Proposed Research for Innovative Solutions for Chickpeas and Beans in a Climate Change Scenario: The Mediterranean Basin
Abstract
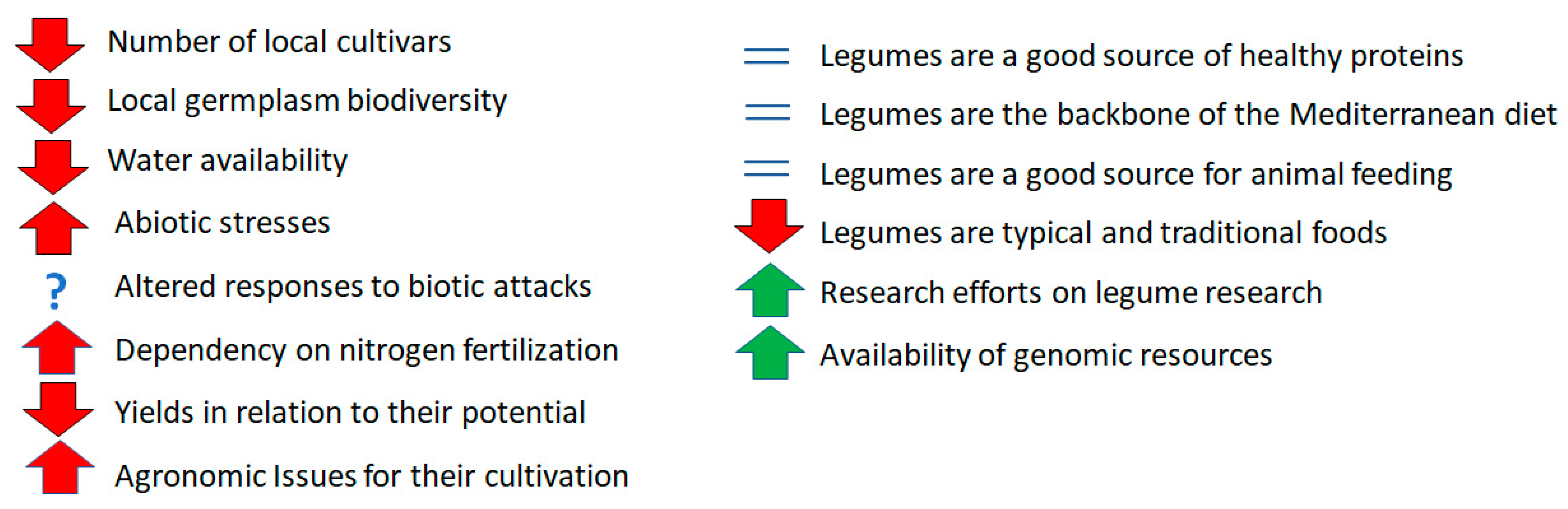
1. Introduction
1.1. Epigenomic Adaptation of Legumes to Climate Change
1.2. Previous -omics Studies Performed in Bean and Chickpea under Drought Conditions
2. Discussion
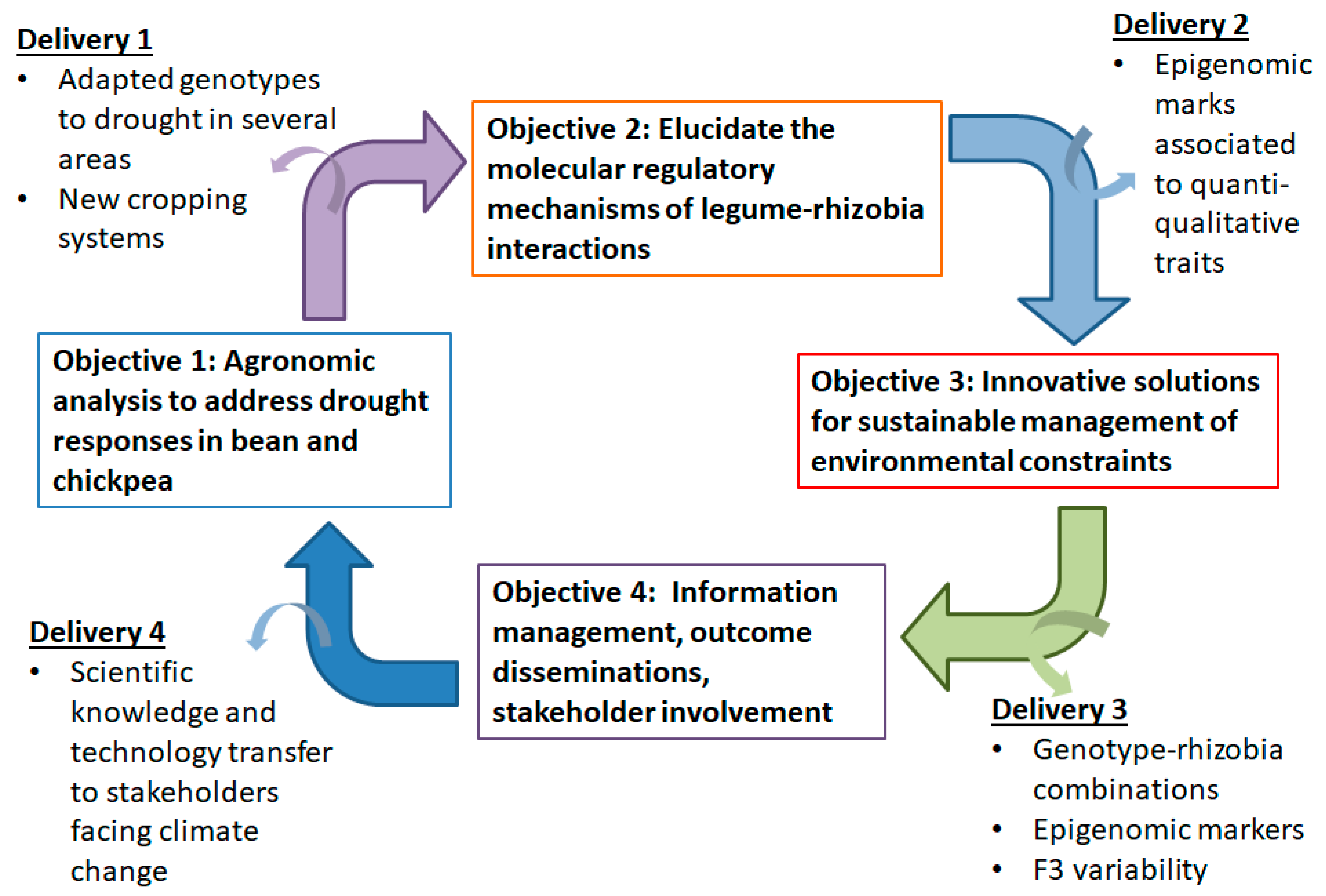
2.1. Proposed Approach
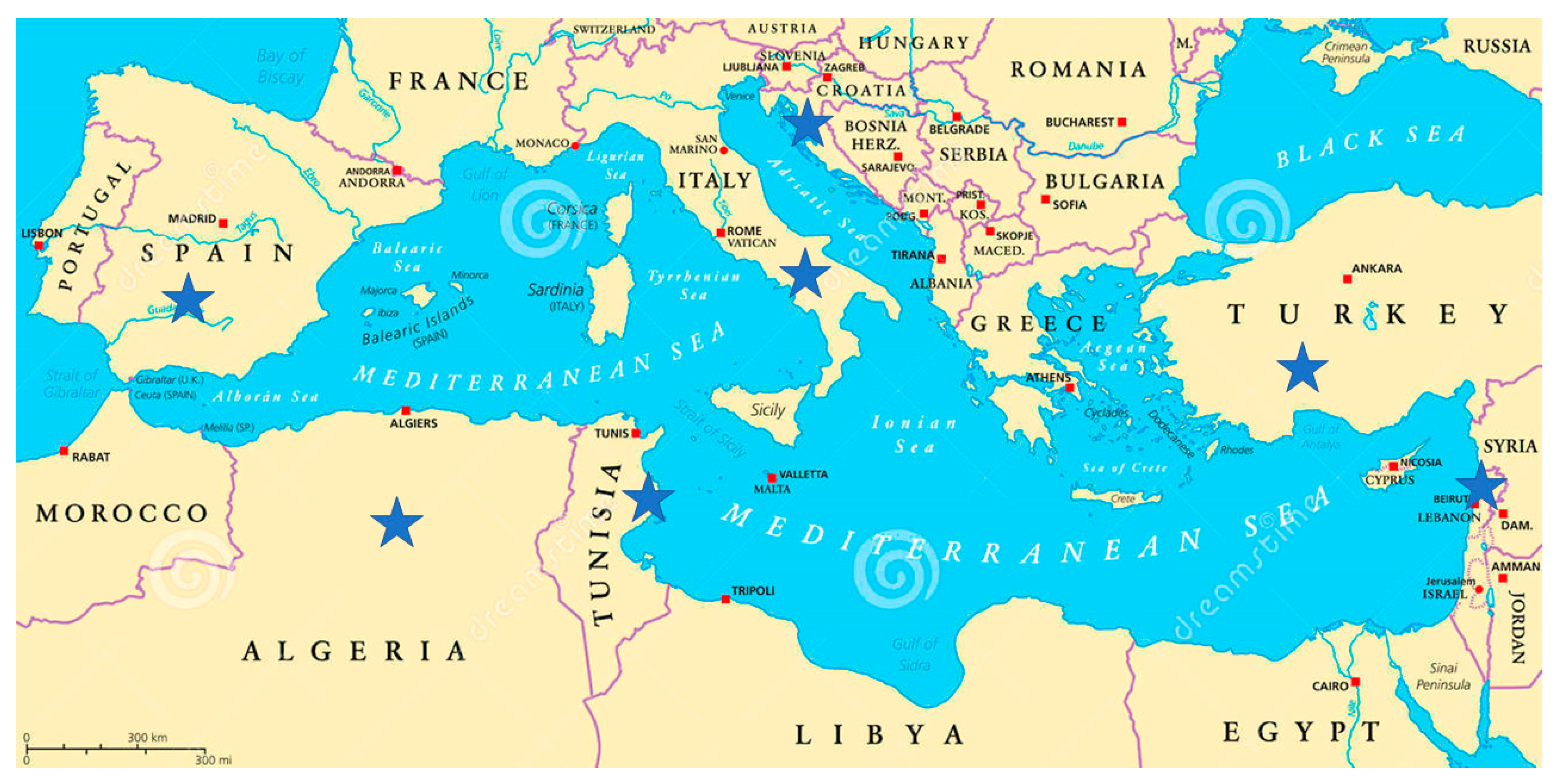
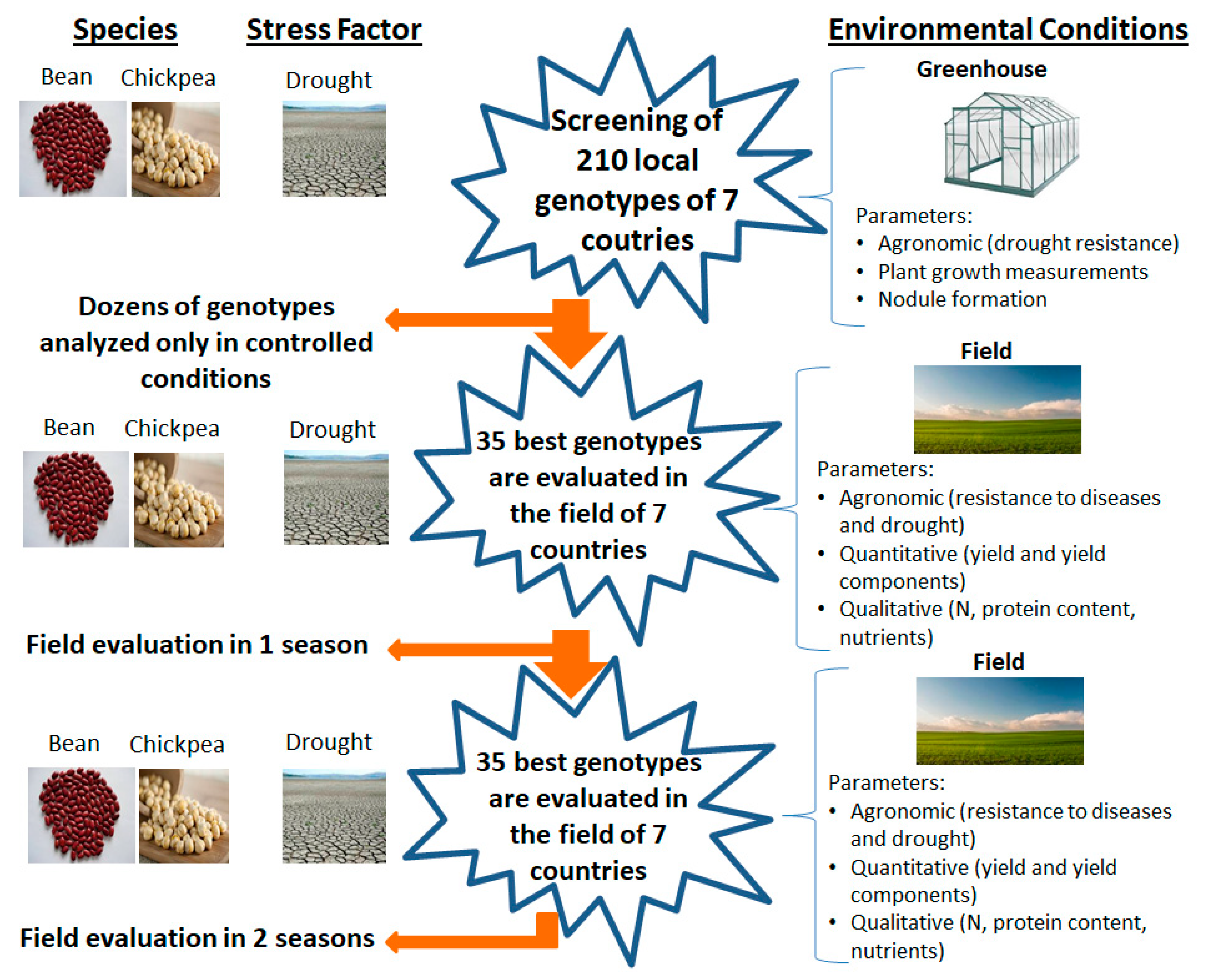
2.1.1. First Objective: Agronomic Characterization of Local Germplasm in the Mediterranean Basin
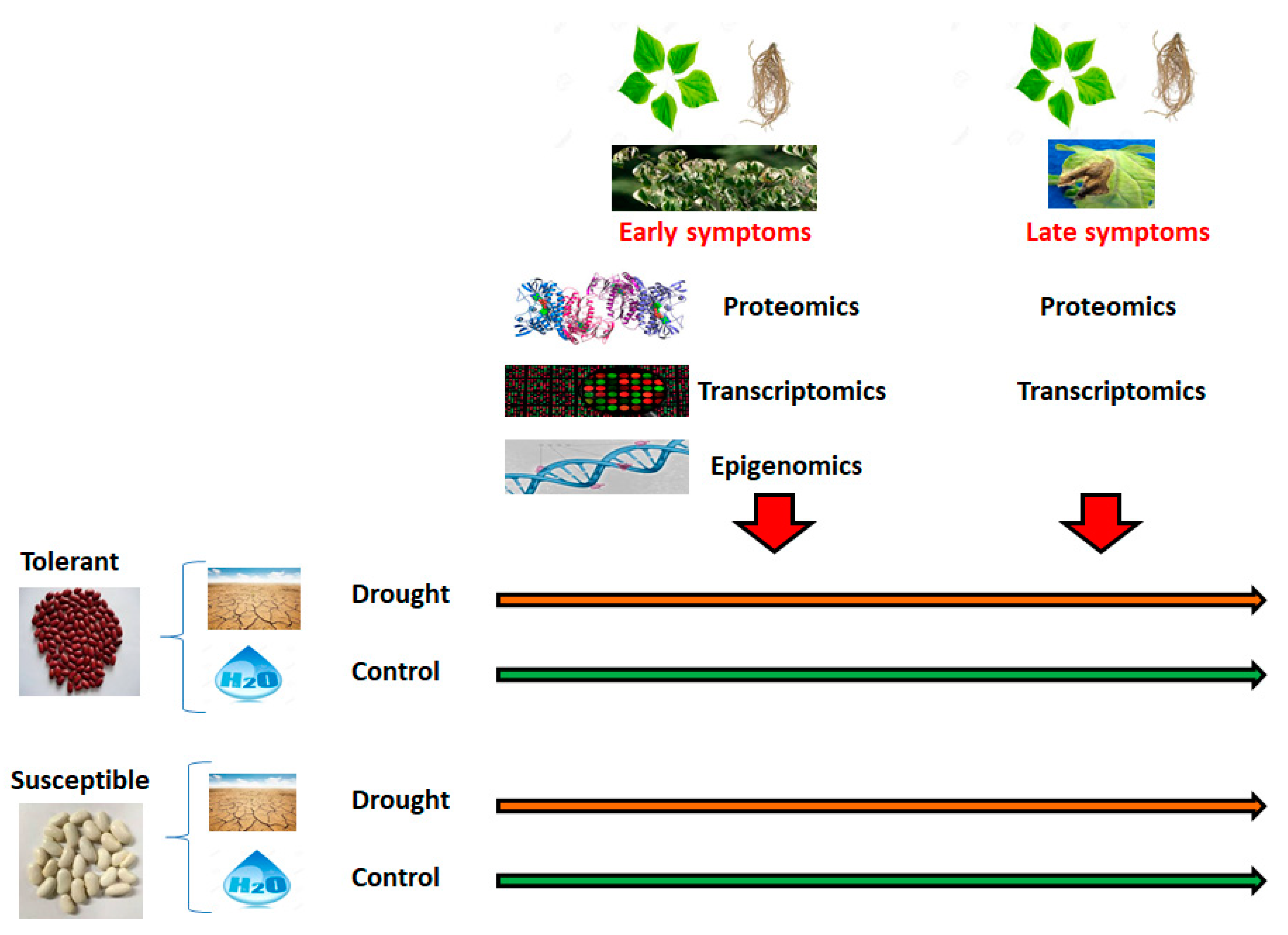
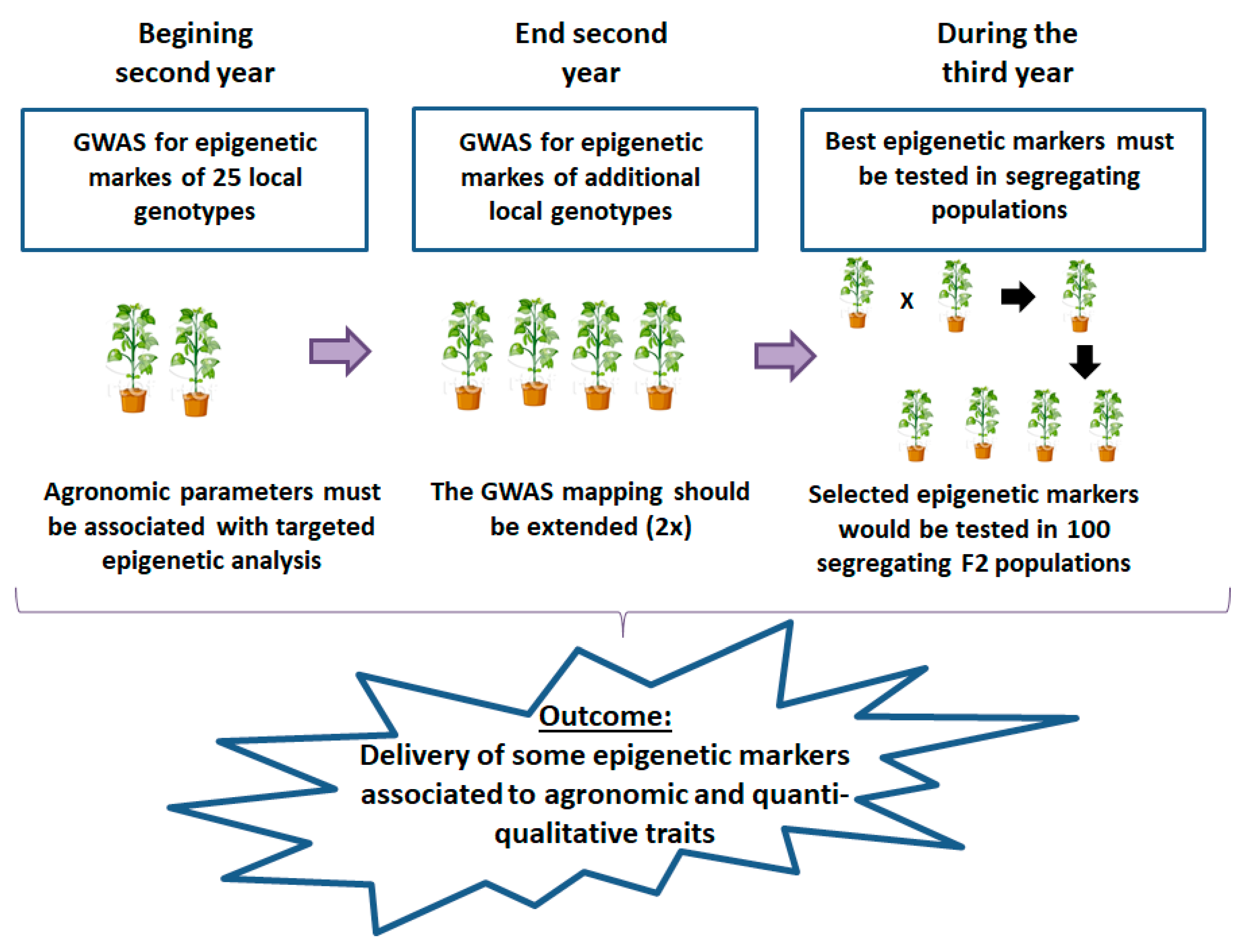
2.1.2. Second Objective: Epigenomic Adaptation to Climate Change
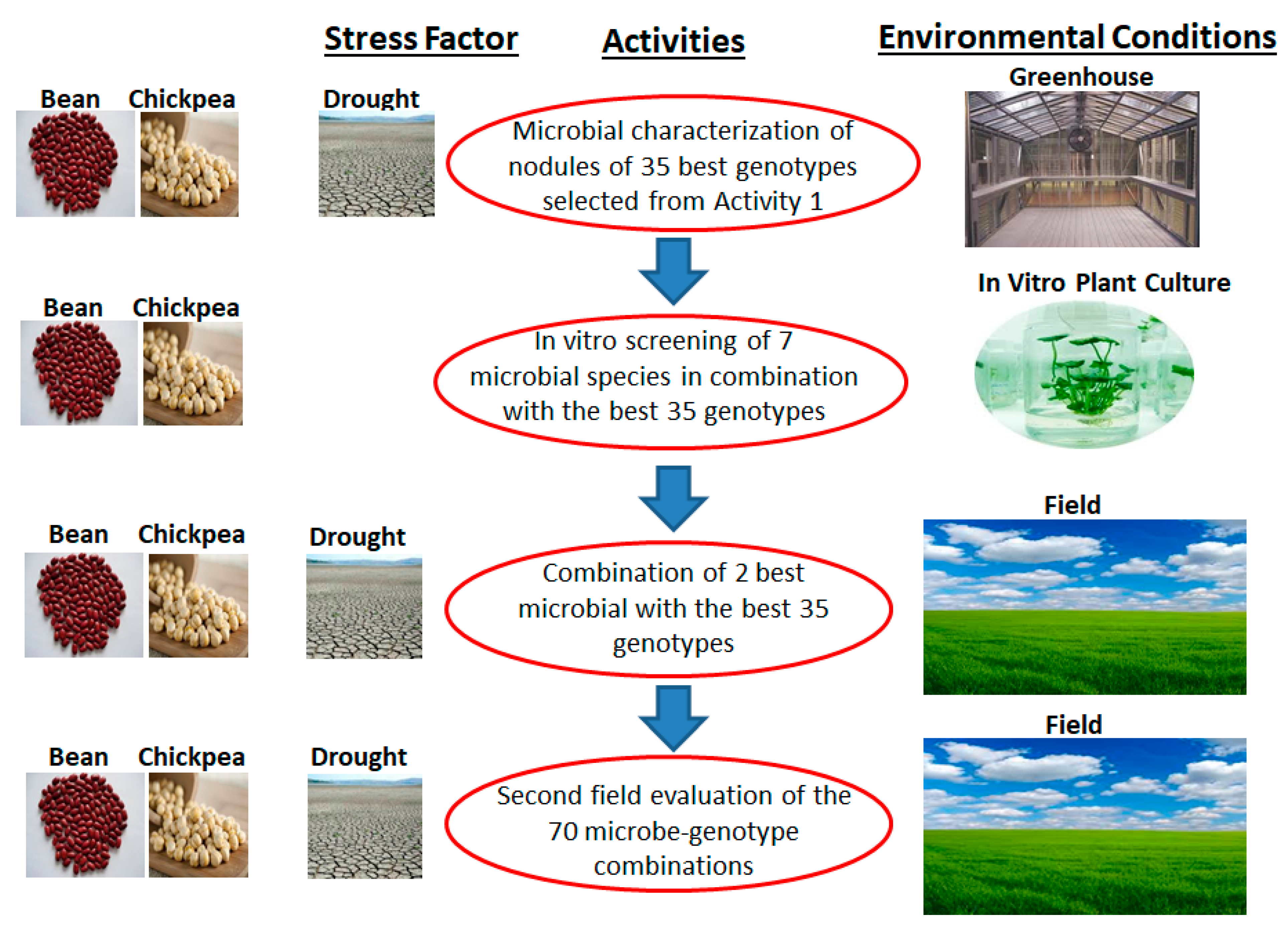
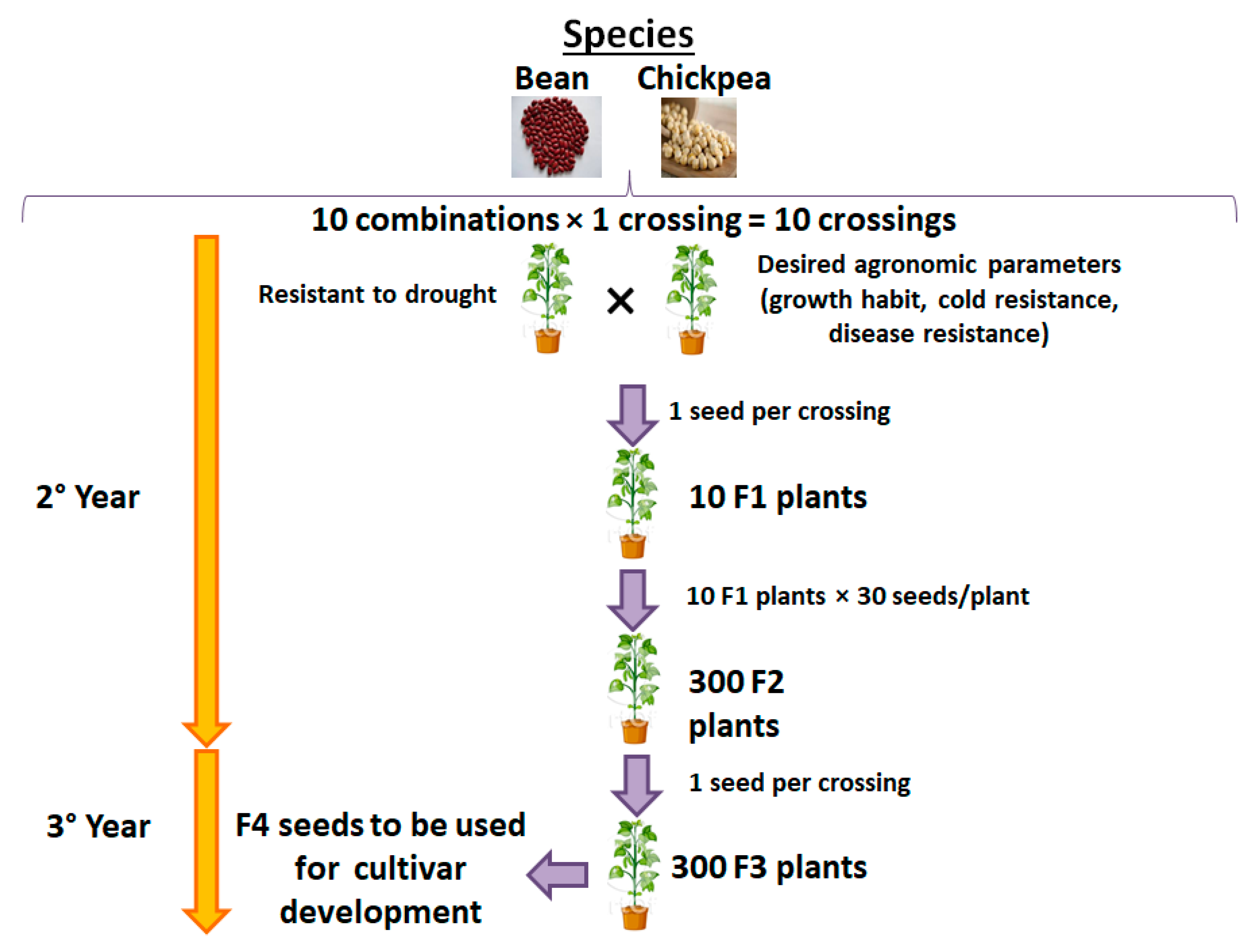
2.1.3. Third Objective: Innovative and Sustainable Biotechnological Solutions to Climate Change
2.1.4. Forth Objective: Information Management, Outcome Dissemination, Stakeholder Involvement
2.2. Deliverables of the Proposed Approach
3. Expected Impacts
Author Contributions
Funding
Conflicts of Interest
References
- FAO. FAOSTAT; Food and Agriculture Organization of the United Nations: Rome, Italy, 2015. [Google Scholar]
- Willett, W.; Rockström, J.; Loken, B.; Springmann, M.; Lang, T.; Vermeulen, S.; Garnett, T.; Tilman, D.; DeClerck, F.; Wood, A.; et al. Food in the Anthropocene: the EAT–Lancet Commission on healthy diets from sustainable food systems. Lancet 2019, 393, 447–492. [Google Scholar] [CrossRef]
- Beebe, S.E.; Idupulapati, M.R.; Blair, M.W.; Acosta-Gallegos, J.A. Phenotyping common beans for adaptation to drought. Front. Physiol. 2013, 4, 35. [Google Scholar] [CrossRef]
- Chavas, J.P.; Di Falco, S.; Adinolfi, F.; Capitanio, F. Weather effects and their long-term impact on the distribution of agricultural yields: Evidence from Italy. Eur. Rev. Agric. Econ. 2019, 46, 29–51. [Google Scholar] [CrossRef]
- Bozzola, M.; Massetti, E.; Mendelsohn, R.; Capitanio, F. A Ricardian analysis of the impact of climate change on Italian agriculture. Eur. Rev. Agric. Econ. 2018, 45, 57–79. [Google Scholar] [CrossRef]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Song, C.; Saxena, R.K.; Azam, S.; Yu, S.; Sharpe, A.G.; Cannon, S.; Baek, J.; Rosen, B.D.; Tar’na, B.; et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 2013, 31, 240. [Google Scholar] [CrossRef]
