Comparative Study on Spatial Digital Mapping Methods of Soil Nutrients Based on Different Geospatial Technologies
Abstract
1. Introduction
2. Materials and Methods
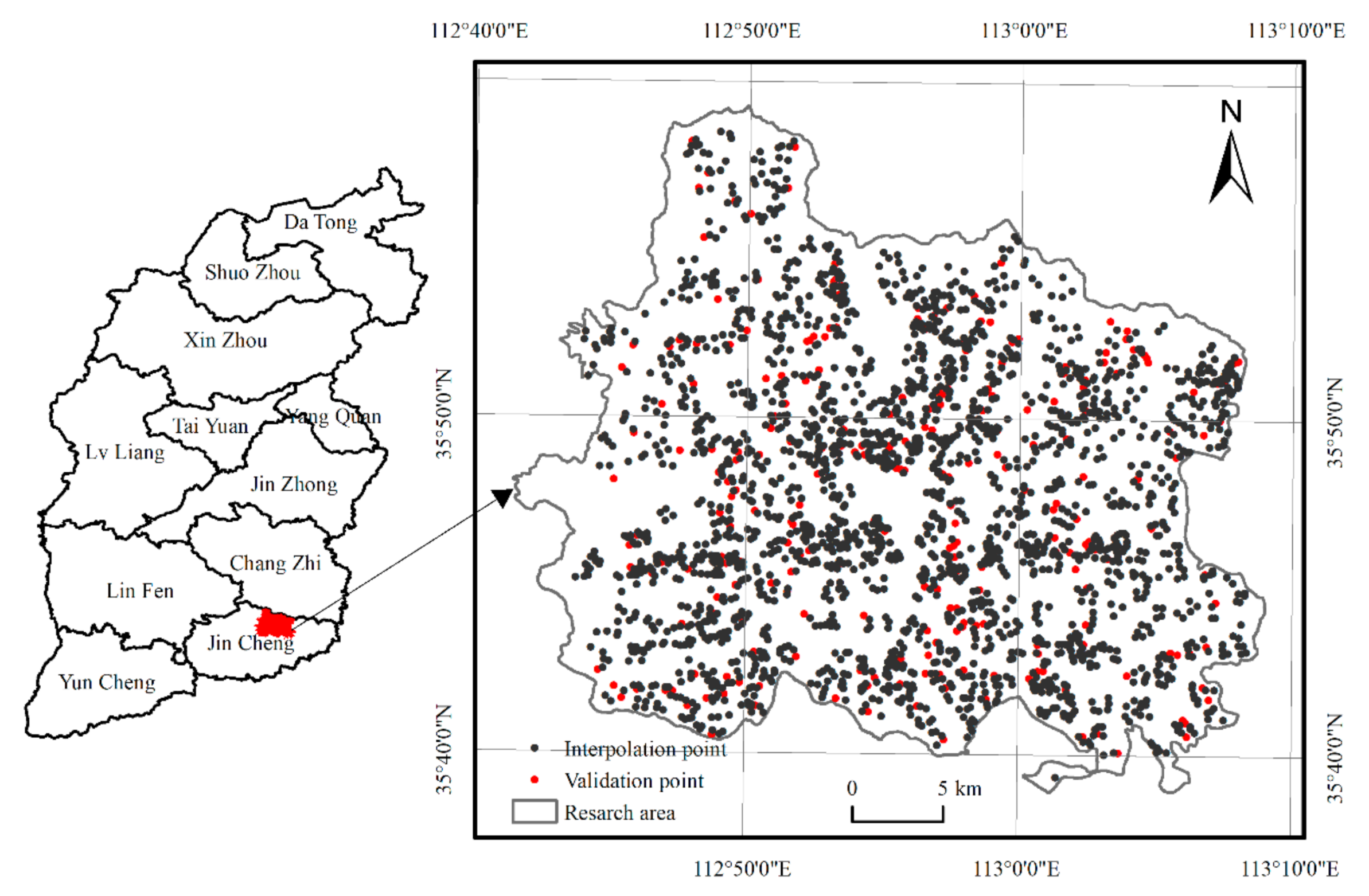
2.1. Study Area
2.2. Data Sources
2.2.1. Soil Sampling and Determination
2.2.2. Environmental Data
2.3. Geospatial Techniques
2.3.1. Ordinary Kriging (OK)
2.3.2. Regression Kriging (RK)
2.3.3. Geographically Weighted Regression Kriging (GWRK)
2.3.4. Multiscale Geographically Weighted Regression Kriging (MGWRK)
2.4. Model Validation and Evaluation
3. Results
3.1. Descriptive Statistics
3.2. Correlation Analysis of Soil Nutrients and Environmental Factors
3.3. Analysis of Spatial Distribution Patterns of Soil Nutrients
3.3.1. Selection of Environmental Covariates and MLR Modeling
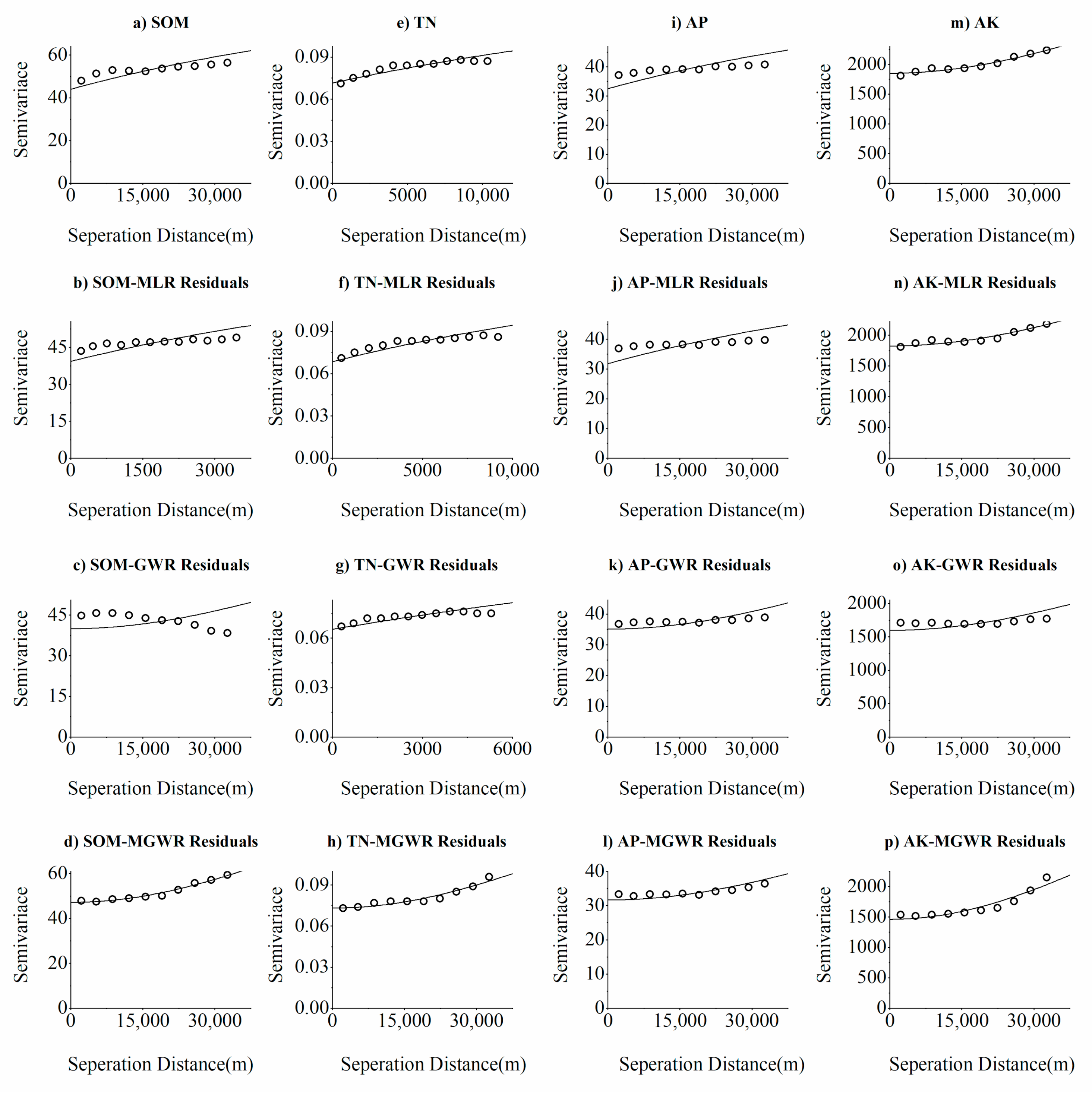
3.3.2. Spatial Variability Characteristics of Soil Nutrients and Residuals of MLR, GWR, and MGWR Models
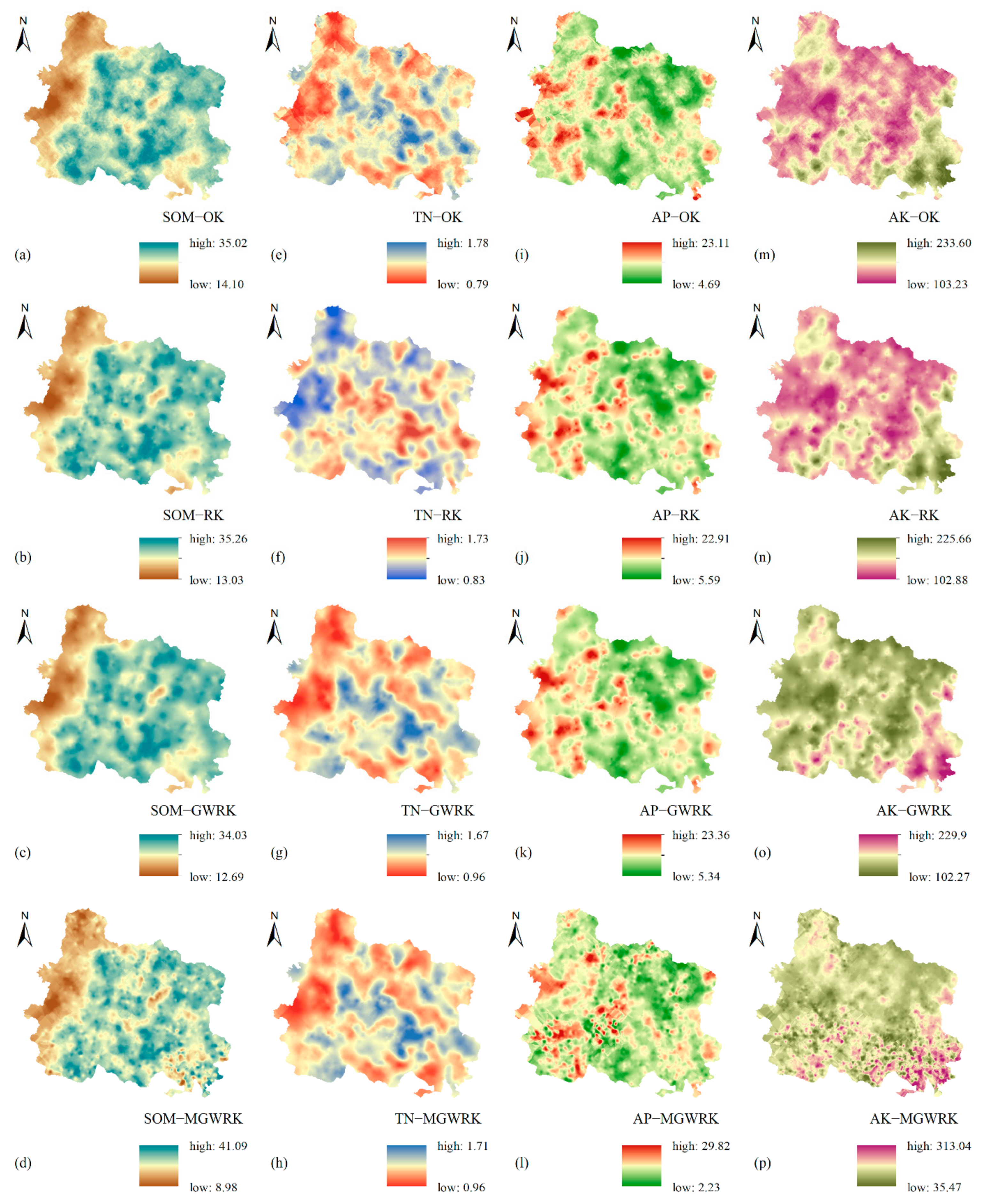
3.3.3. Results of Soil Nutrient Mapping Based on Different Models
3.3.4. Evaluation of Model Accuracy
4. Discussion
4.1. Spatial Non-Stationary Relationship between Soil Nutrients and Environmental Variables
4.2. Comparison of Model Performance in Soil Nutrient Mapping
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mcbratney, A.B.; Odeh, I.O.A.; Bishop, T.F.A.; Dunbar, M.S.; Shatar, T.M. An overview of pedometric techniques for use in soil survey. Geoderma 2000, 97, 293–327. [Google Scholar] [CrossRef]
- Sumfleth, K.; Duttmann, R. Prediction of soil property distribution in paddy soil landscapes using terrain data and satellite information as indicators. Ecol. Indic. 2008, 8, 485–501. [Google Scholar] [CrossRef]
- John, K.; Isong, I.A.; Kebonye, N.M.; Ayito, E.O.; Agyeman, P.C.; Afu, S.M. Using Machine Learning Algorithms to Estimate Soil Organic Carbon Variability with Environmental Variables and Soil Nutrient Indicators in an Alluvial Soil. Land 2020, 9, 487. [Google Scholar] [CrossRef]
- Liu, D.; Wang, Z.; Zhang, B.; Song, K.; Duan, H. Spatial distribution of soil organic carbon and analysis of related factors in croplands of the black soil region, Northeast China. Agric. Ecosyst. Environ. 2006, 113, 73–81. [Google Scholar] [CrossRef]
- Liu, E.; Yan, C.; Mei, X.; He, W.; Bing, S.H.; Ding, L.; Qin, L.; Shuang, L.; Fan, T. Long-term effect of chemical fertilizer, straw, and manure on soil chemical and biological properties in northwest China. Geoderma 2010, 158, 173–180. [Google Scholar] [CrossRef]
- Cambardella, C.; Moorman, T.B.; Novak, J.M.; Parkin, T.B.; Konopka, A. Field-Scale Variability of Soil Properties in Central Iowa Soils. Soilence Soc. Am. J. 1994, 58, 1501–1511. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, C.; Li, W. Comparison of Geographically Weighted Regression and Regression Kriging for Estimating the Spatial Distribution of Soil Organic Matter. Mapp. Sci. Remote Sens. 2012, 49, 915–932. [Google Scholar] [CrossRef]
