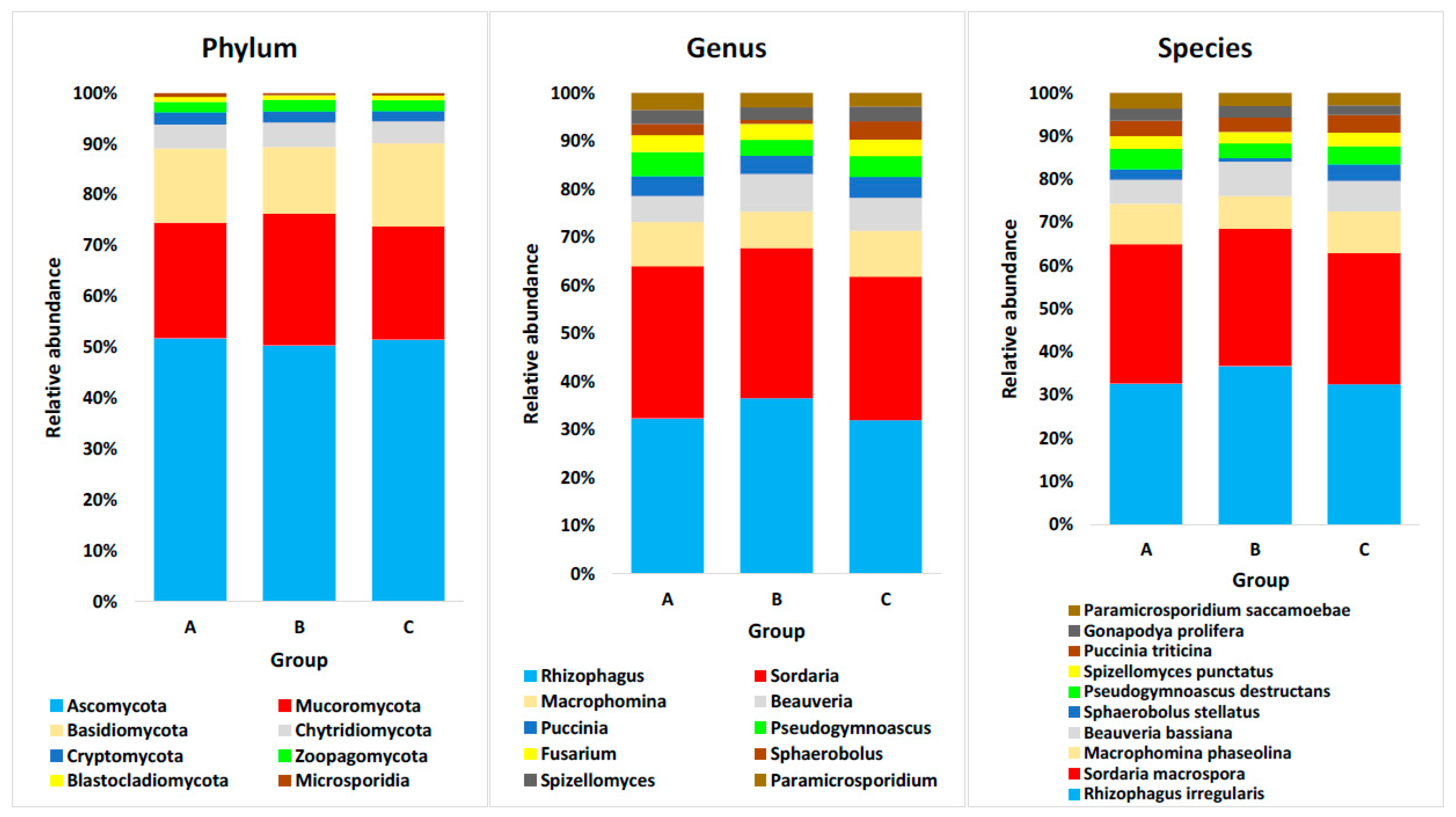
4.3. Domain Eukaryota
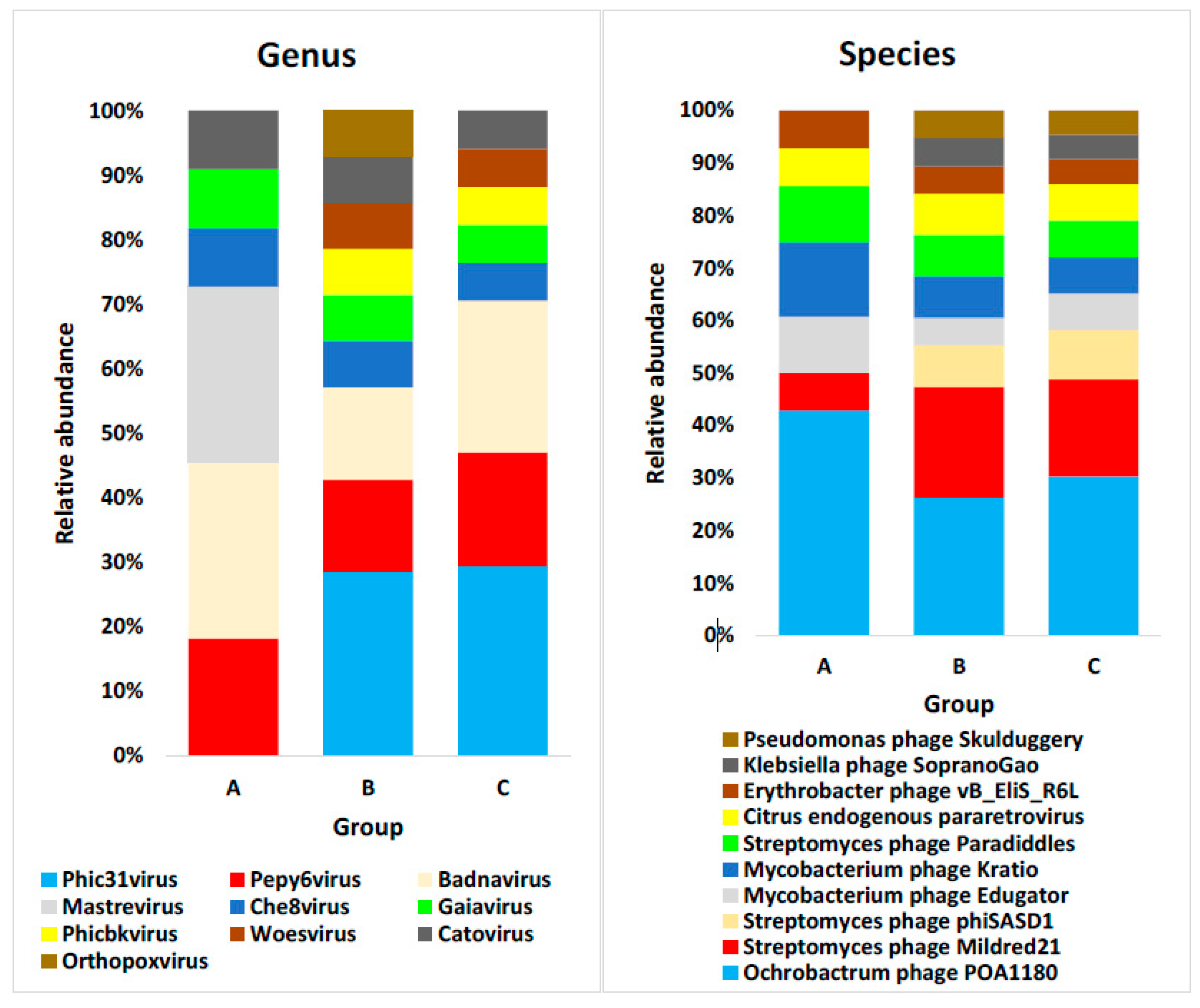
4.4. Domain Viruses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mehmood, K.; Mehmood, S.; Ramzan, M. Biochemical and phyto chemical analysis of dipterygium glaucum collected from Cholistan desert. J. Sci. Res. 2010, 40, 13–18. [Google Scholar]
- Ahmad, S.; Alam, K.; Wariss, H.; Anjum, S.; Mukhtar, M. Ethnobotanical studies of plant resources of Cholistan desert, Pakistan. Int. J. Sci. Res. 2014, 3, 1782–1788. [Google Scholar]
- Sherwani, N.; Farooq, S.A. Impact of habitat heterogeneity on growth dynamics and physiological responses of Dipterygium glaucum. Asian J. Plant Sci. 2019, 18, 75–84. [Google Scholar] [CrossRef][Green Version]
- Moussa, S.A.; Taia, W.K.; Al-Ghamdy, F.G. Acclimation of Dipterygium glaucum Decne grown in the Western Coastal part of Saudi Arabia to different water supplies. Int. J. Res. Chem. Environ. 2012, 2, 301–309. [Google Scholar]
- Abdel-Mogib, M.; Ezmirly, S.; Basaif, S. Phytochemistry of Dipterygium glaucum and Capparis decidua. J. Saudi Chem. Soc. 2000, 4, 103–108. [Google Scholar]
- Hameed, M.; Ashraf, M.; Al-Quriany, F.; Nawaz, T.; Ahmad, M.S.A.; Younis, A.; Naz, N. Medicinal flora of the Cholistan desert: A review. Pak. J. Bot. 2011, 43, 39–50. [Google Scholar]
- Gülçin, I. Antioxidant activity of food constituents: An overview. Arch. Toxicol. 2012, 86, 345–391. [Google Scholar] [CrossRef]
- Barnard, R.L.; Osborne, C.A.; Firestone, M.K. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013, 7, 2229–2241. [Google Scholar] [CrossRef]
- Vogel, T.M.; Simonet, P.; Jansson, J.K.; Hirsch, P.R.; Tiedje, J.M.; Van Elsas, J.D.; Bailey, M.J.; Nalin, R.; Philippot, L. TerraGenome: A consortium for the sequencing of a soil metagenome. Nat. Rev. Microbiol. 2009, 7, 252. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Liu, K.; McInroy, J.A.; Hu, C.H.; Kloepper, J.W. Mixtures of Plant-Growth-Promoting Rhizobacteria Enhance Biological Control of Multiple Plant Diseases and Plant-Growth Promotion in the Presence of Pathogens. Plant Dis. 2018, 102, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Schnitzer, S.A.; Klironomos, J.N.; HilleRisLambers, J.; Kinkel, L.L.; Reich, P.B.; Xiao, K.; Rillig, M.C.; Sikes, B.A.; Callaway, R.M.; Mangan, S.A. Soil microbes drive the classic plant diversity–productivity pattern. Ecology 2011, 92, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Buée, M.; Murat, C.; Frey-Klett, P.; Martin, F. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ. Microbiol. Rep. 2010, 2, 281–288. [Google Scholar] [CrossRef]