- Maxted, N.; Bennett, S.J. (Eds.) Plant Genetic Resources of Legumes in the Mediterranean; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2001; Volume 39. [Google Scholar]
- Cernay, C.; Ben-Ari, T.; Pelzer, E.; Meynard, J.M.; Makowski, D. Estimating variability in grain legume yields across Europe and the Americas. Sci. Rep. 2015, 5, 11171. [Google Scholar] [CrossRef] [PubMed]
- Briñez, B.; Perseguini, J.M.K.C.; Rosa, J.S.; Bassi, D.; Gonçalves, J.G.R.; Almeida, C.; Paulino, J.F.C.; Blair, M.W.; Chioratto, A.F.; Carbonell, S.A.M.; et al. Mapping QTLs for drought tolerance in a SEA 5 x AND 277 common bean cross with SSRs and SNP markers. Gen. Mol. Biol. 2017, 40, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Jaganathan, D.; Thudi, M.; Kale, S.; Azam, S.; Roorkiwal, M.; Gaur, P.M.; Kishor, P.B.; Nguyen, H.; Sutton, T.; Varshney, R.K. Genotyping-by-sequencing based intra-specific genetic map refines a “QTL-hotspot” region for drought tolerance in chickpea. Mol. Genet. Genom. 2015, 290, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Sivasakthi, K.; Thudi, M.; Tharanya, M.; Kale, S.M.; Kholová, J.; Halime, M.H.; Jaganathan, D.; Baddam, R.; Thirunalasundari, T.; Gaur, P.M.; et al. Plant vigour QTLs co-map with an earlier reported QTL hotspot for drought tolerance while water saving QTLs map in other regions of the chickpea genome. BMC Plant Biol. 2018, 18, 29. [Google Scholar] [CrossRef]
- Polania, J.; Poschenrieder, C.; Idupulapati, R.; Beebe, S. Estimation of phenotypic variability in symbiotic nitrogen fixation ability of common bean under drought stress using 15N natural abundance in grain. Eur. J. Agron. 2016, 79, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Cuellar-Ortiz, S.M.; de la Paz Arrieta-Montiel, M.; Acosta-Gallegos, J.; Covarrubias, A.A. Relationship between carbohydrate partitioning and drought resistance in common bean. Plant Cell Environ. 2008, 31, 1399–1409. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U.; Beckers, G.J.; Langenbach, C.J.; Jaskiewicz, M.R. Priming for enhanced defense. Ann. Rev. Phytopathol. 2015, 53. [Google Scholar] [CrossRef]