- Emadi, M.; Baghernejad, M. Comparison of spatial interpolation techniques for mapping soil pH and salinity in agricultural coastal areas, northern Iran. Arch. Agron. Soil Sci. 2014, 60, 1315–1327. [Google Scholar] [CrossRef]
- Ye, H.; Huang, W.; Huang, S.; Huang, Y.; Zhang, S.; Dong, Y.; Chen, P. Effects of different sampling densities on geographically weighted regression kriging for predicting soil organic carbon. Spat. Stat. 2017, 20, 76–91. [Google Scholar] [CrossRef]
- Shen, Q.; Wang, Y.; Wang, X.; Liu, X.; Zhang, X.; Zhang, S. Comparing interpolation methods to predict soil total phosphorus in the Mollisol area of Northeast China. Catena 2019, 174, 59–72. [Google Scholar] [CrossRef]
- Eldeiry, A.A.; Garcia, L.A. Comparison of Ordinary Kriging, Regression Kriging, and Cokriging Techniques to Estimate Soil Salinity Using LANDSAT Images. J. Irrig. Drain. Eng. 2010, 136, 355–364. [Google Scholar] [CrossRef]
- Kumar, A.; Lal, R.; Liu, D. A geographically weighted regression kriging approach for mapping soil organic carbon stock. Geoderma 2012, 189–190, 627–634. [Google Scholar] [CrossRef]
- Watt, M.S.; Palmer, D.J. Use of regression kriging to develop a Carbon: Nitrogen ratio surface for New Zealand. Geoderma 2012, 183–184, 49–57. [Google Scholar] [CrossRef]
- Bangroo, S.; Najar, G.; Achin, E.; Truong, P.N. Application of predictor variables in spatial quantification of soil organic carbon and total nitrogen using regression kriging in the North Kashmir forest Himalayas. Catena 2020, 193, 104632. [Google Scholar] [CrossRef]
- Mondal, A.; Khare, D.; Kundu, S.; Mondal, S.; Mukherjee, S.; Mukhopadhyay, A. Spatial soil organic carbon (SOC) prediction by regression kriging using remote sensing data. Egypt. J. Remote Sens. Space Sci. 2017, 20, 61–70. [Google Scholar] [CrossRef]
- Odeh, I.O.; McBratney, A.B.; Chittleborough, D.J. Further results on prediction of soil properties from terrain attributes: Heterotopic cokriging and regression-kriging. Geoderma 1995, 67, 215–226. [Google Scholar] [CrossRef]
- Hengl, T.; Heuvelink, G.B.; Stein, A. A generic framework for spatial prediction of soil variables based on regression-kriging. Geoderma 2004, 120, 75–93. [Google Scholar] [CrossRef]
- Hengl, T.; Heuvelink, G.B.M.; Rossiter, D.G. About regression-kriging: From equations to case studies. Comput. Geosci. 2007, 33, 1301–1315. [Google Scholar] [CrossRef]
- Walter, C.; Mcbratney, A.B.; Donuaoui, A.; Minasny, B. Spatial prediction of topsoil salinity in the Chelif Valley, Algeria, using local ordinary kriging with local variograms versus whole-area variogram. Soil Res. 2001, 39, 259–272. [Google Scholar] [CrossRef][Green Version]