- Duineveld, B.M.; Kowalchuk, G.A.; Keijzer, A.; van Elsas, J.D.; van Veen, J.A. Analysis of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Appl. Environ. Microbiol. 2001, 67, 172–178. [Google Scholar] [CrossRef]
- Gonzalez-Franco, A.; Robles-Hernandez, L.; Nuñez-Barrios, A.; Strap, J.; Crawford, D. Molecular and cultural analysis of seasonal actinomycetes in soils from Artemisia tridentata habitat. Phyton 2009, 78, 83. [Google Scholar]
- He, S.; Guo, L.; Niu, M.; Miao, F.; Jiao, S.; Hu, T.; Long, M. Ecological diversity and co-occurrence patterns of bacterial community through soil profile in response to long-term switchgrass cultivation. Sci. Rep. 2017, 7, 3608. [Google Scholar] [CrossRef]
- Donn, S.; Kirkegaard, J.A.; Perera, G.; Richardson, A.E.; Watt, M. Evolution of bacterial communities in the wheat crop rhizosphere. Environ. Microbiol. 2015, 17, 610–621. [Google Scholar] [CrossRef]
- Shi, S.; Nuccio, E.E.; Shi, Z.J.; He, Z.; Zhou, J.; Firestone, M.K. The interconnected rhizosphere: High network complexity dominates rhizosphere assemblages. Ecol. Lett 2016, 19, 926–936. [Google Scholar] [CrossRef]
- Walters, W.A.; Jin, Z.; Youngblut, N.; Wallace, J.G.; Sutter, J.; Zhang, W.; Gonzalez-Pena, A.; Peiffer, J.; Koren, O.; Shi, Q.; et al. Large-scale replicated field study of maize rhizosphere identifies heritable microbes. Proc. Natl. Acad. Sci. USA 2018, 115, 7368–7373. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, A.; Ueda, Y.; Zushi, T.; Takase, H.; Yazaki, K. Changes in the bacterial community of soybean rhizospheres during growth in the field. PLoS ONE 2014, 9, e100709. [Google Scholar] [CrossRef] [PubMed]
- Novello, G.; Gamalero, E.; Bona, E.; Boatti, L.; Mignone, F.; Massa, N.; Cesaro, P.; Lingua, G.; Berta, G. The Rhizosphere Bacterial Microbiota of Vitis vinifera cv. Pinot Noir in an Integrated Pest Management Vineyard. Front. Microbiol. 2017, 8, 1528. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zeng, Y.; Guo, C.; Bao, Y.; Lu, G.; Reinfelder, J.R.; Dang, Z. Bacterial, archaeal, and fungal community responses to acid mine drainage-laden pollution in a rice paddy soil ecosystem. Sci. Total Environ. 2018, 616–617, 107–116. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Lu, J.; Salzberg, S.L. A review of methods and databases for metagenomic classification and assembly. Brief Bioinform. 2019, 20, 1125–1136. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef]