- Crampton, M.; Sipathi, V.R.; Hossain, K.; Kalavacharla, V. Analyses of Methylomes Derived from Meso-American Common Bean (Phaseolus vulgaris L.) Using MeDIP-Seq and Whole Genome Sodium Bisulfite-Sequencing. Front. Plant Sci. 2016, 7, 447. [Google Scholar] [CrossRef]
- Kim, K.D.; El Baidouri, M.; Abernathy, B.; Iwata-Otsubo, A.; Chavarro, C.; Gonzales, M.; Libault, M.; Grimwood, J.; Jackson, S.A. A Comparative Epigenomic Analysis of Polyploidy-Derived Genes in Soybean and Common Bean. Plant Physiol. 2015, 168, 1433–1447. [Google Scholar] [CrossRef]
- Richard, M.M.S.; Gratia, A.; Thareau, V.; Kim, K.D.; Balzergue, S.; Joets, J.; Jackson, S.A.; Gefrroy, V. Genomic and epigenomic immunity in common bean: The unusual features of NB-LRR gene family. DNA Res. 2018, 25, 161–172. [Google Scholar] [CrossRef]
- Wu, J.; Wang, L.; Li, L.; Wang, S. De novo assembly of the common bean transcriptome using short reads for the discovery of drought-responsive genes. PLoS ONE 2014, 9, e109262. [Google Scholar]
- Pereira, W.J.; Melo, A.T.D.O.; Coelho, A.S.G.; Rodrigues, F.A.; Mamidi, S.; Alencar, S.A.D.; Lanna, A.C.; Valdisser, P.A.M.R.; Brondani, C.; Nascimento-Júnior, I.R.D.; et al. Genome-wide analysis of the transcriptional response to drought stress in root and leaf of common bean. Genet. Mol. Biol. 2019. [Google Scholar] [CrossRef]
- Zadražnik, T.; Hollung, K.; Egge-Jacobsen, W.; Meglič, V.; Šuštar-Vozlič, J. Differential proteomic analysis of drought stress response in leaves of common bean (Phaseolus vulgaris L.). J. Proteom. 2013, 78, 254–272. [Google Scholar]
- Zadražnik, T.; Egge-Jacobsen, W.; Meglič, V.; Šuštar-Vozlič, J. Proteomic analysis of common bean stem under drought stress using in-gel stable isotope labeling. J. Plant Physiol. 2017, 209, 42–50. [Google Scholar] [CrossRef]
- Y Teran, J.C.B.M.; Konzen, E.R.; Palkovic, A.; Tsai, S.M.; Rao, I.M.; Beebe, S.; Gepts, P. Effect of drought stress on the genetic architecture of photosynthate allocation and remobilization in pods of common bean (Phaseolus vulgaris L.), a key species for food security. BMC Plant Biol. 2019, 19, 171. [Google Scholar] [CrossRef] [PubMed]
- Villordo-Pineda, E.; González-Chavira, M.M.; Giraldo-Carbajo, P.; Acosta-Gallegos, J.A.; Caballero-Pérez, J. Identification of novel drought-tolerant-associated SNPs in common bean (Phaseolus vulgaris). Front. Plant Sci. 2015, 6, 546. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.J.; Blair, M.W. Genotyping by Sequencing and Genome–Environment Associations in Wild Common Bean Predict Widespread Divergent Adaptation to Drought. Front. Plant Sci. 2018, 9, 128. [Google Scholar] [CrossRef] [PubMed]
