Abstract
Contamination of surface waters with pathogens as well as all diseases associated with such events are a significant concern worldwide. In recent decades, there has been a growing interest in developing analytical methods with good performance for the detection of this category of contaminants. The most important analytical methods applied for the determination of bacteria in waters are traditional ones (such as bacterial culturing methods, enzyme-linked immunoassay, polymerase chain reaction, and loop-mediated isothermal amplification) and advanced alternative methods (such as spectrometry, chromatography, capillary electrophoresis, surface-enhanced Raman scattering, and magnetic field-assisted and hyphenated techniques). In addition, optical and electrochemical sensors have gained much attention as essential alternatives for the conventional detection of bacteria. The large number of available methods have been materialized by many publications in this field aimed to ensure the control of water quality in water resources. This study represents a critical synthesis of the literature regarding the latest analytical methods covering comparative aspects of pathogen contamination of water resources. All these aspects are presented as representative examples, focusing on two important bacteria with essential implications on the health of the population, namely Pseudomonas aeruginosa and Escherichia coli.
1. Introduction
The safety of drinking water represents one of the major issues in today’s society. Water production and distribution need to meet some microbiological criteria to avoid becoming a serious health problem.
The quality of water has been thoroughly monitored in the last decades because the presence of different pathogens or elements in water could seriously pose issues regarding human and environmental health. Water contamination could lead to serious environmental, health, and implicit economic problems that could continue over the decades, and the impact of contaminated water, whether contaminated directly or indirectly via alimentary products, can affect generations.
An important issue at the international, national, regional, and local levels is access to safe drinking water, which is an issue of health and fundamental human rights. The importance of sanitation and access to drinking water could lead to economic benefits by reducing adverse effects and directly influencing overall healthcare costs [].
“Water is essential to sustain life, and a satisfactory (adequate, safe, and accessible) supply must be available to all” []. The World Health Organization (WHO) International Standards were published between 1993 and 1997 after the Guidelines for Drinking-water Quality first and second editions (1983–1984).
The WHO guidelines state that safe drinking water will not represent a risk for health over a lifetime of consumption, except for infants, young children, and the elderly living under unsanitary conditions.
Extra care must be taken in drinking water intended for medical use, such as renal dialysis and rinsing solution for contact lenses, or certain purposes in the food and pharmaceutical industries. For immunocompromised patients, boiling drinking water is advised so that even microorganisms that would typically not be of concern in drinking water can be avoided [].
The established regulations do not promote international standards for drinking water quality but encourage establishing national standards and regulations as a function of the national situation related to water resources and policies. Furthermore, there is no single strategy that can be applied globally. The same guideline underlined the fact that each country must evaluate its status and create a personalized (tailored to its needs) regulatory framework.
However, there are some basic and essential requirements for ensuring the safety of drinking water comprising health-based targets established by a competent health authority, adequate and properly managed systems (adequate infrastructure, proper monitoring, effective planning and management), and a system of independent surveillance.
One crucial health consequence is microbial contamination, and this type of contamination should be avoided. The most common bacteria that are associated with a high and moderate risk for human health are Escherichia coli (pathogenic and enterohemorrhagic) (E. coli), Legionella spp., Pseudomonas aeruginosa (P. aeruginosa), Salmonella typhi, Shigella spp., Vibrio cholerae, and Yersinia enterocolitica (see Table 1 for details). The majority have a moderate or high degree of multiplication and infectivity. To be specific, P. aeruginosa can multiply in water supplies and is resistant to chlorine but has a low risk of infection, the main route of infection being, in this case, the skin contact with immunosuppressed patients (elders, children, patients with burns/extensive wounds, or those with immunosuppressive therapy or AIDS). In the cases described above, if the water source has a significant number of waterborne pathogens, the risk is proportional, and the exposed regions, such as skin or mucous membranes (eye, nose, and throat), are gateways for systemic infections [].
To ensure the microbial safety of drinking water supplies, several barriers are used, such as protection of water resources, proper selection and operation of a series of treatment steps, and management of distribution systems (piped or otherwise) to preserve water quality. Avoiding or reducing the entry of pathogens into water resources and adapting methods to accurately detect pathogens are envisaged by these guidelines [].
About 750 million people in the 21st century lack access to potable water sources/infrastructures, and one-fifth of the world population inhabits areas facing water scarcity simultaneously with global water demand. Consequently, improving the policy coherence area is one of the top priorities in the water sector in the forthcoming period [].
The water contamination problem becomes more stringent in the case of calamities where the water source becomes compromised, and the population needs to be protected and supplied with potable water by the respective authorities. In addition, it is of major importance for military ops undergoing force tasks in uncharted territories and for overcrowded immigration camps where the conditions are poor and have a negative impact on human life in general.
Shallow lakes are the most susceptible to contamination, where alteration depends on the water temperature and alkaline pH, but sewage discharge determined by heavy rainfall has been shown to spread waterborne pathogens within surface waters.
The main safety measures against microbiological hazards should ensure primary sanitation and safe drinking water infrastructure; however, dramatic accidents have occurred in recent years:
- Contamination of groundwater determined outbreaks in Walkerton, Canada in 2000 with Campylobacter and E. coli, in Southern Finland in 2001, and Ohio, the USA in 2004 with Campylobacter. The contamination of surface water via heavy rain-led mud, for which protections were inadequate after heavy rainfall conditions, was mentioned in Spain in 2002 with Shigella and in Oregon, USA in 2005 with Campylobacter and E. coli.
- Outbreaks generated by deficiencies in the disinfection treatment procedures in water treatment plants and from failures/malfunctions in the distribution network systems are more frequent, with Campylobacter (Gourdon, France in 2000; South Wales, the UK in 2000; New Zealand in 2000 and 2012; Spain in 2001; Indiana, the USA in 2006; Ohio, the USA in 2004; Koge, Denmark in 2007 and 2010; Nokia, Finland in 2007; Zurich, Switzerland in 2008; and Utah, the USA in 2010), Shigella (New Zealand in 2004 and Valencia d’Aneu, Spain in 2006), Salmonella (Montana, USA in 2004 and Colorado, USA in 2008), and E. coli (Ahus, Sweden in 2010; Ohio, the USA in 2000; Darcy le Fort, France in 2001; Koge, Denmark in 2007; and Vuorela, Finland in 2012). Between contamination from animal barns, the filtering of wastewater in drinking water systems due to human/technical errors, and broken pipes, the causes of drinking water are multiple and difficult to assess in a short timespan to avoid contamination of the targeted population with all the implied consequences [].
The methods for detecting waterborne pathogens need to be adapted to the necessities of the situation and community. However, as a general requirement, they need to be specific, sensitive, rapid, and easy to use without having advanced knowledge. A quick solution can be represented by rapid tests that can be implemented as point-of-use (POU) or point-of-care (POC) devices and allow rapid and specific detection without any complicated sampling or testing protocol depending on the selected technology. Testing the microbiological quality of water and unravelling the presence of waterborne pathogens is an essential goal in this domain.
An important example of rapid testing, currently highly mediatized, is represented by a rapid test for the early detection of COVID-19 antigens that replaced the classic polymerase chain reaction (PCR) assessment. Based on affinity reactions, with high sensitivity and following a simple protocol, the rapid antigen test allows the detection of the antigen at home and provides a credible response in 10–20 min. This fact changed the testing protocols and enabled a larger perspective of the real infection rate in real-time.
The field of printed wearable sensors has seen enormous progress in the last decade, as determined by previewed performance devices and a broad range of applications. In particular, they can offer real-time information regarding either human body parameters or the monitoring of hazardous chemicals, with applications in security, defense, and environmental monitoring [,]. Initially impossible to achieve, this research area has focused on designing different formats that are important for the development of POC/POU devices based on innovations in materials engineering, platforms based on nanomaterials, polymers, or other composites.
Researchers have channeled their efforts into designing wearable devices embedded in textiles, medical gloves, bandages, and mouthguards, which were previously impossible to achieve because they can play a vital role in the field of personalized POC devices [,,].
This review aims to present the most recent analytical technologies developed to detect various bacteria in the surface- and wastewater, focusing on studies that targeted E. coli and P. aeruginosa. The literature study was done using the well-known scientific databases (ScienceDirect, Scopus, SpringerLink, PubMed), using as criteria research papers and reviews published in the last 10 years on the detection of waterborne pathogens. The next step was to limit the target bacteria to the two previously mentioned and the real samples considered to drinking water, tap water, rivers, lakes, and wastewater. The main topic of this review is schematically illustrated in Figure 1.
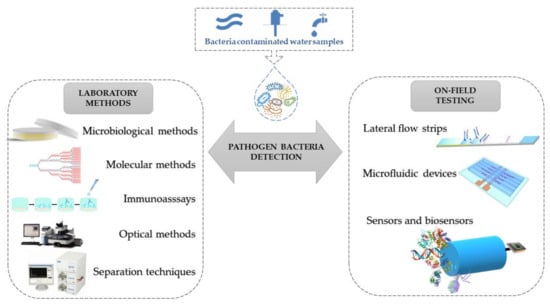
Figure 1.
Schematic representation of the analytical methods applied for the detection of bacteria in surface- and wastewaters.
2. Waterborne Bacteria Short Overview
Waterborne illnesses are conditions caused by recreational or drinking water contaminated with disease-causing microbes or pathogenic microorganisms. These diseases can be contracted while bathing in, washing with, or drinking contaminated water. Other important sources of contamination and spread of diseases related to waterborne pathogens can be the consumption of contaminated food or beverages, contact with animals or their environment, and person-to-person transmission. Waterborne illnesses can cause various symptoms. While diarrhea and vomiting are the most commonly reported symptoms of waterborne illnesses, other symptoms can include skin, ear, respiratory, or eye problems.
Waterborne diseases are a global burden estimated to cause more than 2.2 million deaths per year and increase cases of illness every day, including diarrhea, gastrointestinal diseases, and systemic diseases []. Approximately 65% of the victims are children. Worldwide, for the healthcare of these diseases, an economic loss of nearly 12 billion US dollars per year is estimated. Waterborne infections are caused by ingestion, inhalation, or contact with contaminated water by various infectious agents, including bacteria, viruses, protozoa, and helminths []. It is estimated that 3.2% of global deaths are attributable to unsafe water consumption caused by poor sanitation and hygiene []. The United Nations has identified improving water quality as one of the eight Millennium Development Goals (MDGs), and its target was to reduce the number of people without access to safe water by 50% in 2015 []. The WHO has reported that improving water quality can reduce the global disease burden by approximately 4%. Thus, there is an urgent need to undertake all possible efforts to achieve this goal.
Commonly recognized waterborne infections are: cryptosporidiosis (Cryptosporidium); cyclosporiasis (Cyclospora spp.); E. coli O157:H7 infection and hemolytic uremic syndrome (HUS) (both caused by E. coli O157); giardiasis (Giardia), harmful algal blooms (HABs); hot tub rash (also known as Pseudomonas dermatitis/folliculitis); legionellosis (Legionella); primary amebic meningoencephalitis (Naegleria fowleri); norovirus infection (Norwalk virus, calicivirus, viral gastroenteritis); shigellosis (Shigella); swimmer’s ear (otitis externa); and swimmer’s itch (cercarial dermatitis). According to WHO drinking water guidelines, more aspects related to the most common waterborne pathogen bacteria are presented in Table 1 [].

Table 1.
Most common waterborne pathogen bacteria, according to WHO Drinking Water Guidelines [].
Table 1.