- Liu, Y.; Guo, L.; Jiang, Q.; Zhang, H.; Chen, Y. Comparing geospatial techniques to predict SOC stocks. Soil Tillage Res. 2015, 148, 46–58. [Google Scholar] [CrossRef]
- Brunsdon, C.; Fotheringham, A.S.; Charlton, M.E. Geographically Weighted Regression: A Method for Exploring Spatial Nonstationarity. Geogr. Anal. 1996, 28, 281–298. [Google Scholar] [CrossRef]
- Kumar, S.; Lal, R. Mapping the organic carbon stocks of surface soils using local spatial interpolator. J. Environ. Monit. JEM 2011, 13, 3128–3135. [Google Scholar] [CrossRef]
- Odhiambo, B.O.; Kenduiywo, B.K.; Were, K. Spatial prediction and mapping of soil pH across a tropical afro-montane landscape. Appl. Geogr. 2020, 114, 102129. [Google Scholar] [CrossRef]
- Kumar, S. Estimating spatial distribution of soil organic carbon for the Midwestern United States using historical database. Chemosphere 2015, 127, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Mitran, T.; Mishra, U.; Lal, R.; Ravisankar, T.; Sreenivas, K. Spatial distribution of soil carbon stocks in a semi-arid region of India. Geoderma Reg. 2018, 15, e00192. [Google Scholar] [CrossRef]
- Yu, H.; Fotheringham, A.S.; Li, Z.; Oshan, T.; Kang, W.; Wolf, L.J. Inference in multiscale geographically weighted regression. Geogr. Anal. 2020, 52, 87–106. [Google Scholar] [CrossRef]
- Mcmaster, R.B.; Sheppard, E. Introduction: Scale and Geographic Inquiry; Blackwell Publishing Ltd.: Hoboken, NJ, USA, 2008. [Google Scholar]
- Lam, N.S.N.; Quattrochi, D.A. On the Issues of Scale, Resolution, and Fractal Analysis in the Mapping Sciences. Prof. Geogr. 1992, 44, 88–98. [Google Scholar] [CrossRef]
- Peng, G.; Ling, B. Scale Effects on Spatially Embedded Contact Networks. Comput. Environ. Urban Syst. 2016, 59, 142–151. [Google Scholar] [CrossRef]
- Cao, C. Detecting the Scale and Resolution Effects in Remote Sensing and GIS; Louisiana State University and Agricultural & Mechanical College: Baton Rouge, LA, USA, 1992. [Google Scholar]
- Pan, Y.; Roth, A.; Yu, Z.; Doluschitz, R. The impact of variation in scale on the behavior of a cellular automata used for land use change modeling. Comput. Environ. Urban Syst. 2010, 34, 400–408. [Google Scholar] [CrossRef]
- Murakami, D.; Lu, B.; Harris, P.; Brunsdon, C.; Charlton, M.; Nakaya, T.; Griffith, D.A. The importance of scale in spatially varying coefficient modeling. Ann. Am. Assoc. Geogr. 2019, 109, 50–70. [Google Scholar] [CrossRef]
- Yang, S.-H.; Liu, F.; Song, X.-D.; Lu, Y.-Y.; Li, D.-C.; Zhao, Y.-G.; Zhang, G.-L. Mapping topsoil electrical conductivity by a mixed geographically weighted regression kriging: A case study in the Heihe River Basin, northwest China. Ecol. Indic. 2019, 102, 252–264. [Google Scholar] [CrossRef]
- Mei, C.L.; He, S.Y.; Fang, K.T. A Note on the Mixed Geographically Weighted Regression Model. J. Reg. Sci. 2004, 44, 143–157. [Google Scholar] [CrossRef]