- Raes, J.; Foerstner, K.U.; Bork, P. Get the most out of your metagenome: Computational analysis of environmental sequence data. Curr. Opin. Microbiol. 2007, 10, 490–498. [Google Scholar] [CrossRef]
- Wilkins, L.G.; Ettinger, C.L.; Jospin, G.; Eisen, J.A. Metagenome-assembled genomes provide new insight into the microbial diversity of two thermal pools in Kamchatka, Russia. Sci. Rep. 2019, 9, 3059. [Google Scholar] [CrossRef]
- Al-Eisawi, D.M.; Al-Ruzayza, S. The flora of holy Mecca district, Saudi Arabia. Int. J. Biodivers. Conserv. 2015, 7, 173–189. [Google Scholar]
- Geng, L.-L.; Shao, G.-X.; Raymond, B.; Wang, M.-L.; Sun, X.-X.; Shu, C.-L.; Zhang, J. Subterranean infestation by Holotrichia parallela larvae is associated with changes in the peanut (Arachis hypogaea L.) rhizosphere microbiome. Microbiol. Res. 2018, 211, 13–20. [Google Scholar] [CrossRef]
- Dai, L.; Zhang, G.; Yu, Z.; Ding, H.; Xu, Y.; Zhang, Z. Effect of drought stress and developmental stages on microbial community structure and diversity in peanut rhizosphere soil. Int. J. Mol. Sci. 2019, 20, 2265. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H.B.; Almeida, M.; Juncker, A.S.; Rasmussen, S.; Li, J.; Sunagawa, S.; Plichta, D.R.; Gautier, L.; Pedersen, A.G.; Le Chatelier, E. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 2014, 32, 822–828. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Zeller, G.; Tap, J.; Voigt, A.Y.; Sunagawa, S.; Kultima, J.R.; Costea, P.I.; Amiot, A.; Bohm, J.; Brunetti, F.; Habermann, N.; et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014, 10, 766. [Google Scholar] [CrossRef]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Ocean plankton. Structure and function of the global ocean microbiome. Science 2015, 348, 1261359. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.-J.; Tappu, R. MEGAN community edition-interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Mitra, S.; Ruscheweyh, H.-J.; Weber, N.; Schuster, S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011, 21, 1552–1560. [Google Scholar] [CrossRef] [PubMed]
- Shaltout, K.H.; Ahmed, M.T.; Alrumman, S.A.; Ahmed, D.A.; Eid, E.M. Evaluation of the carbon sequestration capacity of arid mangroves along nutrient availability and salinity gradients along the Red Sea coastline of Saudi Arabia. Oceanologia 2020, 62, 56–69. [Google Scholar] [CrossRef]