- Garg, R.; Shankar, R.; Thakkar, B.; Kudapa, H.; Krishnamurthy, L.; Mantri, N.; Varshney, R.K.; Bhatia, S.; Jain, M. Transcriptome analyses reveal genotype-and developmental stage-specific molecular responses to drought and salinity stresses in chickpea. Sci. Rep. 2016, 6, 19228. [Google Scholar] [CrossRef] [PubMed]
- Mashaki, K.M.; Garg, V.; Ghomi, A.A.N.; Kudapa, H.; Chitikineni, A.; Nezhad, K.Z.; Yamchi, A.; Soltanloo, H.; Varshney, R.K.; Thudi, M. RNA-Seq analysis revealed genes associated with drought stress response in kabuli chickpea (Cicer arietinum L.). PLoS ONE 2016, 13, e0199774. [Google Scholar] [CrossRef]
- Çevik, S.; Akpinar, G.; Yildizli, A.; Kasap, M.; Karaosmanoğlu, K.; Ünyayar, S. Comparative physiological and leaf proteome analysis between drought-tolerant chickpea Cicer reticulatum and drought-sensitive chickpea C. arietinum. J. Biosci. 2019, 44, 20. [Google Scholar] [CrossRef]
- Varshney, R.K.; Thudi, M.; Nayak, S.N.; Gaur, P.M.; Kashiwagi, J.; Krishnamurthy, L.; Jaganathan, D.; Koppolu, J.; Bohra, A.; Tripathi, S.; et al. Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.). Theor. Appl. Genet. 2014, 127, 445–462. [Google Scholar] [CrossRef]
- Kale, S.M.; Jaganathan, D.; Ruperao, P.; Chen, C.; Punna, R.; Kudapa, H.; Thudi, M.; Roorkiwal, M.; Katta, M.A.; Doddamani, D.; et al. Prioritization of candidate genes in “QTL-hotspot” region for drought tolerance in chickpea (Cicer arietinum L.). Sci. Rep. 2015, 5, 15296. [Google Scholar] [CrossRef]
- Li, Y.; Ruperao, P.; Batley, J.; Edwards, D.; Khan, T.; Colmer, T.D.; Pang, J.; Siddique, K.H.M.; Sutton, T. Investigating drought tolerance in chickpea using genome-wide association mapping and genomic selection based on whole-genome resequencing data. Front. Plant Sci. 2018, 9, 190. [Google Scholar] [CrossRef]
- Radin, J.W.; Ackerson, R. Water relations of cotton plants under nitrogen deficiency: III. Stomatal conductance, photosynthesis, and abscisic acid accumulation during drought. Plant Physiol. 1981, 67, 115–119. [Google Scholar] [CrossRef]