- McMillen, D.P. Geographically Weighted Regression: The Analysis of Spatially Varying Relationships. Am. J. Agric. Econ. 2004, 86, 554–556. [Google Scholar] [CrossRef]
- Nakaya, T.; Fotheringham, S.; Charlton, M.; Brunsdon, C. Semiparametric Geographically Weighted Generalised Linear Modelling in GWR 4.0. 2009. Available online: http://www.geocomputation.org/2009/PDF/nakaya_et_al.pdf (accessed on 15 October 2020).
- Wu, C.; Ren, F.; Hu, W.; Du, Q. Multiscale geographically and temporally weighted regression: Exploring the spatiotemporal determinants of housing prices. Int. J. Geogr. Inf. Sci. 2019, 33, 489–511. [Google Scholar] [CrossRef]
- Fotheringham, A.S.; Yang, W.; Wei, K. Multiscale Geographically Weighted Regression (MGWR). Ann. Am. Assoc. Geogr. 2017, 107, 1247–1265. [Google Scholar] [CrossRef]
- Qiao, L.; Zhang, W.; Huang, M.; Wang, G.; Ren, J. Mapping of soil organic matter and its driving factors study based on MGWRK. Sci. Agric. Sin. 2020, 53, 1830–1844. [Google Scholar] [CrossRef]
- Carter, M.R.; Gregorich, E.G. Soil Sampling and Methods of Analysis; CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar]
- Yang, S.; Zhang, H.; Zhang, C.; Li, W.; Guo, L.; Chen, J. Predicting soil organic matter content in a plain-to-hill transition belt using geographically weighted regression with stratification. Arch. Agron. Soil Sci. 2019, 65, 1745–1757. [Google Scholar] [CrossRef]
- Zeng, C.; Yang, L.; Zhu, A.X.; Rossiter, D.G.; Liu, J.; Liu, J.; Qin, C.; Wang, D. Mapping soil organic matter concentration at different scales using a mixed geographically weighted regression method. Geoderma 2016, 281, 69–82. [Google Scholar] [CrossRef]
- National Meteorological Information Center. Available online: http://data.cma.cnr (accessed on 15 October 2020).
- Geospatial Data Cloud. Available online: http://www.gscloud.cn (accessed on 15 October 2020).
- Shabrina, Z.; Buyuklieva, B.; Ng, M.K.M. Short-Term Rental Platform in the Urban Tourism Context: A Geographically Weighted Regression (GWR) and a Multiscale GWR (MGWR) Approaches. Geogr. Anal. 2020, 1–22. [Google Scholar] [CrossRef]
- Oshan, T.M.; Li, Z.; Kang, W.; Wolf, L.J.; Fotheringham, A.S. MGWR: A Python Implementation of Multiscale Geographically Weighted Regression for Investigating Process Spatial Heterogeneity and Scale. Int. J. Geo Inf. 2019, 8, 269. [Google Scholar] [CrossRef]
- Sun, Y.S.; Wang, W.F.; Li, G.C. Spatial distribution of forest carbon storage in Maoershan region, Northeast China based on geographically weighted regression kriging model. Chin. J. Appl. Ecol. 2019, 30, 1642–1650. [Google Scholar] [CrossRef]
- Wang, S.; Hu, K.; Lu, P.; Yu, T. Spatial variability of soil available phosphorus and environmental risk analysis of soil phosphorus in Pinggu County of Beijing. Sci. Agric. Sin. 2009, 42, 1290–1298. [Google Scholar] [CrossRef]