- Bomberg, M.; Timonen, S. Distribution of Cren-and Euryarchaeota in Scots pine mycorrhizospheres and boreal forest humus. Microb. Ecol. 2007, 54, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Alves, R.J.E.; Kerou, M.; Zappe, A.; Bittner, R.; Abby, S.S.; Schmidt, H.A.; Pfeifer, K.; Schleper, C. Ammonia oxidation by the arctic terrestrial thaumarchaeote Candidatus Nitrosocosmicus arcticus is stimulated by increasing temperatures. Front. Microbiol. 2019, 10, 1571. [Google Scholar] [CrossRef] [PubMed]
- Alves, R.J.E.; Minh, B.Q.; Urich, T.; von Haeseler, A.; Schleper, C. Unifying the global phylogeny and environmental distribution of ammonia-oxidising archaea based on amoA genes. Nat. Commun. 2018, 9, 1517. [Google Scholar] [CrossRef] [PubMed]
- Nelkner, J.; Henke, C.; Lin, T.W.; Pätzold, W.; Hassa, J.; Jaenicke, S.; Grosch, R.; Pühler, A.; Sczyrba, A.; Schlüter, A. Effect of long-term farming practices on agricultural soil microbiome members represented by metagenomically assembled genomes (MAGs) and their predicted plant-beneficial genes. Genes 2019, 10, 424. [Google Scholar] [CrossRef]
- Sathya, A.; Vijayabharathi, R.; Gopalakrishnan, S. Plant growth-promoting actinobacteria: A new strategy for enhancing sustainable production and protection of grain legumes. 3 Biotech 2017, 7, 102. [Google Scholar] [CrossRef]
- Lazcano, C.; Boyd, E.; Holmes, G.; Hewavitharana, S.; Pasulka, A.; Ivors, K. The rhizosphere microbiome plays a role in the resistance to soil-borne pathogens and nutrient uptake of strawberry cultivars under field conditions. Sci. Rep. 2021, 11, 3188. [Google Scholar] [CrossRef]
- Kalam, S.; Basu, A.; Ahmad, I.; Sayyed, R.; El Enshasy, H.A.; Dailin, D.J.; Suriani, N. Recent understanding of soil Acidobacteria and their ecological significance: A critical review. Front. Microbiol. 2020, 11, 2712. [Google Scholar] [CrossRef]
- Walker, L.R.; Wardle, D.A.; Bardgett, R.D.; Clarkson, B.D. The use of chronosequences in studies of ecological succession and soil development. J. Ecol. 2010, 98, 725–736. [Google Scholar] [CrossRef]
- Yang, N.; Nesme, J.; Roder, H.L.; Li, X.; Zuo, Z.; Petersen, M.; Burmolle, M.; Sorensen, S.J. Emergent bacterial community properties induce enhanced drought tolerance in Arabidopsis. NPJ Biofilms Microbiomes 2021, 7, 82. [Google Scholar] [CrossRef] [PubMed]