Most common waterborne pathogen bacteria, according to WHO Drinking Water Guidelines [].
| Pathogen Bacteria | Taxonomic Family | Localization | Disease | Symptoms | Infectivity | Resistance to Chlorine [] |
|---|---|---|---|---|---|---|
| Burkholderia pseudomallei | Burkholderiaceae | Southeast Asia and Northern Australia [] | Melioidosis [] | Septic shock, pulmonary infection, acute suppurative parotitis, prostatic abscesses, brainstem encephalitis | Low | Low [] |
| Campylobacter jejuni, C. coli | Campylobacteraceae | Worldwide (increasing incidence in North America, Europe, Australia, Asia) | Campylobacteriosis [] | Gastroenteritis, extraintestinal infection, postinfection complications (reactive arthritis, Guillain–Barré syndrome, irritable bowel syndrome) | Moderate | Low |
| E. coli—pathogenic (ETEC, EPEC, EAEC, EIEC, DAEC) | Enterobacteriaceae | Worldwide | Gastroenteritis [] | Acute and chronic diarrhea, chronic gut inflammation | Low | Low |
| E. coli O157:H7—enterohemorrhagic | Enterobacteriaceae | Worldwide | Gastroenteritis, hemolytic uremia [] | Bloody diarrhea, hemolytic uremic syndrome | High | Low |
| Legionella pneumophila | Legionellaceae | Worldwide | Legionnaires’ disease [] | Fever, nonproductive cough, headache, muscle pain, dyspnea, diarrhea, delirium | Moderate | Low |
| Mycobacterium avium complex (non-tuberculous) | Mycobacteriaceae | Worldwide | Pulmonary disease, skin infection [] | Dyspnea, cough, bronchiectasis, lymphadenitis | Low | High |
| Pseudomonas aeruginosa | Pseudomonadaceae | Worldwide | Pulmonary disease, skin infection [] | Acute, chronic lung infection, soft tissue infections | Low [] | Moderate |
| Salmonella typhi | Enterobacteriaceae | High incidence in southeast Asia, sub-Saharan Africa | Typhoid fever [] | Fever, headache, muscle pain, constipation, diarrhea | Low | Low |
| Salmonella enterica | Enterobacteriaceae | Highest incidence in sub-Saharan Africa | Salmonellosis [] | Diarrheal disease, bacteremia, meningitis | Low | Low |
| Shigella spp. | Enterobacteriaceae | Globally | Shigellosis [] | Acute watery diarrhea, dysentery, bloody stools, fever | High | Low |
| Vibrio cholera | Vibrionaceae | Endemic in Asia and Africa | Cholera [] | Acute watery diarrhea, vomiting, hypotensive shock | Low | Low |
3. Methods for Bacterial Detection
In order to avoid contamination of the environment with pathogenic bacteria or with bacteria that can become pathogenic after reaching living organisms, their detection must be done quickly, in the field, with high sensitivity and specificity. The quantitative evaluation of bacteria or the assessment of microbial content of water samples present a limiting factor, especially in the case of drinking water, namely the often small number of such microorganisms present. Thus, it is important to mention that most of the analytical techniques applied to detect and quantify bacteria present in water samples require laborious preconcentration steps performed before detection. These preconcentration steps are limited when rapid, decentralized on-field detection is desired and may be necessary when the efficient concentration of a certain target microorganism fails, even before being subjected to the detection system.
The most common approaches for the detection of microorganisms are based on various bacterial culturing methods (e.g., agar plate cultures and other liquid cultivation techniques), enzyme-linked immunosorbent assay (ELISA) based on specific antigen–antibody interactions, PCR technology, and loop-mediated isothermal amplification (LAMP) [,].
These traditional detection methods have a significant impact and contribute to the detection of pathogenic bacteria as well as the prevention of water contamination. Nevertheless, they present some disadvantages, the most important of which is related to the analysis duration, which ranges from a few hours to a few days. A comparative table presenting the main advantages and disadvantages of different methods applied in the detection of bacteria is provided at the end of this section (see Table 2).
3.1. Bacterial Culturing Methods
Bacterial culturing methods involve the use of detection strategies that are time-consuming and labor-intensive. These detection strategies are usually performed on colonies, involve several tests performed using biochemical methods, and require approximately three days or even longer to complete the analysis and obtain the result. These detection methods have been intensively applied for bacterial detection using three different techniques: multiple tube fermentation (MTF), membrane filtration, and bacterial cell culture. The results obtained in all strategies are expressed as the most probable number (MPN) or as colony-forming units (CFU), both of which provide a statistical estimation of the concentration of microorganisms in a sample [].
3.2. ELISA
ELISA tests can be applied to identify and quantify a plethora of bacteria, but they do not show sensitivity, and antigen cross-reactions are likely to occur. ELISA requires a shorter detection time than cell culture-based methods; however, the synthesis of suitable antibodies for the envisaged target bacteria is difficult and time-consuming because it requires living organisms as hosts. The principle of ELISA is based on the immobilization of antigens on a substrate containing the respective antibodies. Detection is provided through an enzyme-labeled antigen or antibody via luminescence after amplification of the signal due to enzymatic catalysis []. Several studies have reported the detection of pathogenic bacteria in water samples, mainly for E. coli O157:H7, which is one of the most dangerous foodborne pathogenic bacteria worldwide. An innovative, fast, and low-cost paper-based enzyme-linked immunosorbent assay (p-ELISA) has been developed, highly sensitive and specific for on-site detection of E. coli. The limit of detection was 1 × 104 CFU/mL for less than 3 h, and 5 μL of the sample was necessary for bacterial detection [].
3.3. LAMP and PCR
Both the LAMP- and PCR-based methods are molecular biological technologies. These methods exhibit high specificity and require low volumes of samples; however, complex pre-steps for sample preparation are involved, and highly qualified and skilled individuals must operate the equipment. The strategies for bacterial detection applied in the case of PCR require the extraction of bacterial DNA and the use of a thermocycler, which is unsuitable for on-field testing [,].
PCR is based on the replication of a DNA sample as an amplification step necessary to reach a detectable amount of target. Using this technique, copies of trace amounts of DNA sequences are exponentially amplified in a succession of steps involving temperature changes. PCR has been chosen as a suitable technique for bacterial detection in water samples during the last decade. For example, E. coli O157:H7 was detected in artificially contaminated irrigation water by qPCR and nanopore sequencing using Shiga toxin-producing E. coli (STEC). This detection strategy also involves a 24-h step after artificial contamination of the samples to enrich the irrigation water sample with bacteria. A limit of detection of 30 CFU was obtained via the proof-of-concept optimized qPCR method (equivalent to 105 CFU/mL in the enriched water sample) []. P. aeruginosa species were isolated and tested from abattoir wastewater and surface water in Eastern Cape, South Africa, by real-time PCR (rPCR), and their antibiotic resistance genes were identified. In this study, the real samples were filtered through a membrane, cultured on a suitable medium, and the molecular characterization of the isolates was confirmed using rPCR. It was found that 55.6% of P. aeruginosa species found in this sample exhibited multiple antibiotic resistance profiles and were labeled as multidrug-resistant (MDR) strands. This study concluded that the bacterial isolates obtained from nonclinical samples are resistant to the majority of first-choice antipseudomonal drugs, which can be an important cause of concern for the authorities [].
An alternative to PCR, recombinase polymerase amplification (RPA), was used in combination with a lateral flow assay to evaluate E. coli O157:H7 in water and food samples. Several bacterial genes were used as targets, such as rfbE, fliC, and stx, and excellent detection limits and specificity were obtained within approximately 38 min with reduced handling and simple equipment (8 min at temperatures between 37 and 42 °C and 5–30 min of incubation for amplification of the target) [].
The impact of DNA extraction and primer choice on microbial evolution and the detection limit of the 16S rRNA gene of E. coli were determined. The PowerWater DNA Isolation Kit and the FastDNA SPIN Kit for Soil in combination with PCR amplification were used. The limit of detection was successfully determined for pyrogen-free water samples spiked with 101–106 E. coli cells/mL [].
A portable nanopore sequencing technology (16s RNA) was used to map the diversity of bacteria and assess the presence of pathogen species in surface water from a river in Cambridge, UK. The optimized experimental procedure can be used to characterize the hydrological core microbiome, acquire relevant data from sewage discharge, and draw taxonomic pathogen maps for the studied locations [].
A multifunctional micropattern array capable of isolating individual cells and lysing and extracting the DNA directly for quantification of E. coli O157:H7, Salmonella enteritidis, and Staphylococcus aureus from aqueous samples was fabricated by the deposition of a copolymer film on a chip surface via a chemical vapor deposition process combined with microcontact printing. The use of this multifunctional micropattern array in the microfluidic chip enabled the signal amplification and detection of DNA templates obtained for a dynamic range of 0.01–2 ng/μL and provides a novel multifunctional strategy for fast and simple evaluation of nucleic acids in samples [].
An alternative method to PCR-based methods, LAMP, has been found to be an innovative gene amplification technology for bacterial testing.
LAMP can be characterized as nucleic amplification under isothermal conditions. Furthermore, the obtained results can be analyzed through the color change of a dye such as SYBR Green I. Another strategy applied for LAMP products is their indirect detection via the turbidity produced in the reaction mixture after magnesium pyrophosphate formation. Because the amount of white precipitate can be correlated with the amount of DNA produced in the primary reaction, the LAMP reaction can be monitored by real-time turbidity measurement [].
Considering all these aspects, sustained efforts have been made in the last decade for the development of advanced alternative methods, such as spectrometric and electrochemical methods.
The detection of bacteria via spectrometric and electrochemical methods was performed mainly indirectly based on the signals obtained for some compounds, the presence of which in the detection environment can be correlated with the presence of bacteria. These compounds are known as biomarkers or bacterial markers.
Another widely used strategy is the comparison of chromatograms as fingerprints for target bacteria. It is well known that the development and multiplication of bacteria is a process accompanied by the release of some compounds into the environment by bacteria, among which are volatile compounds that can be subjected to profiling and used for their speciation and detection. However, there are uncertainties and concerns about the reliability of biomarker-based and fingerprint analysis of bacteria, which are usually prone to show variations in terms of the conditions of bacterial growth, cell age, and the origin from which the samples were collected. These variations cannot be controlled; thus, the strategies based on the use of fingerprinting do not have many applications reported thus far [].
3.4. Chromatography
Chromatography methods require high technical complexity, such as the large dimensions of the equipment, high consumption, and the need for low-pressure and thermostatting systems, which can only be provided in the laboratory; therefore, they do not qualify them for on-field testing and monitoring. A specific and fast method was developed for the detection of E. coli based on GC–differential mobility spectrometry (GC–DMS) and the interaction of bacteria with o-nitrophenyl-β-D-galactopyranoside. In this case, the required equipment was relatively small and portable, namely a complex gas detector that requires a low-power source, a GC-type column [].
Water supply biofilms have a high potential for waterborne diseases; one of the most common bacteria known to form colonies in water supply pipelines is Pseudomonas sp. In a study on these biofilms, metabolomic techniques were applied. A proof-of-concept application was obtained after coupling liquid chromatography with time-of-flight mass spectrometry (LC–TOF-MS) to detect three samples of P. putida. The evaluation of the samples in the +ESI and −ESI modes allowed the detection of 887 and 1789 metabolites, respectively. Metabolite features were then subjected to the Metabolomics Standard Initiative (MSI), and promising results were obtained [].
3.5. Capillary Electrophoresis
Capillary electrophoresis (CE) is an important and highly efficient method for identifying and separating charged compounds that provide low electrolyte consumption and low sample amount and usually does not require laborious steps of processing the sample before separation. CE could offer relevant insights into the bacterial metabolome; given the ionic character of many molecules belonging to the bacterial metabolism and cellular structures, such as peptides, amino acids, carboxylic acids, nucleotides, and lipopolysaccharides, CE appears to be the most suitable method for the investigation of the bacterial metabolome. Bacteria are also charged microorganisms, thus having electrophoretic mobility and being suitable for identification and quantification by CE. Recently, several studies have been published on some important aspects such as CE use for the evaluation of biological processes, optimization of bacterial growth process, detection of some metabolites, and purification and identification of bacterial endotoxins in various samples [].