- Wang, S.; Adhikari, K.; Wang, Q.; Jin, X.; Li, H. Role of environmental variables in the spatial distribution of soil carbon (C), nitrogen (N), and C:N ratio from the northeastern coastal agroecosystems in China. Ecol. Indic. Integr. Monit. Assess. Manag. 2018, 84, 263–272. [Google Scholar] [CrossRef]
- Tu, C.; He, T.; Lu, X.; Luo, Y.; Smith, P. Extent to which pH and topographic factors control soil organic carbon level in dry farming cropland soils of the mountainous region of Southwest China. Catena 2018, 163, 204–209. [Google Scholar] [CrossRef]
- Song, X.D.; Brus, D.J.; Liu, F.; Li, D.C.; Zhao, Y.G.; Yang, J.L.; Zhang, G.L. Mapping soil organic carbon content by geographically weighted regression: A case study in the Heihe River Basin, China. Geoderma 2016, 261, 11–12. [Google Scholar] [CrossRef]
- Naipal, V.; Ciais, P.; Wang, Y.; Lauerwald, R.; Oost, K.V. Global soil organic carbon removal by water erosion underclimate change and land use change during 1850–2005 AD. Biogeosci. Discuss. 2018, 4459–4480. [Google Scholar] [CrossRef]
- Davidson, E.A.; Janssens, I.A. Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 2006, 440, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Schillaci, C.; Acutis, M.; Lombardo, L.; Lipani, A.; Fantappiè, M.; Märker, M.; Saia, S. Spatio-temporal topsoil organic carbon mapping of a semi-arid Mediterranean region: The role of land use, soil texture, topographic indices and the influence of remote sensing data to modelling. Sci. Total Environ. 2017, 601–602, 821–832. [Google Scholar] [CrossRef]
- Agostinelli, C. Robust stepwise regression. J. Appl. Stat. 2003, 48, 557–558. [Google Scholar] [CrossRef]
- Behera, S.K.; Singh, M.V.; Singh, K.N.; Todwal, S. Distribution variability of total and extractable zinc in cultivated acid soils of India and their relationship with some selected soil properties. Geoderma 2012, 162, 242–250. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhao, G.; Jiang, H. Study on spatial variability of soil nutrients and reasonable sampling number at county scale. J. Nat. Resour. 2012, 27, 1382–1391. [Google Scholar] [CrossRef]
- Qu, M.; Li, W.; Zhang, C.; Huang, B.; Zhao, Y. Spatial assessment of soil nitrogen availability and varying effects of related main soil factors on soil available nitrogen. Environ. Sci. Process. Impacts 2016, 18, 1449–1457. [Google Scholar] [CrossRef] [PubMed]
- Hengl, T. A Practical Guide to Geostatistical Mapping of Environmental Variables; European Commission, Joint Research Centre, Institute for Environment and Sustainability: Ispra, Italy, 2007. [Google Scholar]
- Zhang, S.; Wang, Z.; Zhang, B.; Song, K.; Liu, D.; Li, F.; Ren, C.; Huang, J.; Zhang, H. Prediction of spatial distribution of soil nutrients using terrain attributes and remote sensing data. Trans. Chin. Soc. Agric. Eng. 2010, 26, 188–194. [Google Scholar] [CrossRef]