- Radin, J.W.; Boyer, J.S. Control of leaf expansion by nitrogen nutrition in sunflower plants. Plant Physiol. 1982, 69, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Staudinger, C.; Mehmeti-Tershani, V.; Gil-Quintana, E.; Gonzalez, E.M.; Hofhansl, F.; Bachmann, G.; Wienkoop, S. Evidence for a rhizobia-induced drought stress response strategy in Medicago truncatula. J. Proteom. 2016, 136, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Giovino, A.; Bertolini, E.; Fileccia, V.; Al_hassan, M.; Labra, M.; Martinelli, F. Transcriptome Analysis of Phoenix canariensis Chabaud in Response to Rhynchophorus ferrugineus Olivier Attacks. Front. Plant Sci. 2015, 6, 817. [Google Scholar] [CrossRef]
- Tosetti, R.; Martinelli, F.; Tonutti, P. Metabolomics Approach to Studying Minimally Processed Peach (Prunus persica) Fruit. Acta Hortic. 2010, 934, 1017–1022. [Google Scholar] [CrossRef]
- Martinelli, F.; Ibanez, A.M.; Reagan, R.L.; Davino, S.; Dandekar, A.M. Stress responses in citrus peel: Comparative analysis of host responses to Huanglongbing disease and puffing disorder. Sci. Hortic. 2015, 192, 409–420. [Google Scholar] [CrossRef]
- Martinelli, F.; Reagan, R.L.; Uratsu, S.L.; Phu, M.L.; Albrecht, U.; Zhao, W.; Davis, C.E.; Bowman, K.D.; Dandekar, A.M. Gene regulatory networks elucidating huanglongbing disease mechanisms. PLoS ONE 2013, 8, e74256. [Google Scholar] [CrossRef]
- Balan, B.; Marra, F.P.; Caruso, T.; Martinelli, F. Transcriptomic responses to biotic stresses in Malus x domestica: A meta-analysis study. Sci. Rep. 2018, 8, 1970. [Google Scholar] [CrossRef]
- Schuster, A.M.; Davies, E. Ribonucleic acid and protein metabolism in pea epicotyls: I. The aging process. Plant Physiol. 1983, 73, 809–816. [Google Scholar] [CrossRef]
| Bean | ||
| Approach | Results | Reference |
| RNA-seq | Identification of genes involved in drought stress determining the differentially expressed genes between terminal drought and optimal irrigation treatments and between tolerant and sensitive genotypes, belonging to the cellular pathways plant hormone signal transduction, protein processing in endoplasmic reticulum, ribosome production, starch and sucrose metabolism and purine metabolism. | [19] |
| RNA-seq | Characterization of genes related to the response to drought stress in leaf and root tissue of drought-susceptible and tolerant genotypes with a predominance of genes involved in oxidative stress, response to stimulus and kinase activity. | [20] |