- Chukwuneme, C.F.; Babalola, O.O.; Kutu, F.R.; Ojuederie, O.B. Characterization of actinomycetes isolates for plant growth promoting traits and their effects on drought tolerance in maize. J. Plant Interact. 2020, 15, 93–105. [Google Scholar] [CrossRef]
- Normand, P.; Daffonchio, D.; Gtari, M. The family geodermatophilaceae. In The Prokaryotes: Actinobacteria; Springer: Berlin/Heidelberg, Germany, 2014; pp. 361–379. [Google Scholar]
- Denner, E.B.M.; Kolari, M.; Hoornstra, D.; Tsitko, I.; Kampfer, P.; Busse, H.J.; Salkinoja-Salonen, M. Rubellimicrobium thermophilum gen. nov., sp. nov., a red-pigmented, moderately thermophilic bacterium isolated from coloured slime deposits in paper machines. Int. J. Syst. Evol. Microbiol. 2006, 56, 1355–1362. [Google Scholar] [CrossRef]
- Dastager, S.G.; Lee, J.C.; Ju, Y.J.; Park, D.J.; Kim, C.J. Rubellimicrobium mesophilum sp. nov., a mesophilic, pigmented bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2008, 58, 1797–1800. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, H.; Sun, L.; Qi, G.; Chen, S.; Zhao, X. Microbial community composition is related to soil biological and chemical properties and bacterial wilt outbreak. Sci. Rep. 2017, 7, 343. [Google Scholar] [CrossRef]
- Msaddak, A.; Rejili, M.; Duran, D.; Rey, L.; Imperial, J.; Palacios, J.M.; Ruiz-Argueso, T.; Mars, M. Members of Microvirga and Bradyrhizobium genera are native endosymbiotic bacteria nodulating Lupinus luteus in Northern Tunisian soils. FEMS Microbiol. Ecol. 2017, 93, fix068. [Google Scholar] [CrossRef]
- Singh, M.; Kumar, A.; Singh, R.; Pandey, K.D. Endophytic bacteria: A new source of bioactive compounds. 3 Biotech 2017, 7, 315. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Chen, J.; Liu, H.Y.; Zhang, Y.Q.; Li, W.J.; Yu, L.Y. Geodermatophilus ruber sp. nov., isolated from rhizosphere soil of a medicinal plant. Int. J. Syst. Evol. Microbiol. 2011, 61, 190–193. [Google Scholar] [CrossRef][Green Version]
- Camarena-Pozos, D.A.; Flores-Núñez, V.M.; López, M.G.; López-Bucio, J.; Partida-Martínez, L.P. Smells from the desert: M icrobial volatiles that affect plant growth and development of native and non-native plant species. Plant Cell Environ. 2019, 42, 1368–1380. [Google Scholar] [CrossRef] [PubMed]
- Flores-Núñez, V.M.; Fonseca-García, C.; Desgarennes, D.; Eloe-Fadrosh, E.; Woyke, T.; Partida-Martínez, L.P. Functional signatures of the epiphytic prokaryotic microbiome of agaves and cacti. Front. Microbiol. 2020, 10, 3044. [Google Scholar] [CrossRef]