- Ren, L.; Yang, L.; Wang, H.; Yang, F.; Chen, W.; Zhang, L.; Jinhao, X.U. Spatial prediction of soil organic matter in apple region based on random forest. Resour. Environ. Arid Region 2018. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, X.Q.; Sun, X.L.; Wang, H.L. Comparing Prediction Accuracies of Ordinary Kriging and Regression Kriging with REML in Soil Properties Mapping. Chin. J. Soil Sci. 2018, 49, 283–292. [Google Scholar] [CrossRef]
- Ziadat, M.F. Analyzing Digital Terrain Attributes to Predict Soil Attributes for a Relatively Large Area. Soil Sci. Soc. Am. J. 2005, 69, 1590–1599. [Google Scholar] [CrossRef]
- Benítez, F.L.; Anderson, L.O.; Formaggio, A.N.R. Evaluation of geostatistical techniques to estimate the spatial distribution of aboveground biomass in the Amazon rainforest using high-resolution remote sensing data. Acta Amaz. 2016, 46, 151–160. [Google Scholar] [CrossRef]
- Imran, M.; Stein, A.; Zurita-Milla, R. Using geographically weighted regression kriging for crop yield mapping in West Africa. Int. J. Geogr. Inf. Syst. 2015, 29, 234–257. [Google Scholar] [CrossRef]
- Kravchenko, A.N.; Robertson, G.P. Can Topographical and Yield Data Substantially Improve Total Soil Carbon Mapping by Regression Kriging? Agron. J. 2007, 99, 12–17. [Google Scholar] [CrossRef]
- Zhang, C.-T.; Yang, Y. Can the spatial prediction of soil organic matter be improved by incorporating multiple regression confidence intervals as soft data into BME method? Catena 2019, 178, 322–334. [Google Scholar] [CrossRef]
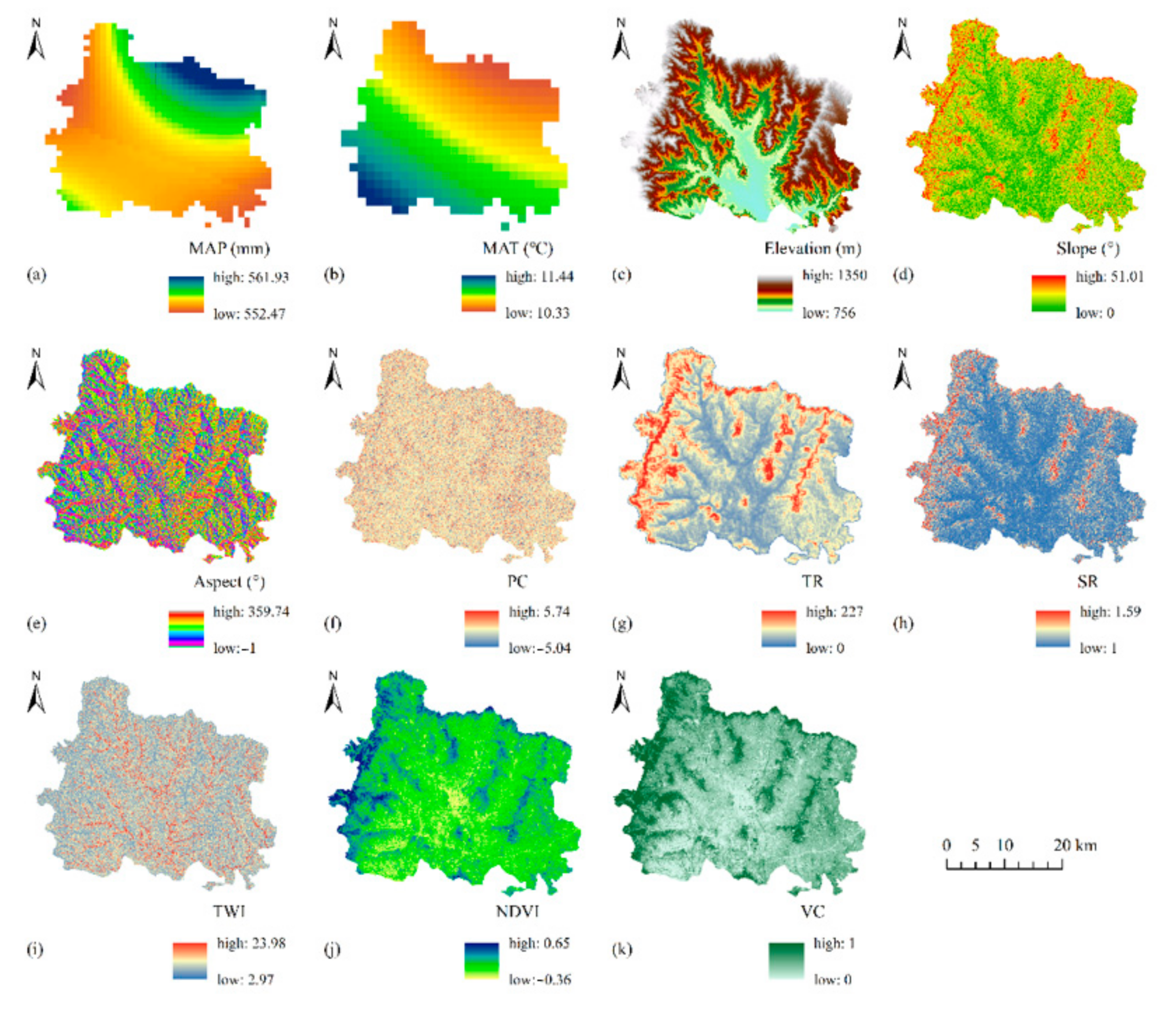

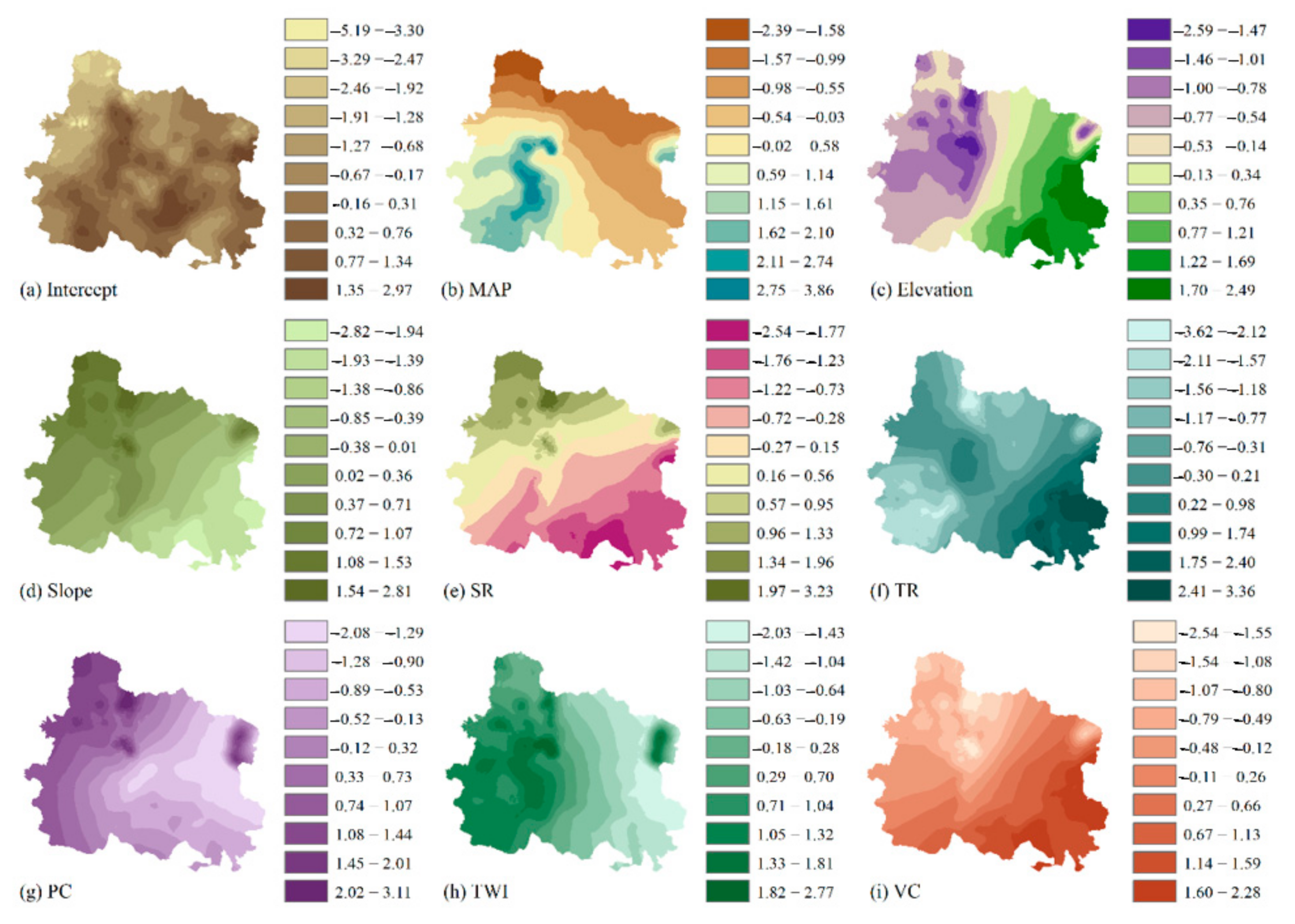
| Data Type | Index |
|---|---|
| Terrain data | Elevation |
| Slope | |
| Aspect | |
| Plane curvature, PC | |
| Topographic relief, TR | |
| Surface roughness, SR | |
| Topographic wetness index, TWI | |
| Meteorological data | Mean average precipitation, MAP |
| Mean average temperature, MAT | |
| Remote sensing data | Normalized differential vegetation index, NDVI |
| Vegetation coverage, VC |
| Index | Unit | Min | Max | Mean | S.D. | Variance | CV (%) |
|---|---|---|---|---|---|---|---|
| Soil organic matter, SOM | g kg−1 | 3.10 | 45.00 | 27.04 | 7.28 | 52.99 | 26.92 |
| Total nitrogen, TN | g kg−1 | 0.11 | 2.51 | 1.33 | 0.29 | 0.08 | 21.65 |
| Avaliable Phosophorus, AP | mg kg−1 | 1.40 | 39.90 | 12.90 | 6.25 | 39.09 | 48.48 |
| Avaliable potassium, AK | mg kg−1 | 38.00 | 388.00 | 153.03 | 44.30 | 1962.28 | 28.95 |
| Index | Unit | Min | Max | Mean | S.D. | Variance | CV (%) |
|---|---|---|---|---|---|---|---|
| Mean Average Precipitation, MAP | mm | 552.52 | 561.93 | 555.35 | 1.67 | 2.79 | 0.30 |
| Mean Average Temperature, MAT | °C | 10.33 | 11.44 | 10.82 | 0.25 | 0.06 | 2.29 |
| Elevation | m | 780.00 | 1273.00 | 914.78 | 68.08 | 4635.17 | 7.44 |
| Slope | ° | 0.00 | 34.20 | 7.38 | 4.46 | 19.87 | 60.41 |