Several recently published studies have referred to the use of CE for the investigation of the bacterial metabolome through the monitoring of processes that take place in the extra- and intracellular environment, the detection and quantification of bacterial metabolites or endotoxins in various samples, and the study of target and non-targeted fingerprinting of microbiota in biological samples [,]. Although this analytical method has significant advantages, it also has disadvantages, mainly related to the low repeatability of retention times, low sensitivity, and the possible adhesion of basic compounds to the capillary walls; these disadvantages make it impossible to use this method for biological sample investigation. A possible solution to these shortcomings was found by using some pre-concentration steps (e.g., extraction of metabolites, enrichment of sample in target analytes, etc.) as well as highly sensitive detection strategies such as mass spectrometry (MS) and laser-induced fluorescence (LIF) [].
3.6. Hyphenated Methods
A group of sophisticated bacterial detection technologies that require MS detection, the so-called hyphenated methods have also been implemented to date. Thus, MS coupled with technologies such as matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) allows the detection of biocomponents via ions generated during the analysis. Electrospray ionization mass spectroscopy (ESI-MS) is another method that uses MS. In this case, ESI is responsible for ion production as an aerosol and microarray testing to investigate the target biomolecules []. These technologies produce rapid and accurate results for analysis and provide information regarding microbial identification and detection in various complex samples such as body fluids (e.g., blood, serum, and urine) and environmental samples (e.g., surface water and wastewater).
Legionella spp. were identified in drinking water in Germany using a MALDI-TOF MS Biotyper system, and the results are promising given the use of this detection strategy as an alternative identification method to assess water quality []. The same method was recently proposed in another study for the clinical and environmental evaluation of the same bacteria. The results obtained after the MALDI-TOF assay were also compared with the standard culture technique and macrophage infectivity potentiator gene sequencing. The proposed method allowed for the correct identification of approximately 45% of Legionella anisa, Legionella rubrilucens, Legionella feeleii, and Legionella jordanis isolates, in good agreement with the control detection results [].
MALDI-TOF MS also proved useful for rapid and accurate (high concordance with 16S rRNA sequencing) coliform identification in wastewater, river water, and groundwater. The results were validated via 16S rRNA gene sequence analysis. Accurate detection of bacteria at the genus level was obtained, and the concordance rate of bacterial identification for the 100 isolates obtained by MALDI-TOF MS analysis and 16S rRNA gene sequence detection in different real samples such as sewage, river water, and groundwater were 96%, 74%, and 62%, respectively [].
Accidental contamination with pathogenic bacteria through ballast water in the marine environment is another critical issue that could be addressed by MALDI-TOF MS testing because Vibrio species, enterococci, and coliforms were found in this environment. Seawater samples from the North Sea were evaluated, and 36 isolates were identified at the genus level using MALDI-TOF MS. Only the opportunistic pathogenic P. aeruginosa was found, and the MALDI-TOF MS results were confirmed by 16S rRNA gene analysis []. MALDI-TOF MS with Biotyper software was also able to classify bacteria from Arctic water []; however, there is still a need to improve the available spectral databases to identify environmental bacteria using this method [,].
A different approach utilized ultra-performance liquid chromatography/electrospray ionization-mass spectrometry (UPLC/ESI-MS) to quantify purine biomarkers released by S. aureus and E. coli in water. Using this method, adenine and hypoxanthine were dramatically released into water from a single S. aureus and E. coli cell within an hour [].
3.7. Magnetic Field-Assisted Methods
Magnetic field-assisted methods are simple and cost-effective detection strategies that do not require the use of sophisticated equipment and have found important applications in the field of pathogen bacterial detection; they are related to the use of antibody-functionalized magnetic nanoparticles (MNPs). Detection with the naked eye is possible in this case and is based on the formation of colored bacteria–magnetic nanoparticle complexes that are further immersed over a filter membrane. A selective preconcentration of the complexes on the membrane and naked eye identification of the bacteria present in the samples are enabled. This detection strategy is often disturbed by high background noise and thus presents poor detection sensitivity. A possible solution to this problem is magnetophoretic chromatography, an analytical method in which a liquid-type filter is used to efficiently separate the complexes. This strategy involves the formation of two liquid layers containing free MNPs and bacteria–MNP complexes, which are added to a polymer solution. The separation of free MNPs is achieved using an external magnet that blocks these particles at the interface between the two layers of liquid with different viscosities. Although magnetophoretic chromatography is easy to use and time- and cost-effective, it often leads to false-negative results. A colorimetric method based on monoclonal antibody-conjugated platinum-coated magnetic nanoparticle clusters (Pt/MNCs) and magnetophoretic chromatography was proposed to detect E. coli (O157:H7) pathogenic bacteria in milk. The colorimetric detection was based on the catalytic oxidation of tetramethylbenzidine by Pt, which causes a color change and enables the naked-eye detection of E. coli up to a limit of detection of 10 CFU/mL, with the total duration of the process being 30 min. Transmission electron microscopy (TEM) images of the Pt/MNCs with and without E. coli bacteria are shown in Figure 2 [].
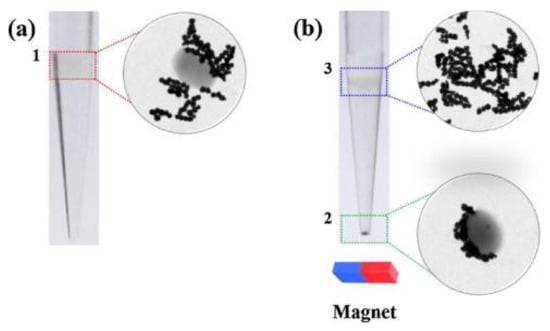
Figure 2.
Optical and TEM images of a Pt/MNC–EC bacterium and free Pt/MNCs (a) before and (b) after the magnetophoretic chromatography. Reprinted with permission from Elsevier [].
Microfluidic systems with various architectures have been employed for the construction of detection instruments that target pathogenic bacteria. Thus, the development of a plastic 3D microfluidic magnetic pre-concentrator via printing technology and antibody-conjugated MNPs was assessed for the selective preconcentration and detection of E. coli O157:H7 in 100 mL within 1 h. The detection limit was 10 CFU/mL in blood, which demonstrates the feasibility of the preconcentration method to be used together with existing bacterial detection systems [].
E. coli O157:H7 bacteria were separated and detected using a microfluidic impedance biosensor combined with immune MNPs. MNPs were functionalized with streptavidin and then conjugated with biotinylated polyclonal antibodies to form immune particles for bacterial separation from the background to form MNP–bacterial complexes. In the next step, these complexes were conjugated with gold nanoparticles (AuNPs) labeled with urease and oligonucleotides against E. coli O157:H7 (Figure 3). A decrease in the impedance spectra was observed upon hydrolysis of urea into ammonium carbonate catalyzed by these complexes. A good correlation between impedance modification and bacterial concentration was obtained (LOD = 12 CFU/mL) [].
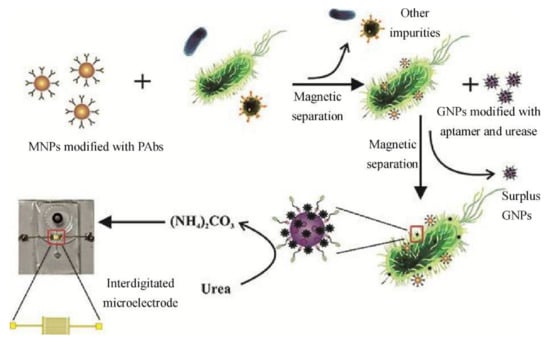
Figure 3.
The principle of the microfluidic biosensor proposed for the evaluation of E. coli O157:H7. Reprinted with permission from Elsevier [].
A biosensor for E. coli O157:H7 was developed based on the optical response of an MNP-specific peptide probe upon bacterial protease action. The detection principle relied on the intensification of color resulting from the dissociation of the self-assembled monolayer, which was correlated with the E. coli O157:H7 level. This sensor demonstrated high sensitivity and applicability and a limit of detection of 12 CFU/mL in broth samples [].
An innovative method was found to selectively and sensitively (LOD = 102 CFU/100 mL) isolate E. coli cells in environmental water samples with no enrichment using gold-coated magnetic microdiscs functionalized with aptamers. Fluorescent markers such as SYTO9, propidium iodide, and carbon quantum dots were immobilized on the microdiscs; only one step was required for both detection and viability tests for E. coli in agricultural water in less than 45 min [].
3.8. Surface-Enhanced Raman Scattering
Surface-enhanced Raman scattering (SERS) is a newer method for the detection, characterization, and quantification of bacteria that comprises an ultrasensitive vibrational spectroscopic technique based on Raman scattering and nanotechnology. Raman spectroscopy can characterize the molecular vibration of the target compound, providing its fingerprint spectrum, but it fails to perform sensitive detection when a detrimental defect appears [].

Table 2.
Comparative table presenting advantages, disadvantages, and other characteristics of analytical methods for pathogen detection.
Table 2.
Comparative table presenting advantages, disadvantages, and other characteristics of analytical methods for pathogen detection.
| Detection Method | Type of Method | Advantages | Disadvantages | Analysis Duration | Estimated Cost * | On-Site Testing | Ref. |
|---|---|---|---|---|---|---|---|
| Microbiological culturing | Conventional | High sensitivity, accuracy, can provide diagnosis of acute infections | Intensive labor, lengthy analysis, requires sterile laboratory conditions | Several days (48–72 h) | Low | No | [] |
| ELISA | Conventional | High specificity and sensitivity | Requires enrichment for quantification, expensive plate reader | >8 h | High | No | [] |
| Paper-based | Lightweight, disposable, biodegradable, chemically compatible, low LOD | 175 min/sample | Low | Yes | |||
| PCR | Conventional | High specificity and sensitivity | Requires sterile laboratory conditions, costly reagents, and highly trained personnel; false negatives due to sample cross-contamination; inhibition of amplification reaction by matrix sample compounds; challenges in differentiating viable from nonviable cells | 1–4 h | High | No | [,] |
| MS | MALDI-TOF | High sensitivity, rapid, robust, high throughput | Lower resolution spectrometer, incompatibility with tandem analysis, lack of sufficient reference spectra | Several hours | High | No | [] |
| Optical | SPR sensors | High-sensitivity, rapid, label-free, real-time analysis with reproducible results | Chance of false results due to fluctuations in refractive index with the temperature or composition of the sample, nonspecific interactions from nontarget or structurally similar molecules to the sensor surface | <30 min | High | No | [] |
| SERS sensors | High sensitivity and high spectra resolution, the possibility of multiplexed detection, label-free SERS also helps in the differentiation of viable and nonviable bacterial cells | Limited usage of label-based SERS for in situ and high-throughput recognition of pathogens because of increased requirements of reactant volumes, preparation steps, and analytical time | 10 min–2 h | High | No | [] | |
| CL sensors | Easy device handling, flexibility, specificity, sensitivity, rapidity, wide dynamic range, relatively simple equipment (no sophisticated optics with excitation source required), low instrumentation costs | Requirement of chemiluminescent labels | A few minutes to a few hours | Low | Yes | [] | |
| Electrochemistry | Simplicity, specificity, low detection limit, ease of use, real-time measurement, multitarget testing and automation, portability, miniaturization, rapid detection | A few minutes to a few hours | Low | Yes | [] | ||
| Paper-based sensors | Low cost, single-use, portable, environmentally friendly | Low | Yes | [] |
Note: * The cost was estimated based on required equipment and reagents.