| Proteomics | Identification of drought-responsive proteins in leaves of two cultivars differing in their response to drought that could be classified into functional categories that include energy metabolism, photosynthesis, ATP interconversion, protein synthesis and proteolysis, stress and defence related proteins. | [21] |
| Proteomics | Plants exposed to drought for 17 days and control plants at the same developmental stage were included in quantitative proteomic analysis. Quantified proteins were grouped into several functional groups, mainly into energy, metabolism, photosynthesis, proteolysis, protein synthesis and proteins related to defence and stress. | [22] |
| QTLs | Identification of QTLs was performed crossing two cultivars (drought sensitive and tolerant), obtaining and genotyping a recombinant inbred population and measuring fourteen traits in both well and water stress conditions. 22 QTLs associated with chlorophyll, leaf and stem fresh biomass, leaf biomass dry weight, leaf temperature, number of pods per plant, number of seeds per plant, seed weight, days to flowering, dry pod weight and total yield were identified under drought stressing conditions. | [10] |
| QTLs | Identification of QTLs was performed crossing two cultivars (drought sensitive and tolerant), obtaining and genotyping a recombinant inbred population and measuring the inheritance of pod harvest index under well-watered conditions and terminal and intermittent drought stress. 7 QTLs associated with the partitioning of pod biomass to seed biomass were identified with drought. | [23] |
| SNPs | Two drought-tolerant parental lines were used to generate a recombinant inbred population characterized by 169SNPs. 83 SNPs were significant associated with quantified phenotypes under drought conditions, specially days to flowering and seed biomass. Thirty-seven out of the 83 SNPs were annotated to a gene with a potential function related to drought tolerance, such as starch or proline biosynthesis. | [24] |
| SNPs | A total of 22,845 SNPs was found across eighty-six American wild bean lines. Allelic associations with a bioclimatic-based drought index were calculated. 115 SNPs were associated with the bioclimatic-based drought index. A gene coding for an ankyrin repeat-containing protein and a phototropic-responsive NPH3 gene were identified as potential candidates involved in drought responses. | [25] |