| Aspect | ° | −1.00 | 359.17 | 176.34 | 96.16 | 9245.94 | 54.53 |
| Surface Roughness, SR | 1.00 | 1.21 | 1.01 | 0.02 | 0.00 | 1.49 | |
| Topographic relief, TR | 14.00 | 157.00 | 46.24 | 18.69 | 349.19 | 40.41 | |
| Plane Curvature, PC | −2.24 | 2.34 | 0.01 | 0.57 | 0.33 | 5870.25 | |
| Topographic wetness index, TWI | 3.81 | 22.38 | 7.44 | 2.68 | 7.16 | 35.95 | |
| Normalized differential vegetation index, NDVI | −0.16 | 0.37 | 0.07 | 0.07 | 0.01 | 100.89 | |
| Vegetation Coverage, VC | 0.00 | 1.00 | 0.29 | 0.18 | 0.03 | 61.33 |
| Index | SOM | TN | AP | AK |
|---|---|---|---|---|
| MAP | 0.114 ** | −0.006 | −0.098 ** | −0.109 ** |
| MAT | −0.015 | 0.018 | 0.082 ** | 0.079 ** |
| Elevation | −0.154 ** | −0.057 ** | 0.039 | −0.025 |
| Slope | −0.094 ** | −0.022 | −0.008 | −0.012 |
| Aspect | 0.016 | −0.002 | 0.012 | 0.015 |
| SR | −0.082 ** | −0.020 | 0.001 | −0.016 |
| TR | −0.214 ** | −0.066 ** | 0.019 | −0.029 |
| PC | −0.028 | −0.018 | 0.025 | 0.001 |
| TWI | 0.018 | 0.006 | −0.031 | −0.043 * |
| NDVI | −0.151 ** | −0.098 ** | −0.004 | 0.010 |
| VC | −0.154 ** | −0.098 ** | −0.006 | 0.006 |
| Index | Regression Equation | R2 |
|---|---|---|
| SOM | SOM = 0.7188 × MAP − 0.1338 × TWI − 0.0123 × Elevation − 0.1669 × Slope − 2.5425 × VC + 52.1433 × SR − 0.05106 × TR − 0.5255 × PC − 408.3766 | 0.075 |
| TN | TN = 0.4060069 × NDVI + 1.354716 | 0.0097 |
| AP | AP = −0.3824 × MAP + 1.5562 × MAT + 0.0123 × Elevation − 0.0170 × TR + 197.9734 | 0.0193 |
| AK | AK = −2.9193 × MAP − 0.8736 × TWI − 0.0909 × TR + 1785.001 | 0.0154 |
| Index | Type | Fitting Model | Nugget (C0) | Sill (C0 + C) | Nugget/Sill [C0/(C0 + C), %] | Range (m) | R2 |
|---|---|---|---|---|---|---|---|
| SOM | OK | Exponential Model | 44.000 | 88.010 | 50.0 | 71,100 | 0.873 |
| MLR residuals | Exponential Model | 39.300 | 78.610 | 50.0 | 8110 | 0.841 | |
| GWR residuals | Gaussion Model | 40.000 | 80.010 | 50.0 | 71,100 | 0.966 | |
| MGWR residuals | Gaussion Model | 47.200 | 109.980 | 57.1 | 71,100 | 0.979 | |
| TN | OK | Exponential Model | 0.0715 | 0.143 | 50.0 | 31,100 | 0.844 |
| MLR residuals | Exponential Model | 0.068 | 0.137 | 50.0 | 21,100 | 0.876 | |
| GWR residuals | Exponential Model | 0.065 | 0.131 | 50.0 | 21,100 | 0.837 | |
| MGWR residuals | Gaussion Model | 0.073 | 0.176 | 58.5 | 71,100 | 0.923 | |
| AP | OK | Exponential Model | 32.500 | 65.010 | 50.0 | 71,100 | 0.941 |
| MLR residuals | Exponential Model | 31.800 | 63.610 | 50.0 | 71,100 | 0.896 | |
| GWR residuals | Gaussion Model | 35.100 | 70.210 | 50.0 | 71,100 | 0.835 | |
| MGWR residuals | Gaussion Model | 31.630 | 63.270 | 50.0 | 71,100 | 0.883 | |
| AK | OK | Gaussion Model | 1851 | 3882 | 52.3 | 71,100 | 0.959 |
| MLR residuals | Gaussion Model | 1826 | 3653 | 50.0 | 71,100 | 0.915 | |
| GWR residuals | Gaussion Model | 1599 | 3199 | 50.0 | 71,100 | 0.850 | |