4. Sensors
Sensors and biosensors are important alternatives to conventional detection techniques for bacteria. They allow real-time analysis of the presence of bacteria and can be applied to any type of real sample, such as biological fluids, food, and environmental samples []. Sensors are devices capable of registering chemical information from a probe and converting it with the help of a transducer into a measurable analytical signal (e.g., optical, electrochemical, electrical, or piezoelectric) []. A biosensor is a sensing device based on a physicochemical detector that converts a chemical process that incorporates a biological or biomimetic material. Thus, a biosensor consists of two main parts: a biological component (e.g., enzyme, antibody, cell, tissue, microbial cells, organelles, or nucleic acid) and a transducer, or an electronic part that transmits and detects signals through optical, electrochemical, piezoelectric, or thermometric methods. Biomimetic compounds, such as molecularly imprinted polymers or synthetic catalysts, are often used to replace recognition elements. The interaction between these two units is obtained in the form of an analytical signal that can be easily quantified []. Biosensors can be classified according to their field of application, e.g., clinical biosensors, food biosensors, biosensors for forensic and military applications, biosensors for water safety, and environmental monitoring biosensors.
4.1. Optical Sensors
Optical sensing of waterborne pathogens represents a sensitive and selective method for accessible in situ analysis based on spectroscopic measurements of absorption, fluorescence, phosphorescence, refraction, and dispersion []. Optical methods such as fluorescence and chemiluminescence spectroscopy, surface plasmon resonance (SPR), and SERS are mostly used for bacterial sensing.
In the era of remarkable advances, microbial culture methods, although still very common, have started to become unpopular as they may take up to several days to identify a specific pathogen for most bacterial strains. The combination of “on-chip” microbial culture and SPR detection allowed faster qualitative and quantitative determination of Salmonella enterica serovar Enteritidis, Streptococcus pneumoniae, and E. coli O157:H7 with an LOD of 2.8 ± 19.6 CFU/mL []. It was demonstrated that the total bacterial and abiotic particles of 0.77–3 μm could be counted in less than 10 min by a 3D image recognition optical sensor with a resolution of 1.6 × 102−1.5 × 106 particles/mL []. Another time-effective approach (20 min) reported the use of T4 bacteriophages to determine the presence of 103 CFU/mL E. coli O157:H7 by SPR [].
Petrovszki et al. [] developed label-free optical microsystem-based dielectrophoretic surface electrodes, a rib waveguide, and a microfluidic channel for E. coli detection. The optimal scattered light pattern, magnification, and frequency utilized in the process of dielectrophoretic cell collection allowed fast and sensitive detection as low as 102 CFU/mL.
Functionalized metallic nanoparticles may also mediate bacterial detection. The electro-optical features of gold nanorods (AuNRs) functionalized with sugars induced photoablation when the targeted bacteria, E. coli, adhered to the modified AuNRs []. In a relevant study, the changes were examined by both spectrophotometric and microscopic techniques, specificity was tested against P. aeruginosa and lectin, and promising results were obtained.
Recently, porous silicon (PSi) was introduced in the spotlight of biosensor development because of its unique optical and chemical features []. Lectins are likely to specifically interact with carbohydrates and may represent a cost-effective alternative to antibodies. Lectin-conjugated PSi-based biosensors were developed for E. coli and S. aureus using reflectometric interference Fourier transform spectroscopy (RIFTS). The authors revealed that lectin affinity changed depending on the type of bacteria: Gram-positive and Gram-negative bacteria were preferentially and sensitively detected in the presence of wheat germ agglutinin and concanavalin A lectins, respectively []. Another interesting approach is the ability to distinguish between Gram-positive and Gram-negative bacteria due to the affinity of the strands to a monodisperse liquid crystal (LC) emulsion droplet that changes conformation from bipolar to radial when in contact with E. coli (Gram-negative). The method was also successfully tested against a virus (A/NWS/Tokyo/67) [].
In addition, the use of MNPs may significantly reduce the LOD to 102–106 cells/mL through magnetic separation. Verbarg et al. [] developed an optical in-built system that performs immunomagnetic separation and sample processing for the simultaneous detection of E. coli O157:H7, Salmonella common structural antigen, Listeria sp. and Shigella sp.
Refractive index sensors, namely SpectroSens™ chips containing high-precision planar Bragg gratings, were used for the selective determination of a plethora of biological targets, including E. coli, employing an antibody-antigen affinity reaction []. Upon optical sensing over the immunoaffinity complex, an increase in the wavelength of light reflected from the Bragg grating was observed.
Fluorescent labeling experiments and RIFTS were performed on a hybrid nanomaterial composed of a Psi optical transducer and a polyacrylamide hydrogel conjugated with IgG antibodies. Exposure of these modified hybrids to E. coli K12 resulted in bacterial entrapment at the biosensor surface and enabled detection within several minutes [].
Another notable approach used an antibody-modified oxidized porous silicon (PSi) thin film to selectively bind E. coli and evaluate the changes in the reflectivity spectra. The analysis was performed in real-time when exposed to water samples from food production pipelines, with no pre-enrichment or prior processing steps []. Yang et al. used carboxyl-functionalized graphene quantum dots (cf-GQDs) instead of a Psi film and reported that the fluorescent sensor selectively detected 102 CFU/mL E. coli O157:H7 in water and food samples []. A chemiluminescent (CL) sandwich immunoassay was developed for E. coli O157:H7 using an enzyme-labeled (glucose oxidase) secondary antibody detection system. The enzymatically catalyzed product, H2O2, undergoes laccase activity over luminol [].
Polyaniline changes color upon H+ and e− depletion []. Electrochemically deposited polyaniline at an indium tin oxide screen-printed electrode was used as an optical readout and antibody immobilization platform for the colorimetric detection of E. coli. When a constant potential was applied throughout the electrochemical cell, the color of the PANI changed according to its redox state. In contrast, in the presence of E. coli, different electrochromic responses were obtained due to the increased resistance in the circuit. The LOD calculated using ImageJ software for data analysis was one order of magnitude lower than that determined by the naked eye (102 CFU/mL) [].
Functionalization of poly(carboxybetaine acrylamide) and immobilization of specific antibodies for E. coli O157:H7 and Salmonella sp., respectively, on a gold SPR surface resulted in sensitive detection down to 17 CFU/mL for E. coli and 11.7 × 103 CFU/mL for Salmonella sp. []. However, using aptamers instead of antibodies, Wu et al. demonstrated good sensitivity against E. coli O157:H7 and S. typhimurium. The approach consisted of a colorimetric assay based on the optical properties of AuNPs functionalized with aptamers that entrap the target at the nanoparticle surface; thus, aggregation of AuNPs occurs upon aptamer–target interaction, which leads to a visible color change from red to purple []. Aptamers generally show superior features over antibodies []; however, AuNPs are widely used for bacterial detection in different real samples, including wastewater []. Hence, gold and silver nanoparticles are noticeable substrates for SERS optical pathogen biosensors [,,]. Another aptasensing approach was realized by Yildirim et al. based on a fluorescent-labeled specific aptamer for indirect determination of the E. coli O157:H7 strain in wastewater samples []. Hybridization with a complementary DNA strand was further realized after target interaction; thus, the higher the fluorescent signal, the lower the levels of E. coli found in the samples.
4.2. Electrochemical Sensors
Electrochemical biosensors are based on the modifications observed in electrical parameters such as current and potential, correlated with the interaction between the sensor and sample, and have gained an important area of research. Electrochemical biosensors are classified into different categories according to the parameters that are important for detection: the current signal in amperometry, impedance in electrochemical impedance spectroscopy, and potential in potentiometry. Electrochemical detection strategies have important advantages, such as low cost, fast response, good sensitivity, and limited selectivity. The shortcomings related to the reduced selectivity can be solved using biological components or biomimetic elements, thus justifying the use of biosensors in this category of detection methods. Among the various types of electrochemical biosensing approaches, amperometric systems are the most commonly used. In this case, the current is directly correlated with the analyte concentration, and this correlation is usually linear. In contrast, potentiometric detection methods are the least common methods. In this case, a bioactive material and an ion-selective membrane are used, and extremely small concentration changes can be observed during the main reaction. Detection methods based on impedance spectroscopy have promising applications in the area of biosensors, including the detection of bacteria [].
For example, a signal-off impedimetric immunosensor for the sensitive detection of E. coli O157:H7 bacteria was developed using AuNPs to mediate the electron transfer through a self-assembled monolayer (SAM). An antibody specific for the selected bacteria was immobilized on the gold electrode surface after functionalization with SAM and used for bacterial capture (Figure 4). The interaction between bacteria and AuNPs deposited on the electrode significantly reduced the charge transfer resistance between the redox probe in the solution and the electrode surface, thus ensuring the amplification of the detected signal [].
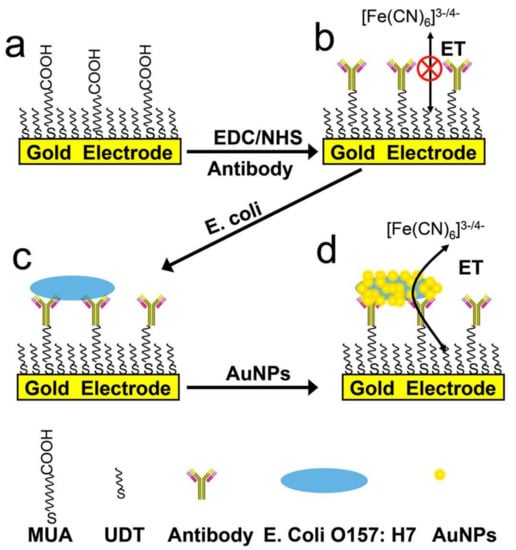
Figure 4.
Design principle and operation of the biosensor for E. coli O157:H7 bacteria detection. Reprinted with permission from [].
Carbon screen-printed electrodes (SPEs) decorated with AuNPs were used for label-free detection of E. coli O157. In this study, the presence of AuNPs at the electrode increased the stability and effectiveness of the biosensor for the detection of target bacteria. The immobilization of antibody molecules at the modified electrode was obtained through NHS cross-linking. After optimization, the electrochemical biosensor enabled the detection of E. coli O157 in the range of 10–106 CFU/mL with a limit of detection of 15 CFU/mL; no labels were necessary to assess the analytical performance, which is promising for the development of portable diagnostic tools [].
E. coli detection with an impressive detection limit of 1.3 × 10−18 M was recently performed using a carbon dot (CD)/ZnO nanorod/PANI composite-based electrochemical sensor [].
The same bacteria were detected using cyclic voltammetry with an innovative electrochemical portable sensor based on mesoporous ZrO2-Ag-G-SiO2 (ZAGS) and In2O3-G-SiO2 (IGS) composites. This sensor was able to detect a single E. coli cell from a small sample volume (1 μL) very quickly (within 30 s) with high specificity, reproducibility, stability, and selectivity, showing promising characteristics for the analysis of complex matrices [].
The electrochemical detection of E. coli without the need for DNA amplification or immunoassay was performed by exploiting the activity of the β-galactosidase enzyme, which hydrolyzes p-aminophenyl-β-d-galactopyranoside into p-aminophenol. E. coli is known to consume p-aminophenyl-β-d-galactopyranoside; thus, the remaining amount of this compound after 30 min of contact with the bacteria is oxidized on a gold electrode using voltammetry. E. coli was detected in a range from 102–104 CFU/mL in nutrient broth buffer in fewer than 100 min, thus proving the utility of electrochemical sensors for the rapid detection of bacteria [].
In recent years, bacteriophages have been increasingly employed as bioprobes in microbial detection studies because of their high affinity and specificity, low cost, robustness, and high stability. A more extensive review of phage technologies for monitoring pathogen bacteria in water and wastewater can be consulted [].