- Zhao, Y.; Fu, W.; Hu, C.; Chen, G.; Xiao, Z.; Chen, Y.; Wang, Z.; Cheng, H. Variation of rhizosphere microbial community in continuous mono-maize seed production. Sci. Rep. 2021, 11, 1544. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, Y.; Tang, Y.; Dai, J.; Zhang, L.; An, H.; Luo, G.; Rahman, E.; Fang, C. Niastella populi sp. nov., isolated from soil of Euphrates poplar (Populus euphratica) forest, and emended description of the genus Niastella. Int. J. Syst. Evol. Microbiol. 2010, 60, 542–545. [Google Scholar] [CrossRef]
- Dzurendova, S.; Losada, C.B.; Dupuy-Galet, B.X.; Fjær, K.; Shapaval, V. Mucoromycota fungi as powerful cell factories for modern biorefinery. Appl. Microbiol. Biotechnol. 2021, 106, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Pawłowska, J.; Okrasińska, A.; Kisło, K.; Aleksandrzak-Piekarczyk, T.; Szatraj, K.; Dolatabadi, S.; Muszewska, A. Carbon assimilation profiles of mucoralean fungi show their metabolic versatility. Sci. Rep. 2019, 9, 11864. [Google Scholar] [CrossRef]
- Money, N.P. Fungal diversity. In The Fungi; Elsevier: Amsterdam, The Netherlands, 2016; pp. 1–36. [Google Scholar]
- Sparrow, F. Aquatic Phycomycetes; University of Michigan Press: Ann Arbor, MI, USA, 1960. [Google Scholar]
- Wu, L.; Chen, J.; Xiao, Z.; Zhu, X.; Wang, J.; Wu, H.; Wu, Y.; Zhang, Z.; Lin, W. Barcoded Pyrosequencing Reveals a Shift in the Bacterial Community in the Rhizosphere and Rhizoplane of Rehmannia glutinosa under Consecutive Monoculture. Int. J. Mol. Sci. 2018, 19, 850. [Google Scholar] [CrossRef]
- Ghosh, T.; Biswas, M.; Guin, C.; Roy, P. A review on characterization, therapeutic approaches and pathogenesis of Macrophomina phaseolina. Plant Cell Biotechnol. Mol. Biol. 2018, 19, 72–84. [Google Scholar]
- Khan, A.N.; Shair, F.; Malik, K.; Hayat, Z.; Khan, M.A.; Hafeez, F.Y.; Hassan, M.N. Molecular Identification and Genetic Characterization of Macrophomina phaseolina Strains Causing Pathogenicity on Sunflower and Chickpea. Front. Microbiol. 2017, 8, 1309. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, M.; Du, J.; Huang, T.; Liu, J.; Dong, T.; Chen, Y. Isolation of Burkholderia sp. HQB-1, a promising biocontrol bacteria to protect banana against Fusarium wilt through phenazine-1-carboxylic acid secretion. Front. Microbiol. 2020, 11, 3156. [Google Scholar] [CrossRef]