| MGWR residuals | Gaussion Model | 1462 | 4453 | 67.2 | 71,100 | 0.896 |
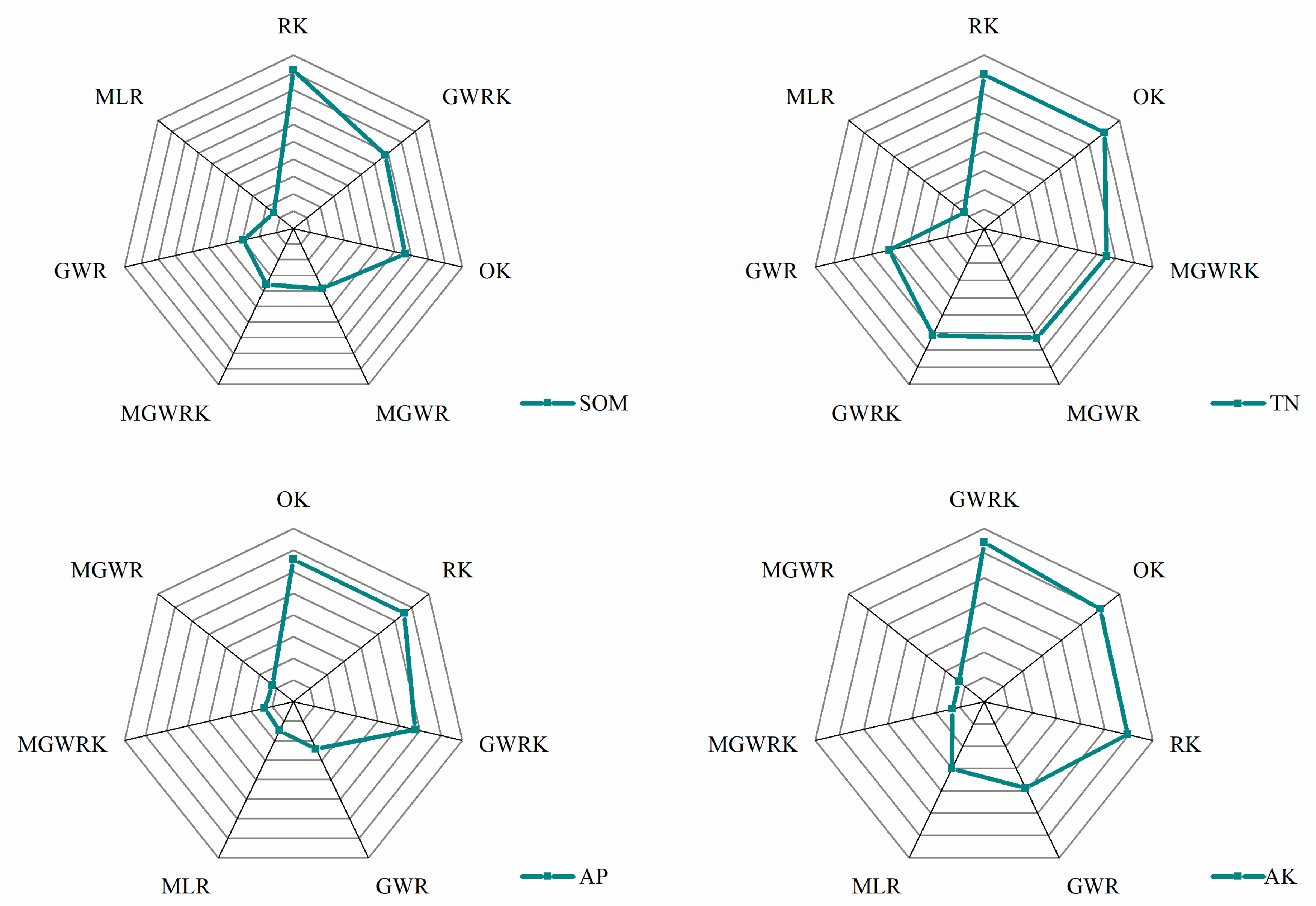
| Index | Methods | ME | MAE | RMSE | ACC |
|---|---|---|---|---|---|
| SOM | OK | 0.0232 | 4.4986 | 5.6669 | 0.8062 |
| MLR | −0.0001 | 5.6720 | 6.9999 | 0.7545 | |
| RK | 0.0160 | 3.9056 | 4.9402 | 0.8317 | |
| GWR | 0.0340 | 5.3505 | 6.6495 | 0.7696 | |
| GWRK | 0.0244 | 4.4597 | 5.6152 | 0.8079 | |
| MGWR | 0.1947 | 5.2317 | 6.6090 | 0.7785 | |
| MGWRK | 0.0186 | 5.2615 | 6.6687 | 0.7759 | |
| TN | OK | 0.0003 | 0.1832 | 0.2392 | 0.8450 |
| MLR | −0.0003 | 0.2234 | 0.2858 | 0.8117 | |
| RK | 0.0004 | 0.1831 | 0.2388 | 0.8450 | |
| GWR | −0.0019 | 0.1997 | 0.2598 | 0.8303 | |
| GWRK | 0.0010 | 0.1942 | 0.2530 | 0.8358 | |
| MGWR | −0.0007 | 0.1928 | 0.2512 | 0.8365 | |
| MGWRK | 0.0007 | 0.1922 | 0.2504 | 0.8377 | |
| AP | OK | −0.0048 | 3.6354 | 4.8464 | 0.6716 |
| MLR | 0.0000 | 4.6657 | 6.1906 | 0.5696 | |
| RK | −0.0069 | 3.6383 | 4.8454 | 0.6712 | |
| GWR | −0.0577 | 4.4876 | 5.9648 | 0.5883 | |
| GWRK | −0.0086 | 3.8141 | 5.0737 | 0.6557 | |
| MGWR | −0.0262 | 4.8092 | 6.3629 | 0.5645 | |
| MGWRK | −0.0137 | 4.8087 | 6.3818 | 0.5674 | |
| AK | OK | 0.0385 | 25.5836 | 33.9677 | 0.8200 |
| MLR | −0.0002 | 33.6282 | 43.9463 | 0.7601 | |
| RK | 0.0419 | 25.7632 | 34.1836 | 0.8188 | |
| GWR | 0.2043 | 31.4605 | 41.5619 | 0.7775 | |
| GWRK | 0.0508 | 24.3765 | 32.4621 | 0.8286 | |
| MGWR | −1.5545 | 39.4682 | 53.3011 | 0.7260 | |
| MGWRK | −0.1635 | 39.4764 | 53.5148 | 0.7264 |
| Model | RSS | AIC | AICC | R2 | Adjust R2 |
|---|---|---|---|---|---|
| MLR | 2347.743 | 7022.766 | 7024.853 | 0.075 | 0.072 |
| GWR | 2118.583 | 6939.399 | 6947.297 | 0.165 | 0.132 |
| MGWR | 2028.812 | 6816.744 | 6823.630 | 0.201 | 0.171 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, L.; Huang, M.; Zhang, W.; Qiao, L.; Wang, G.; Zhang, X. Comparative Study on Spatial Digital Mapping Methods of Soil Nutrients Based on Different Geospatial Technologies. Sustainability 2021, 13, 3270. https://doi.org/10.3390/su13063270
Gao L, Huang M, Zhang W, Qiao L, Wang G, Zhang X. Comparative Study on Spatial Digital Mapping Methods of Soil Nutrients Based on Different Geospatial Technologies. Sustainability. 2021; 13(6):3270. https://doi.org/10.3390/su13063270
Chicago/Turabian StyleGao, Li, Mingjing Huang, Wuping Zhang, Lei Qiao, Guofang Wang, and Xumeng Zhang. 2021. "Comparative Study on Spatial Digital Mapping Methods of Soil Nutrients Based on Different Geospatial Technologies" Sustainability 13, no. 6: 3270. https://doi.org/10.3390/su13063270
APA StyleGao, L., Huang, M., Zhang, W., Qiao, L., Wang, G., & Zhang, X. (2021). Comparative Study on Spatial Digital Mapping Methods of Soil Nutrients Based on Different Geospatial Technologies. Sustainability, 13(6), 3270. https://doi.org/10.3390/su13063270