For example, an electrochemical sensor was developed based on bacteriophages, AuNRs, and electrochemical impedance spectroscopy as a detection method. E. coli K12 was used as a model to successfully evaluate the propagation of its specific T4-phages through changes that occur at the level of interfacial charge transfer resistance. An LOD of 103 CFU/mL was obtained for E. coli K12 in approximately 100 μL of the bacterial suspension []. The same strategy, based on T7lacZ bacteriophages with β-galactosidase enzyme involvement, was applied for the detection of E. coli. This type of phage can infect bacteria, triggering the overexpression of the enzyme caused by infection. Electrochemical detection was based on p-aminophenol as the electroactive compound and catalyst product of 4-aminophenyl-β-galactopyranoside used as an enzymatic substrate at the electrode. The optimized sensor was applied for the sensitive and selective detection of E. coli in drinking water, apple juice, and skim milk in approximately 3 h at a concentration of 105 CFU/mL (102 CFU/mL after 7 h) []. Engineered phages were conjugated in another study with magnetic beads and used for the separation, preconcentration, and detection of E. coli in drinking water through the expression of gold-binding peptides fused to alkaline phosphatase. This protein exhibits enzymatic activity and the ability to directly attach to an electrode made of gold. A limit of detection of 105 CFU/mL was obtained for the target bacteria after 4 h [].
Electrochemical devices allow the specific, rapid detection of one or multiple targets in the case of multiplexed detection. An advantage is that the sensor can be integrated into portable devices, which, coupled with a miniaturized potentiostat, allows for rapid on-site detection performed either by first responders or by the population. The testing protocol is simple and does not require any special conditions or specific knowledge. The development of nanotechnology coupled with different techniques for printing electrodes has boosted the development of specific sensors for pathogenic bacteria from different samples, such as biological fluids, surfaces, or culture plates.
The literature mentions different sensor configurations for pathogenic bacteria based on the detection of specific targets that do not involve any risk to users. One category of these sensors is represented by nanoplatforms capable of detecting virulence factors specific to P. aeruginosa, such as pyocyanin or a specific siderophore, namely pyoverdine for P. aeruginosa and enterobactin for E. coli. Both factors are electrochemically active and thus suitable for use as targets for sensor development. The approaches presented in the literature generally use screen-printed electrodes because they are suitable for the development of portable devices and because they do not pose significant risks regarding contamination procedures [,].
The literature mentions several examples of composite gold- and carbon-based nanoplatforms for the detection of pyoverdine. The deposition of carboxylic polypyrrole by multipulse amperometry followed by electrochemical generation of AuNPs was performed on a graphene-based screen-printed electrode that allowed the detection of pyoverdine in serum and saliva samples with excellent analytical performance []. Graphene AuNPs were deposited on carbon-based screen-printed electrodes, and the configuration had similar performance as in the case where AuNPs were electrochemically generated on a reduced graphene sheet topography [,]. These configurations have similar analytical performances, but the results in real samples and in the presence of common interfering agents are promising and oriented toward the development of portable sensing devices.
Pyocyanin, a virulence factor specific to the metabolism of P. aeruginosa, is also a redox-active compound that can be directly analyzed using electrochemical sensors. A nanograss-based sensor was fabricated by etching a nanograss on the electrode surface. The 3D structure was then decorated with 200 nm of gold. The amperometric quantification of P. aeruginosa via pyocyanin in spiked hypertonic saline samples and in airway samples was performed with a limit of detection of 172 nM in 60 s without the need for any pretreatment [].
Pyocyanin was detected in whole blood without any pretreatment using screen-printed electrodes modified with an agar and Au/Ag NP mixture, offering perspectives for the development of POC devices. The technology was brought one step further with the development of flexible dual sensors for the detection of the same targets. P. aeruginosa on different surfaces, such as furniture, sinks, and medical scrapper surfaces. This strategy enables the development of portable sensing devices that can be used as control tools when screening nosocomial agents in hospital environments [].
Examples of sensors and biosensors reported in the last decade for the detection of E. coli and P. aeruginosa in water samples, along with their principle of detection and analytical performance descriptors, are presented below in Table 3.

Table 3.
Comparative presentation of the analytical parameters for the sensors applied for bacteria detection in water samples.
5. Challenges
It is essential to mention and understand that the detection of microorganisms present in food, water, soil, air, and even animal and human bodies that are possible pathogens is a mandatory prerogative. It is necessary to detect infectious agents (e.g., bacteria, fungi, protozoa, and viruses). Finding fast and cheap methods to monitor water quality, food, and the environment has become a constant concern of authorities and a topic of high interest to researchers. It has been demonstrated that most microorganisms are non-pathogenic, with only a very small percentage, less than 0.1%, of microorganisms being responsible for disease. Thus, before the need for clinical treatment, it is important to develop prevention strategies that ensure a high quality of life, namely, to detect life-threatening bacteria on time. These strategies must be based primarily on robust, smart, rapid, sensitive, selective, and reliable methods of detection.
In the development and optimization of new approaches for microorganism detection, several challenges need to be overcome. For example, traditional analytical techniques that are currently applied to detect bacteria and microorganisms from water, wastewater, and other liquid samples, which restrict the noncultivable microbial information from complex environmental samples, are often slow and time-consuming, although they display high sensitivity and good accuracy.
As the field is of great interest and in continuous development, new analytical techniques are constantly appearing to eliminate these limitations. Sensors and immunological techniques are part of this category of methods. These are also novel, and most of them combine immunological assays and different types of sensors. Many research efforts have been made to develop automatic, combined, and miniaturized approaches; however, challenges still exist in practical applications.
Despite being very promising sensing tools, electrochemical biosensors face a few challenges that need to be overcome. One such issue is the fact that the developed sensors usually allow the detection of one bacterial species. For real applications, it is essential to consider multiple detections of pathogenic microorganisms, as well as simultaneous detection. This could be achieved by designing biosensors with multiple recognition elements bound to the electrode surface. Another challenge is the numerous interferences existing in real samples that do not undergo pretreatment steps.
Another factor to be considered is the stability and robustness of the developed sensor. Enzymes, antibodies, and aptamers are sensitive to extreme temperature conditions, which could be met in warm climate environments and limit their utility.
Another critical obstacle is related to the analysis of experimental data, which often has a high degree of difficulty and introduces new problems. Attempts have been made to find solutions for this issue; thus, molecular dynamics, chemometrics, and many other statistical tools have provided interdisciplinary and cutting-edge scientific support to these emerging detection techniques by overcoming associated limitations and facilitating the accomplishment of a notable steady revolution in science and technology.
6. Perspectives
6.1. Commercial Rapid Tests
Currently, there are very few commercially available rapid tests for testing the presence of pathogenic bacteria in water samples.
There are rapid tests on the market for assessing bacterial contamination in food, such as Singlepath® E. coli O157 (an immunochromatographic assay based on gold-labeled antibodies). The test provides high sensitivity and efficiency (>99%) and is able to detect as low as 1 CFU of E. coli O157 (including H7) in 25 g of food sample, with a response time of 20 min; however, prior to applying the test, an enrichment step of 18 h is recommended in the product specifications. Related immunological lateral flow tests for detecting the presence or absence of pathogenic bacteria from food matrices are Singlepath® Salmonella and Singlepath® Campylobacter.
A similar product is RapidChek® E. coli O157 (including H7), which is also intended to identify pathogenic bacteria in foodstuffs with simplicity and high accuracy. The testing kit contains a bottle of media for sample incubation and lateral flow immunochemical test strips. The time necessary to obtain the result is 8–18 h, depending on the analyzed sample.
It would also be desirable for rapid testing tools to fit the ASSURED criteria, which means to have the following characteristics: affordability, sensitivity, specificity, user-friendliness, rapidity and robustness, equipment-free functionality, and deliverability to end-users. These criteria are especially important when the purpose is on-site testing in remote and resource-limited settings [].
6.2. Future Perspectives
Industrial fields (water treatment plants, wastewater disinfection, drinking water quality control) still rely on other parameters (such as turbidity and dissolved organic matter) to screen for microbial water quality. The development of rapid and accurate methods for pathogen detection could hopefully improve the online real-time monitoring of bacterial load in the water.
Artificial intelligence and machine learning have opened future perspectives in many scientific fields. Progress has been registered in the field of smart technologies for monitoring environmental water based on the Internet of Things (IoT) and wireless sensor networks.
For example, sensors for real-time monitoring of E. coli concentration in wastewater are based on the prediction of several conventional physical and chemical parameters, as reported by Foschi et al. []. The soft sensor developed in this study was paired with an artificial neural network to assess the bacterial concentration in the analyzed water samples and optimize the disinfectant dosage used in the treatment process. This method could be a potential solution for a more efficient disinfection process in wastewater treatment plants.
In the near future, methods to rapidly and accurately detect the presence and concentration of bacteria would be available to collect data from water sources and utilize them together with machine learning algorithms in POU/POC applications (steps towards this direction have already been made [,]) to achieve better and faster decisions concerning water quality management.
7. Conclusions
This review covers the latest analytical findings on the detection of two waterborne pathogen bacteria, comparing the aspects of different methods and their figures of merit.
Several aspects, such as water quantity and quality, available resources, temperature and pressure of water, resource aging, large fluctuations in water consumption and need, operation and maintenance hazards, losses, unforeseen pollution, soil problems regarding stability, additional costs, and supply standards should be recognized and analyzed to meet the required objectives and goals set by the WHO guidelines. To this end, new methods and techniques must be applied. According to the WHO, water sources can be contaminated with specific pathogenic bacteria that pose a serious threat to human health.
The most common bacteria associated with high and moderate risk for human health are E. coli (pathogenic and enterohemorrhagic), Legionella spp., P. aeruginosa, Salmonella typhi, Shigella spp., Vibrio cholerae, and Yersinia enterocolitica. The majority of these have a moderate or high degree of multiplication and infectivity. To be specific, P. aeruginosa can multiply in water supplies and is resistant to chlorine but has a low risk of infection, with the main route of infection being skin contact with immunosuppressed patients (elders, children, patients with burns/extensive wounds, and those with immunosuppressive therapy and AIDS.
An exhaustive description of the most recent studies published between 2011 and 2021 was presented focusing on the detection of E. coli and P. aeruginosa.
Despite the variety of the existing analytical methods applied for bacterial detection, several issues remain, and simultaneous analysis is significantly lower in performance than that of single bacterial strands. Considering the remaining challenges, researchers still need to refine current investigation tools to ensure water quality monitoring.
Furthermore, newly developed methods must be improved and validated in real scenarios to be utilized for routine on-field analysis.
Author Contributions
Conceptualization, M.T., O.H., A.C. (Andreea Cernat), and C.C.; investigation, A.C. (Alexandra Canciu); resources, A.C. (Alexandra Canciu), M.T. and O.H.; data curation, A.C. (Andreea Cernat) and F.G.; writing—original draft preparation, A.C. (Alexandra Canciu), M.T., O.H., A.C. (Andreea Cernat) and C.C.; writing—review and editing, A.C. (Andreea Cernat), M.T., O.H., F.G. and C.C.; funding acquisition, C.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the European Union’s Horizon 2020 Research and Innovation Program under grant agreement No 883484, PathoCERT.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kanakoudis, V.; Papadopoulou, A.; Tsitsifli, S.; Curk, B.C.; Karleusa, B.; Matic, B.; Altran, E.; Banovec, P. Policy recommendation for drinking water supply cross-border networking in the Adriatic region. J. Water Supply Res. Technol. AQUA 2017, 66, 489–508. [Google Scholar] [CrossRef]
- Water Sanitation and Health. Available online: https://www.who.int/teams/environment-climate-change-and-health/water-sanitation-and-health/water-safety-and-quality/drinking-water-quality-guidelines (accessed on 26 April 2021).