| Chickpea | ||
| Approach | Results | Reference |
| RNA-seq | RNA-seq of the roots of drought and salinity related genotypes was carried out under control and stress conditions at vegetative and/or reproductive stages. A total of 4954 genes exclusively regulated in drought-tolerant genotypes were identified, including genes coding for enzymes involved in metabolic pathways, photosynthesis, lipid metabolism, generation of precursor metabolites/energy, protein modification, redox homeostasis, and cell wall component biogenesis. | [26] |
| RNA-seq | RNA-seq of roots and shoots of two contrasting chickpea genotypes at early flowering stage under controlled and drought conditions. A total of 4572 differentially expressed genes were identified. Of these, 261 and 169 drought stress responsive genes were identified in the shoots and the roots, respectively, and 17 genes were common in the shoots and the roots. Several genes coding for proteins involved in response to stress, defense response, and response to stimulus were specific from the tolerant genotype under drought stressing conditions. | [27] |
| Proteomics | Leaf proteome analysis between drought-tolerant and drought-sensitive chickpeas under controlled and drought conditions. A total of 24 differently expressed proteins were identified in response to drought in both plants. The proteins involved in photosynthesis and energy mechanisms were up-regulated in tolerant chickpea but down-regulated in sensitive chickpea under drought, suggesting that photosynthesis capacity is higher in the tolerant chickpea. | [28] |
| QTLs | Identification of QTLs was performed crossing four cultivars, obtaining and genotyping two recombinant inbred populations and measuring a total of 20 drought root traits in several seasons and locations under field conditions. Analysis of extensive genotypic and precise phenotypic data revealed 45 QTLs. Some QTLs for several drought tolerance traits appeared clustered. Among these clusters, one cluster harboring most QTLs for 12 traits and explaining most phenotypic variation was identified and referred as “QTL-hotspot.” | [29] |