- Nieto-Jacobo, M.F.; Steyaert, J.M.; Salazar-Badillo, F.B.; Nguyen, D.V.; Rostas, M.; Braithwaite, M.; De Souza, J.T.; Jimenez-Bremont, J.F.; Ohkura, M.; Stewart, A.; et al. Environmental Growth Conditions of Trichoderma spp. Affects Indole Acetic Acid Derivatives, Volatile Organic Compounds, and Plant Growth Promotion. Front. Plant Sci. 2017, 8, 102. [Google Scholar] [CrossRef]
- Russ, C.; Lang, B.F.; Chen, Z.; Gujja, S.; Shea, T.; Zeng, Q.; Young, S.; Cuomo, C.A.; Nusbaum, C. Genome Sequence of Spizellomyces punctatus. Genome Announc. 2016, 4, e00849-00816. [Google Scholar] [CrossRef]
- Urbina, J.; Chestnut, T.; Allen, J.M.; Levi, T. Pseudogymnoascus destructans growth in wood, soil and guano substrates. Sci. Rep. 2021, 11, 763. [Google Scholar] [CrossRef]
- Barbarin, A.M.; Jenkins, N.E.; Rajotte, E.G.; Thomas, M.B. A preliminary evaluation of the potential of Beauveria bassiana for bed bug control. J. Invertebr. Pathol. 2012, 111, 82–85. [Google Scholar] [CrossRef]
- Reznikov, S.; Chiesa, M.A.; Pardo, E.M.; De Lisi, V.; Bogado, N.; Gonzalez, V.; Ledesma, F.; Morandi, E.N.; Ploper, L.D.; Castagnaro, A.P. Soybean-Macrophomina phaseolina-Specific Interactions and Identification of a Novel Source of Resistance. Phytopathology 2019, 109, 63–73. [Google Scholar] [CrossRef]
- Marquez, N.; Giachero, M.L.; Gallou, A.; Debat, H.J.; Declerck, S.; Ducasse, D.A. Transcriptome analysis of mycorrhizal and nonmycorrhizal soybean plantlets upon infection with Fusarium virguliforme, one causal agent of sudden death syndrome. Plant Pathol. 2019, 68, 470–480. [Google Scholar] [CrossRef]
- Zaynab, M.; Fatima, M.; Abbas, S.; Sharif, Y.; Umair, M.; Zafar, M.H.; Bahadar, K. Role of secondary metabolites in plant defense against pathogens. Microb. Pathog. 2018, 124, 198–202. [Google Scholar] [CrossRef]
- Lima-Mendez, G.; Toussaint, A.; Leplae, R. A modular view of the bacteriophage genomic space: Identification of host and lifestyle marker modules. Res. Microbiol. 2011, 162, 737–746. [Google Scholar] [CrossRef]
- Jäckel, C.; Hertwig, S.; Scholz, H.C.; Nöckler, K.; Reetz, J.; Hammerl, J.A. Prevalence, host range, and comparative genomic analysis of temperate Ochrobactrum phages. Front. Microbiol. 2017, 8, 1207. [Google Scholar] [CrossRef]
- Branco, R.; Morais, P.V. Identification and characterization of the transcriptional regulator ChrB in the chromate resistance determinant of Ochrobactrum tritici 5bvl1. PLoS ONE 2013, 8, e77987. [Google Scholar] [CrossRef]