- International Water Association. Available online: https://iwa-network.org/ (accessed on 26 April 2021).
- World Health Organization. WHO Microbial Aspects. WHO Guidel. Drink. Qual. 2011, 38, 117–153. [Google Scholar]
- Coalition for Water Security. Available online: https://www.coalitionforwatersecurity.org/ (accessed on 26 April 2021).
- UN World Water Development Report Archives|UN-Water. Available online: https://www.unwater.org/publication_categories/world-water-development-report/ (accessed on 26 April 2021).
- Moreira, N.A.; Bondelind, M. Safe drinking water and waterborne outbreaks. J. Water Health 2017, 15, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Matzeu, G.; Florea, L.; Diamond, D. Advances in wearable chemical sensor design for monitoring biological fluids. Sens. Actuators B Chem. 2015, 211, 403–418. [Google Scholar] [CrossRef]
- Kim, J.; Kumar, R.; Bandodkar, A.J.; Wang, J. Advanced Materials for Printed Wearable Electrochemical Devices: A Review. Adv. Electron. Mater. 2017, 3, 1600260. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Water Sanitation and Health. Available online: https://www.who.int/teams/environment-climate-change-and-health/water-sanitation-and-health (accessed on 26 April 2021).
- Ramírez-Castillo, F.Y.; Loera-Muro, A.; Jacques, M.; Garneau, P.; Avelar-González, F.J.; Harel, J.; Guerrero-Barrera, A.L. Waterborne pathogens: Detection methods and challenges. Pathogens 2015, 4, 307–334. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO|Progress on Sanitation and Drinking-Water. Available online: https://apps.who.int/iris/bitstream/handle/10665/81245/9789241505390_eng.pdf;jsessi (accessed on 26 April 2021).
- WHO|Millennium Development Goals (MDGs). Available online: https://www.who.int/news-room/fact-sheets/detail/millennium-development-goals-(mdgs) (accessed on 26 April 2021).
- World Health Organization. Guidelines for Drinking-Water Quality: Fourth Edition Incorporating the First Addendum; WHO—World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Munakata, N.; Kuo, J. Disinfection Processes. Water Environ. Res. 2015, 87, 1127–1146. [Google Scholar] [CrossRef]
- Thaipadungpanit, J.; Chierakul, W.; Pattanaporkrattana, W.; Phoodaeng, A.; Wongsuvan, G.; Huntrakun, V.; Amornchai, P.; Chatchen, S.; Kitphati, R.; Wuthiekanun, V.; et al. Burkholderia pseudomallei in water supplies, Southern Thailand. Emerg. Infect. Dis. 2014, 20, 1947–1949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiersinga, W.J.; van der Poll, T.; White, N.J.; Day, N.P.; Peacock, S.J. Melioidosis: Insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 2006, 4, 272–282. [Google Scholar] [CrossRef]
- Howard, K.; Inglis, T.J.J. Disinfection of Burkholderia pseudomallei in potable water. Water Res. 2005, 39, 1085–1092. [Google Scholar] [CrossRef] [Green Version]
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global epidemiology of campylobacter infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef] [Green Version]
- Jang, J.; Hur, H.-G.; Sadowsky, M.J.; Byappanahalli, M.N.; Yan, T.; Ishii, S. Environmental Escherichia coli: Ecology and public health implications—A review. J. Appl. Microbiol. 2017, 123, 570–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fields, B.S.; Benson, R.F.; Besser, R.E. Legionella and legionnaires’ disease: 25 Years of investigation. Clin. Microbiol. Rev. 2002, 15, 506–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loret, J.F.; Dumoutier, N. Non-tuberculous mycobacteria in drinking water systems: A review of prevalence data and control means. Int. J. Hyg. Environ. Health 2019, 222, 628–634. [Google Scholar] [CrossRef] [PubMed]
- Caskey, S.; Stirling, J.; Moore, J.E.; Rendall, J.C. Occurrence of Pseudomonas aeruginosa in waters: Implications for patients with cystic fibrosis (CF). Lett. Appl. Microbiol. 2018, 66, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Dhiman, A.; Kapil, A.; Bansal, V.; Sharma, T.K. Aptamer-mediated colorimetric and electrochemical detection of Pseudomonas aeruginosa utilizing peroxidase-mimic activity of gold NanoZyme. Anal. Bioanal. Chem. 2019, 411, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.; Mylona, E.; Frankel, G. Typhoidal Salmonella: Distinctive virulence factors and pathogenesis. Cell. Microbiol. 2018, 20, e12939. [Google Scholar] [CrossRef] [Green Version]
- Stanaway, J.D.; Parisi, A.; Sarkar, K.; Blacker, B.F.; Reiner, R.C.; Hay, S.I.; Nixon, M.R.; Dolecek, C.; James, S.L.; Mokdad, A.H.; et al. The global burden of non-typhoidal salmonella invasive disease: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2019, 19, 1312–1324. [Google Scholar] [CrossRef] [Green Version]
- Chompook, P. Shigellosis. In Encyclopedia of Environmental Health; Elsevier: Amsterdam, The Netherlands, 2019; Volume 43, pp. 626–632. [Google Scholar] [CrossRef]
- Harris, J.B.; LaRocque, R.C.; Qadri, F.; Ryan, E.T.; Calderwood, S.B. Cholera. In The Lancet; Lancet Publishing Group: London, UK, 2012; Volume 379, pp. 2466–2476. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Zhang, Y.; Gao, Z.F.; Ye, Y.; Wu, Q.; Chen, H.; Xu, J. Recent advances in nanotechnology for simultaneous detection of multiple pathogenic bacteria. Nano Today 2021, 38, 101121. [Google Scholar] [CrossRef]
- Wang, P.; Sun, Y.; Li, X.; Wang, L.; Xu, Y.; He, L.; Li, G. Recent advances in dual recognition based surface enhanced Raman scattering for pathogenic bacteria detection: A review. Anal. Chim. Acta 2021, 338279. [Google Scholar] [CrossRef]
- Deshmukh, R.A.; Joshi, K.; Bhand, S.; Roy, U. Recent developments in detection and enumeration of waterborne bacteria: A retrospective minireview. Microbiologyopen 2016, 5, 901–922. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Ali, Z.; Zou, J.; Jin, G.; Zhu, J.; Yang, J.; Dai, J. Detection methods for: Pseudomonas aeruginosa: History and future perspective. RSC Adv. 2017, 7, 51789–51800. [Google Scholar] [CrossRef] [Green Version]
- Pang, B.; Zhao, C.; Li, L.; Song, X.; Xu, K.; Wang, J.; Liu, Y.; Fu, K.; Bao, H.; Song, D.; et al. Development of a low-cost paper-based ELISA method for rapid Escherichia coli O157:H7 detection. Anal. Biochem. 2018, 542, 58–62. [Google Scholar] [CrossRef]
- Saptalena, L.G.; Kuklya, A.; Telgheder, U. Gas Chromatography-Differential Mobility Spectrometry and Gas Chromatography-Mass Spectrometry for the detection of coliform bacteria. Int. J. Mass Spectrom. 2015, 388, 17–25. [Google Scholar] [CrossRef]
- Ma, X.; Ding, W.; Wang, C.; Wu, H.; Tian, X.; Lyu, M.; Wang, S. DNAzyme biosensors for the detection of pathogenic bacteria. Sens. Actuators B Chem. 2021, 331, 129422. [Google Scholar] [CrossRef]
- Maguire, M.; Kase, J.A.; Roberson, D.; Muruvanda, T.; Brown, E.W.; Allard, M.; Musser, S.M.; González-Escalona, N. Precision long-read metagenomics sequencing for food safety by detection and assembly of Shiga toxin-producing Escherichia coli in irrigation water. PLoS ONE 2021, 16, e0245172. [Google Scholar] [CrossRef]
- Hosu, M.C.; Vasaikar, S.; Okuthe, G.E.; Apalata, T. Molecular Detection of Antibiotic-Resistant Genes in Pseudomonas aeruginosa from Nonclinical Environment: Public Health Implications in Mthatha, Eastern Cape Province, South Africa. Int. J. Microbiol. 2021, 2021, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rani, A.; Ravindran, V.B.; Surapaneni, A.; Shahsavari, E.; Haleyur, N.; Mantri, N.; Ball, A.S. Evaluation and comparison of recombinase polymerase amplification coupled with lateral-flow bioassay for Escherichia coli O157:H7 detection using diifeerent genes. Sci. Rep. 2021, 11, 1881. [Google Scholar] [CrossRef]
- Brandt, J.; Albertsen, M. Investigation of Detection Limits and the Influence of DNA Extraction and Primer Choice on the Observed Microbial Communities in Drinking Water Samples Using 16S rRNA Gene Amplicon Sequencing. Front. Microbiol. 2018, 9, 2140. [Google Scholar] [CrossRef] [PubMed]
- Urban, L.; Holzer, A.; Baronas, J.J.; Hall, M.B.; Braeuninger-Weimer, P.; Scherm, M.J.; Kunz, D.J.; Perera, S.N.; Martin-Herranz, D.E.; Tipper, E.T.; et al. Freshwater monitoring by nanopore sequencing. eLife 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Song, Y.; Kim, Y.T.; Lee, S.J.; Lee, K.G.; Im, S.G. Multifunctional Printable Micropattern Array for Digital Nucleic Acid Assay for Microbial Pathogen Detection. ACS Appl. Mater. Interfaces 2021. [Google Scholar] [CrossRef]
- Li, Y.; Bai, C.; Yang, L.; Fu, J.; Yan, M.; Chen, D.; Zhang, L. High flux isothermal assays on pathogenic, virulent and toxic genetics from various pathogens. Microb. Pathog. 2018, 116, 68–72. [Google Scholar] [CrossRef]
- Kouremenos, K.A.; Beale, D.J.; Antti, H.; Palombo, E.A. Liquid chromatography time of flight mass spectrometry based environmental metabolomics for the analysis of Pseudomonas putida Bacteria in potable water. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 966, 179–186. [Google Scholar] [CrossRef]
- Kartsova, L.A.; Makeeva, D.V.; Kravchenko, A.V.; Moskvichev, D.O.; Polikarpova, D.A. Capillary electrophoresis as a powerful tool for the analyses of bacterial samples. TrAC Trends Anal. Chem. 2021, 134, 116110. [Google Scholar] [CrossRef]
- Buszewski, B.; Rogowska, A.; Pomastowski, P.; Złoch, M.; Railean-Plugaru, V. Identification of microorganisms by modern analytical techniques. J. AOAC Int. 2017, 100, 1607–1623. [Google Scholar] [CrossRef] [PubMed]
- Fiori, J.; Turroni, S.; Candela, M.; Gotti, R. Assessment of gut microbiota fecal metabolites by chromatographic targeted approaches. J. Pharm. Biomed. Anal. 2020, 177, 112867. [Google Scholar] [CrossRef] [PubMed]
- Glenn, T.C. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 2011, 11, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Dilger, T.; Melzl, H.; Gessner, A. Rapid and reliable identification of waterborne Legionella species by MALDI-TOF mass spectrometry. J. Microbiol. Methods 2016, 127, 154–159. [Google Scholar] [CrossRef]
- Pascale, M.R.; Mazzotta, M.; Salaris, S.; Girolamini, L.; Grottola, A.; Simone, M.L.; Cordovana, M.; Bisognin, F.; Dal Monte, P.; Bucci Sabattini, M.A.; et al. Evaluation of MALDI–TOF Mass Spectrometry in Diagnostic and Environmental Surveillance of Legionella Species: A Comparison With Culture and Mip-Gene Sequencing Technique. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Suzuki, Y.; Niina, K.; Matsuwaki, T.; Nukazawa, K.; Iguchi, A. Bacterial flora analysis of coliforms in sewage, river water, and ground water using MALDI-TOF mass spectrometry. J. Environ. Sci. Health Part A Toxic Subst. Environ. Eng. 2018, 53, 160–173. [Google Scholar] [CrossRef]
- Emami, K.; Askari, V.; Ullrich, M.; Mohinudeen, K.; Anil, A.C.; Khandeparker, L.; Burgess, J.G.; Mesbahi, E. Characterization of bacteria in Ballast water using MALDI-TOF mass spectrometry. PLoS ONE 2012, 7, e0038515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timperio, A.M.; Gorrasi, S.; Zolla, L.; Fenice, M. Evaluation of MALDI-TOF mass spectrometry and MALDI BioTyper in comparison to 16S rDNA sequencing for the identification of bacteria isolated from Arctic sea water. PLoS ONE 2017, 12, e0181860. [Google Scholar] [CrossRef] [PubMed]
- Sala-Comorera, L.; Caudet-Segarra, L.; Galofré, B.; Lucena, F.; Blanch, A.R.; García-Aljaro, C. Unravelling the composition of tap and mineral water microbiota: Divergences between next-generation sequencing techniques and culture-based methods. Int. J. Food Microbiol. 2020, 334, 108850. [Google Scholar] [CrossRef]
- Horká, M.; Šalplachta, J.; Karásek, P.; Ružička, F.; Roth, M. Online Concentration of Bacteria from Tens of Microliter Sample Volumes in Roughened Fused Silica Capillary with Subsequent Analysis by Capillary Electrophoresis and Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. ACS Infect. Dis. 2020, 6, 355–365. [Google Scholar] [CrossRef]
- Chiu, S.W.Y.; Cheng, H.W.; Chen, Z.X.; Wang, H.H.; Lai, M.Y.; Wang, J.K.; Wang, Y.L. Quantification of biomolecules responsible for biomarkers in the surface-enhanced Raman spectra of bacteria using liquid chromatography-mass spectrometry. Phys. Chem. Chem. Phys. 2018, 20, 8032–8041. [Google Scholar] [CrossRef]
- Kwon, D.; Lee, S.; Ahn, M.M.; Kang, I.S.; Park, K.H.; Jeon, S. Colorimetric detection of pathogenic bacteria using platinum-coated magnetic nanoparticle clusters and magnetophoretic chromatography. Anal. Chim. Acta 2015, 883, 61–66. [Google Scholar] [CrossRef]
- Park, C.; Lee, J.; Kim, Y.; Kim, J.; Lee, J.; Park, S. 3D-printed microfluidic magnetic preconcentrator for the detection of bacterial pathogen using an ATP luminometer and antibody-conjugated magnetic nanoparticles. J. Microbiol. Methods 2017, 132, 128–133. [Google Scholar] [CrossRef]
- Yao, L.; Wang, L.; Huang, F.; Cai, G.; Xi, X.; Lin, J. A microfluidic impedance biosensor based on immunomagnetic separation and urease catalysis for continuous-flow detection of E. coli O157:H7. Sens. Actuators B Chem. 2018, 259, 1013–1021. [Google Scholar] [CrossRef]
- Suaifan, G.A.R.Y.; Alhogail, S.; Zourob, M. Paper-based magnetic nanoparticle-peptide probe for rapid and quantitative colorimetric detection of Escherichia coli O157:H7. Biosens. Bioelectron. 2017, 92, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Torres, K.Y.; Arnold, D.P.; McLamore, E.S. Rapid isolation of Escherichia coli from water samples using magnetic microdiscs. Sens. Actuators B Chem. 2019, 291, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Nehra, M.; Mehta, J.; Dilbaghi, N.; Marrazza, G.; Kaushik, A. Point-of-care strategies for detection of waterborne pathogens. Sensors 2019, 19, 4476. [Google Scholar] [CrossRef] [Green Version]
- Simoska, O.; Stevenson, K.J. Electrochemical sensors for rapid diagnosis of pathogens in real time. Analyst 2019, 144, 6461–6478. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.S.; Kim, Y.H. Rapid and robust MALDI-TOF MS techniques for microbial identification: A brief overview of their diverse applications. J. Microbiol. 2018, 56, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, N.; Bhardwaj, S.K.; Bhatt, D.; Kwon, D.; Kim, K.; Deep, A. Trends in Analytical Chemistry Optical detection of waterborne pathogens using nanomaterials. Trends Anal. Chem. 2019, 113, 280–300. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, C.; Fei, R.; Liu, X.; Zhou, Y.; Chen, J.; Chen, H.; Zhou, R.; Hu, Y. Sensitive chemiluminescence immunoassay for E. coli O157:H7 detection with signal dual-amplification using glucose oxidase and laccase. Anal. Chem. 2014, 86, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Razmi, N.; Hasanzadeh, M.; Willander, M.; Nur, O. Recent progress on the electrochemical biosensing of Escherichia coli O157:H7: Material and methods overview. Biosensors 2020, 10, 54. [Google Scholar] [CrossRef]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef] [PubMed]
- Hameed, S.; Xie, L.; Ying, Y. Conventional and emerging detection techniques for pathogenic bacteria in food science: A review. Trends Food Sci. Technol. 2018, 81, 61–73. [Google Scholar] [CrossRef]
- Hulanicki, A.; Glab, S.; Ingman, F. Chemical sensors definitions and classification. Pure Appl. Chem. 1991, 63, 1247–1250. [Google Scholar] [CrossRef]
- Baig, N.; Sajid, M.; Saleh, A. Recent trends in nanomaterial-modified electrodes for electroanalytical applications. TrAC Trends Anal. Chem. 2019, 111. [Google Scholar] [CrossRef]
- Hosu, O.; Florea, A.; Cristea, C.; Sandulescu, R. Functionalized Advanced Hybrid Materials for Biosensing Applications. In Advanced Biosensors for Health Care Applications; Elsevier: Amsterdam, The Netherlands, 2019; pp. 171–207. [Google Scholar] [CrossRef]
- Bouguelia, S.; Roupioz, Y.; Slimani, S.; Mondani, L.; Casabona, M.G.; Durmort, C.; Vernet, T.; Calemczuk, R.; Livache, T. On-chip microbial culture for the specific detection of very low levels of bacteria. Lab Chip 2013, 13, 4024–4032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Højris, B.; Christensen, S.C.B.; Albrechtsen, H.J.; Smith, C.; Dahlqvist, M. A novel, optical, on-line bacteria sensor for monitoring drinking water quality. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Surface plasmon resonance detection of E. coli and methicillin-resistant S. aureus using bacteriophages. Biosens. Bioelectron. 2012, 37, 24–29. [Google Scholar] [CrossRef]
- Petrovszki, D.; Valkai, S.; Gora, E.; Tanner, M.; Bányai, A.; Fürjes, P.; Dér, A. An integrated electro-optical biosensor system for rapid, low-cost detection of bacteria. Microelectron. Eng. 2021, 239–240, 111523. [Google Scholar] [CrossRef]
- Kaushal, S.; Priyadarshi, N.; Pinnaka, A.K.; Soni, S.; Deep, A.; Singhal, N.K. Glycoconjugates coated gold nanorods based novel biosensor for optical detection and photothermal ablation of food borne bacteria. Sens. Actuators B Chem. 2019, 289, 207–215. [Google Scholar] [CrossRef]
- Massad-Ivanir, N.; Shtenberg, G.; Raz, N.; Gazenbeek, C.; Budding, D.; Bos, M.P.; Segal, E. Porous Silicon-Based Biosensors: Towards Real-Time Optical Detection of Target Bacteria in the Food Industry. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yaghoubi, M.; Rahimi, F.; Negahdari, B.; Rezayan, A.H.; Shafiekhani, A. A lectin-coupled porous silicon-based biosensor: Label-free optical detection of bacteria in a real-time mode. Sci. Rep. 2020, 10, 16017. [Google Scholar] [CrossRef]
- Sivakumar, S.; Wark, K.L.; Gupta, J.K.; Abbott, N.L.; Caruso, F. Liquid Crystal Emulsions as the Basis of Biological Sensors for the Optical Detection of Bacteria and Viruses. Adv. Funct. Mater. 2009, 19, 2260–2265. [Google Scholar] [CrossRef]
- Verbarg, J.; Plath, W.D.; Shriver-Lake, L.C.; Howell, P.B.; Erickson, J.S.; Golden, J.P.; Ligler, F.S. Catch and release: Integrated system for multiplexed detection of bacteria. Anal. Chem. 2013, 85, 4944–4950. [Google Scholar] [CrossRef] [Green Version]
- Bhatta, D.; Stadden, E.; Hashem, E.; Sparrow, I.J.G.; Emmerson, G.D. Multi-purpose optical biosensors for real-time detection of bacteria, viruses and toxins. Sens. Actuators B Chem. 2010, 149, 233–238. [Google Scholar] [CrossRef]
- Massad-Ivanir, N.; Shtenberg, C.; Zeidman, T.; Segal, E. Construction and characterization of porous SiO2/hydrogel hybrids as optical biosensors for rapid detection of bacteria. Adv. Funct. Mater. 2010, 20, 2269–2277. [Google Scholar] [CrossRef]
- Yang, X.; Feng, L.; Qin, X. Preparation of the Cf-GQDs-Escherichia coli O157: H7 Bioprobe and Its Application in Optical Imaging and Sensing of Escherichia coli O157: H7. Food Anal. Methods 2018, 11, 2280–2286. [Google Scholar] [CrossRef]
- Hosu, O.; Lettieri, M.; Papara, N.; Ravalli, A.; Sandulescu, R.; Cristea, C.; Marrazza, G. Colorimetric multienzymatic smart sensors for hydrogen peroxide, glucose and catechol screening analysis. Talanta 2019, 204, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, S.; Nejad, M.A.F.; Parolo, C.; Shahrokhian, S.; Merkoçi, A. Smart Chip for Visual Detection of Bacteria Using the Electrochromic Properties of Polyaniline. Anal. Chem. 2019, 91, 14960–14966. [Google Scholar] [CrossRef]
- Vaisocherová-Lísalová, H.; Víšová, I.; Ermini, M.L.; Špringer, T.; Song, X.C.; Mrázek, J.; Lamačová, J.; Scott Lynn, N.; Šedivák, P.; Homola, J. Low-fouling surface plasmon resonance biosensor for multi-step detection of foodborne bacterial pathogens in complex food samples. Biosens. Bioelectron. 2016, 80, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.H.; Li, M.; Wang, Y.; Ouyang, H.X.; Wang, L.; Li, X.C.; Cao, Y.C.; Meng, Q.H.; Lu, J.X. Aptasensors for rapid detection of Escherichia coli O157: H7 and Salmonella typhimurium. Nanoscale Res. Lett. 2012, 7. [Google Scholar] [CrossRef] [Green Version]
- Stefan, G.; Hosu, O.; De Wael, K.; Lobo-Castañón, M.J.; Cristea, C. Aptamers in Biomedicine: Selection Strategies and Recent Advances. Electrochim. Acta 2021, 376, 137994. [Google Scholar] [CrossRef]