- Piłsyk, S.; Paszewski, A. Sulfate permeasesphylogenetic diversity of sulfate transport. Acta Biochim. Pol. 2009, 56, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Imran, A.; Saadalla, M.J.A.; Khan, S.-U.; Mirza, M.S.; Malik, K.A.; Hafeez, F.Y. Ochrobactrum sp. Pv2Z2 exhibits multiple traits of plant growth promotion, biodegradation and N-acyl-homoserine-lactone quorum sensing. Ann. Microbiol. 2014, 64, 1797–1806. [Google Scholar] [CrossRef]
- Matsumura, E.E.; Coletta-Filho, H.D.; Nouri, S.; Falk, B.W.; Nerva, L.; Oliveira, T.S.; Dorta, S.O.; Machado, M.A. Deep Sequencing Analysis of RNAs from Citrus Plants Grown in a Citrus Sudden Death-Affected Area Reveals Diverse Known and Putative Novel Viruses. Viruses 2017, 9, 92. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shami, A.; Jalal, R.S.; Ashy, R.A.; Abuauf, H.W.; Baz, L.; Refai, M.Y.; Barqawi, A.A.; Baeissa, H.M.; Tashkandi, M.A.; Alshareef, S.; et al. Use of Metagenomic Whole Genome Shotgun Sequencing Data in Taxonomic Assignment of Dipterygium glaucum Rhizosphere and Surrounding Bulk Soil Microbiomes, and Their Response to Watering. Sustainability 2022, 14, 8764. https://doi.org/10.3390/su14148764
Shami A, Jalal RS, Ashy RA, Abuauf HW, Baz L, Refai MY, Barqawi AA, Baeissa HM, Tashkandi MA, Alshareef S, et al. Use of Metagenomic Whole Genome Shotgun Sequencing Data in Taxonomic Assignment of Dipterygium glaucum Rhizosphere and Surrounding Bulk Soil Microbiomes, and Their Response to Watering. Sustainability. 2022; 14(14):8764. https://doi.org/10.3390/su14148764
Chicago/Turabian StyleShami, Ashwag, Rewaa S. Jalal, Ruba A. Ashy, Haneen W. Abuauf, Lina Baz, Mohammed Y. Refai, Aminah A. Barqawi, Hanadi M. Baeissa, Manal A. Tashkandi, Sahar Alshareef, and et al. 2022. "Use of Metagenomic Whole Genome Shotgun Sequencing Data in Taxonomic Assignment of Dipterygium glaucum Rhizosphere and Surrounding Bulk Soil Microbiomes, and Their Response to Watering" Sustainability 14, no. 14: 8764. https://doi.org/10.3390/su14148764
APA StyleShami, A., Jalal, R. S., Ashy, R. A., Abuauf, H. W., Baz, L., Refai, M. Y., Barqawi, A. A., Baeissa, H. M., Tashkandi, M. A., Alshareef, S., & Abulfaraj, A. A. (2022). Use of Metagenomic Whole Genome Shotgun Sequencing Data in Taxonomic Assignment of Dipterygium glaucum Rhizosphere and Surrounding Bulk Soil Microbiomes, and Their Response to Watering. Sustainability, 14(14), 8764. https://doi.org/10.3390/su14148764