- Jyoti, A.; Tomar, R.S.; Shanker, R. Nanosensors for the Detection of Pathogenic Bacteria. In Sustainable Agriculture Reviews; Springer: Cham, Switzerland, 2016; Volume 20, pp. 129–150. [Google Scholar]
- Wang, C.; Meloni, M.M.; Wu, X.; Zhuo, M.; He, T.; Wang, J.; Wang, C.; Dong, P. Magnetic plasmonic particles for SERS-based bacteria sensing: A review. AIP Adv. 2019, 9, 10701. [Google Scholar] [CrossRef] [Green Version]
- Tripp, R.A.; Dluhy, R.A.; Zhao, Y. Novel nanostructures for SERS biosensing. Nano Today 2008, 3, 31–37. [Google Scholar] [CrossRef]
- Mosier-Boss, P.A. Review on SERS of bacteria. Biosensors 2017, 7, 51. [Google Scholar] [CrossRef] [Green Version]
- Yildirim, N.; Long, F.; Gu, A.Z. Aptamer based E-coli detection in waste waters by portable optical biosensor system. In Proceedings of the IEEE Annual Northeast Bioengineering Conference, NEBEC, Boston, MA, USA, 25–27 April 2014. [Google Scholar] [CrossRef]
- Wan, J.; Ai, J.; Zhang, Y.; Geng, X.; Gao, Q.; Cheng, Z. Signal-off impedimetric immunosensor for the detection of Escherichia coli O157:H7. Sci. Rep. 2016, 6, 1–6. [Google Scholar] [CrossRef]
- Vu, Q.K.; Tran, Q.H.; Vu, N.P.; Anh, T.; Dang, T.T.; Matteo, T.; Nguyen, T.H.H. A label-free electrochemical biosensor based on screen-printed electrodes modified with gold nanoparticles for quick detection of bacterial pathogens. Mater. Today Commun. 2020, 26, 101726. [Google Scholar] [CrossRef]
- Pangajam, A.; Theyagarajan, K.; Dinakaran, K. Highly sensitive electrochemical detection of E. coli O157:H7 using conductive carbon dot/ZnO nanorod/PANI composite electrode. Sens. Bio-Sensing Res. 2020, 29, 100317. [Google Scholar] [CrossRef]
- Fatema, K.N.; Liu, Y.; Cho, K.Y.; Oh, W.C. Comparative study of electrochemical biosensors based on highly efficient mesoporous ZrO2-Ag-G-SiO2 and In2O3-G-SiO2 for rapid recognition of E. coli O157:H7. ACS Omega 2020, 5, 22719–22730. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.J.; Hsu, Y.C.; Gu, B.C.; Wu, C.C. Voltammetric measurement of Escherichia coli concentration through p-APG hydrolysis by endogenous β-galactosidase. Microchem. J. 2020, 154, 104641. [Google Scholar] [CrossRef]
- Bayat, F.; Didar, T.F.; Hosseinidoust, Z. Emerging investigator series: Bacteriophages as nano engineering tools for quality monitoring and pathogen detection in water and wastewater. Environ. Sci. Nano 2021. [Google Scholar] [CrossRef]
- Moghtader, F.; Congur, G.; Zareie, H.M.; Erdem, A.; Piskin, E. Impedimetric detection of pathogenic bacteria with bacteriophages using gold nanorod deposited graphite electrodes. RSC Adv. 2016, 6, 97832–97839. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Chen, J.; Nugen, S.R. Electrochemical Detection of Escherichia coli from Aqueous Samples Using Engineered Phages. Anal. Chem. 2017, 89, 1650–1657. [Google Scholar] [CrossRef]
- Singh, S.; Hinkley, T.; Nugen, S.R.; Talbert, J.N. Colorimetric detection of Escherichia coli using engineered bacteriophage and an affinity reporter system. Anal. Bioanal. Chem. 2019, 411, 7273–7279. [Google Scholar] [CrossRef]
- Webster, T.A.; Sismaet, H.J.; Conte, J.L.; Chan, I.; Ping, J.; Goluch, E.D. Electrochemical detection of Pseudomonas aeruginosa in human fluid samples via pyocyanin. Biosens. Bioelectron. 2014, 60, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Sismaet, H.J.; Pinto, A.J.; Goluch, E.D. Electrochemical sensors for identifying pyocyanin production in clinical Pseudomonas aeruginosa isolates. Biosens. Bioelectron. 2017, 97, 65–69. [Google Scholar] [CrossRef]
- Cernat, A.; Tertis, M.; Gandouzi, I.; Bakhrouf, A.; Suciu, M.; Cristea, C. Electrochemical sensor for the rapid detection of Pseudomonas aeruginosa siderophore based on a nanocomposite platform. Electrochem. Commun. 2018, 88, 5–9. [Google Scholar] [CrossRef]
- Gandouzi, I.; Tertis, M.; Cernat, A.; Bakhrouf, A.; Coros, M.; Pruneanu, S.; Cristea, C. Sensitive detection of pyoverdine with an electrochemical sensor based on electrochemically generated graphene functionalized with gold nanoparticles. Bioelectrochemistry 2018, 120, 94–103. [Google Scholar] [CrossRef]
- Gandouzi, I.; Tertis, M.; Cernat, A.; Saidane-Mosbahi, D.; Ilea, A.; Cristea, C. A Nanocomposite Based on Reduced Graphene and Gold Nanoparticles for Highly Sensitive Electrochemical Detection of Pseudomonas aeruginosa through Its Virulence Factors. Materials 2019, 12, 1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alatraktchi, F.A.Z.; Dimaki, M.; Støvring, N.; Johansen, H.K.; Molin, S.; Svendsen, W.E. Nanograss sensor for selective detection of Pseudomonas aeruginosa by pyocyanin identification in airway samples. Anal. Biochem. 2020, 593, 113586. [Google Scholar] [CrossRef]
- Ciui, B.; Tertiş, M.; Cernat, A.; Sǎndulescu, R.; Wang, J.; Cristea, C. Finger-Based Printed Sensors Integrated on a Glove for On-Site Screening of Pseudomonas aeruginosa Virulence Factors. Anal. Chem. 2018, 90, 7761–7768. [Google Scholar] [CrossRef]
- Wu, Z.; He, D.; Cui, B.; Jin, Z. A bimodal (SERS and colorimetric) aptasensor for the detection of Pseudomonas aeruginosa. Microchim. Acta 2018, 185, 1–7. [Google Scholar] [CrossRef]
- Jia, F.; Xu, L.; Yan, W.; Wu, W.; Yu, Q.; Tian, X.; Dai, R.; Li, X. A magnetic relaxation switch aptasensor for the rapid detection of Pseudomonas aeruginosa using superparamagnetic nanoparticles. Microchim. Acta 2017, 184, 1539–1545. [Google Scholar] [CrossRef]
- Mejri, M.B.; Baccar, H.; Baldrich, E.; Del Campo, F.J.; Helali, S.; Ktari, T.; Simonian, A.; Aouni, M.; Abdelghani, A. Impedance biosensing using phages for bacteria detection: Generation of dual signals as the clue for in-chip assay confirmation. Biosens. Bioelectron. 2010, 26, 1261–1267. [Google Scholar] [CrossRef] [Green Version]
- Vinay, M.; Franche, N.; Grégori, G.; Fantino, J.-R.; Pouillot, F.; Ansaldi, M. Phage-Based Fluorescent Biosensor Prototypes to Specifically Detect Enteric Bacteria Such as E. coli and Salmonella enterica Typhimurium. PLoS ONE 2015, 10, e0131466. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Luan, T.; Yang, X.; Wang, S.; Zheng, Y.; Huang, T.; Zhu, S.; Yan, X. Trace detection of specific viable bacteria using tetracysteine-tagged bacteriophages. Anal. Chem. 2014, 86, 907–912. [Google Scholar] [CrossRef]
- Sedki, M.; Chen, X.; Chen, C.; Ge, X.; Mulchandani, A. Non-lytic M13 phage-based highly sensitive impedimetric cytosensor for detection of coliforms. Biosens. Bioelectron. 2020, 148, 111794. [Google Scholar] [CrossRef]
- Peng, H.; Chen, I.A. Rapid Colorimetric Detection of Bacterial Species through the Capture of Gold Nanoparticles by Chimeric Phages. ACS Nano 2019, 13, 1244–1252. [Google Scholar] [CrossRef] [PubMed]
- Alcaine, S.D.; Law, K.; Ho, S.; Kinchla, A.J.; Sela, D.A.; Nugen, S.R. Bioengineering bacteriophages to enhance the sensitivity of phage amplification-based paper fluidic detection of bacteria. Biosens. Bioelectron. 2016, 82, 14–19. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Hinkley, T.; Chen, J.; Talbert, J.N.; Nugen, S.R. Phage based electrochemical detection of: Escherichia coli in drinking water using affinity reporter probes. Analyst 2019, 144, 1345–1352. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wang, D.; Kinchla, A.J.; Sela, D.A.; Nugen, S.R. Rapid screening of waterborne pathogens using phage-mediated separation coupled with real-time PCR detection. Anal. Bioanal. Chem. 2016, 408, 4169–4178. [Google Scholar] [CrossRef]
- Burnham, S.; Hu, J.; Anany, H.; Brovko, L.; Deiss, F.; Derda, R.; Griffiths, M.W. Towards rapid on-site phage-mediated detection of generic Escherichia coli in water using luminescent and visual readout. Anal. Bioanal. Chem. 2014, 406, 5685–5693. [Google Scholar] [CrossRef]
- Hinkley, T.; Garing, S.; Jain, P.; Williford, J.; Le Ny, A.-L.; Nichols, K.; Peters, J.; Talbert, J.; Nugen, S. A Syringe-Based Biosensor to Rapidly Detect Low Levels of Escherichia coli (ECOR13) in Drinking Water Using Engineered Bacteriophages. Sensors 2020, 20, 1953. [Google Scholar] [CrossRef] [Green Version]
- Hinkley, T.C.; Garing, S.; Singh, S.; Le Ny, A.L.M.; Nichols, K.P.; Peters, J.E.; Talbert, J.N.; Nugen, S.R. Reporter bacteriophage T7NLC utilizes a novel NanoLuc::CBM fusion for the ultrasensitive detection of: Escherichia coli in water. Analyst 2018, 143, 4074–4082. [Google Scholar] [CrossRef] [Green Version]
- Hinkley, T.C.; Singh, S.; Garing, S.; Le Ny, A.L.M.; Nichols, K.P.; Peters, J.E.; Talbert, J.N.; Nugen, S.R. A phage-based assay for the rapid, quantitative, and single CFU visualization of E. coli (ECOR #13) in drinking water. Sci. Rep. 2018, 8, 14630. [Google Scholar] [CrossRef]
- Chen, J.; Alcaine, S.D.; Jiang, Z.; Rotello, V.M.; Nugen, S.R. Detection of Escherichia coli in Drinking Water Using T7 Bacteriophage-Conjugated Magnetic Probe. Anal. Chem. 2015, 87, 8977–8984. [Google Scholar] [CrossRef]
- Zurier, H.S.; Duong, M.M.; Goddard, J.M.; Nugen, S.R. Engineering Biorthogonal Phage-Based Nanobots for Ultrasensitive, in Situ Bacteria Detection. ACS Appl. Bio Mater. 2020, 3, 5824–5831. [Google Scholar] [CrossRef]
- Kosack, C.S.; Page, A.L.; Klatser, P.R. A guide to aid the selection of diagnostic tests. Bull. World Health Organ. 2017, 95, 639–645. [Google Scholar] [CrossRef]
- Foschi, J.; Turolla, A.; Antonelli, M. Soft sensor predictor of E. coli concentration based on conventional monitoring parameters for wastewater disinfection control. Water Res. 2021, 191, 116806. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Zhao, Z.; Han, S.; Liu, Z. Lagoon water quality monitoring based on digital image analysis and machine learning estimators. Water Res. 2020, 172, 115471. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, M.H.; Wiwasuku, T.; Day, A.S.; Youngme, S.; Hwang, D.S.; Yoon, J.-Y. Human sensor-inspired supervised machine learning of smartphone-based paper microfluidic analysis for bacterial species classification. Biosens. Bioelectron. 2021, 188, 113335. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).