| QTLs | QTL analysis was used to identify candidate genes in the “QTL-hotspot” region for drought tolerance in chickpeas and was performed crossing two cultivars (drought sensitive and tolerant), obtaining and genotyping a recombinant inbred population and measuring 17 drought tolerance related traits obtained over five seasons and five locations under field conditions. Data split the “QTL-hotspot” region into two subregions namely “QTL-hotspot_a” (harboring 15 genes) and “QTL-hotspot_b” (harboring 11 genes). Functional validation using qRT-PCR indicated four promising candidate genes having functional implications for drought tolerance in chickpea. | [30] |
| QTLs | Identification of QTLs was performed crossing two cultivars (drought sensitive and tolerant), obtaining and genotyping a recombinant inbred population and measuring traits contributing to plant water use under well-watered conditions. Twenty-one QTLs were identified for plant vigor and canopy conductance traits, some of them located in the “QTL-hotspot” region involved in drought tolerance. | [12] |
| SNPs | Identification of 828 novel SNPs in a chickpea recombinant inbred line mapping population. Analysis using the genetic map along with phenotyping data for 20 traits collected over seven seasons under field conditions identified 49 SNP markers in the “QTL-hotspot” region, which harbors most drought tolerance QTL in chickpea. | [11] |
| SNPs | Sequencing of 132 chickpea varieties and advanced breeding lines allowed the identification of more than 144,000 SNPs. Thirteen yield and yield-related traits were correlated with identified SNPs in three drought-prone environments to find putative genes involved in drought tolerance, such as genes coding for auxin production proteins, p-glycoproteins, and nodulin transporters. | [31] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinelli, F.; Ollero, F.J.; Giovino, A.; Perrone, A.; Bekki, A.; Sikora, S.; El Nabbout, R.; Bouhadida, M.; Yucel, D.; Bazzicalupo, M.; et al. Proposed Research for Innovative Solutions for Chickpeas and Beans in a Climate Change Scenario: The Mediterranean Basin. Sustainability 2020, 12, 1315. https://doi.org/10.3390/su12041315
Martinelli F, Ollero FJ, Giovino A, Perrone A, Bekki A, Sikora S, El Nabbout R, Bouhadida M, Yucel D, Bazzicalupo M, et al. Proposed Research for Innovative Solutions for Chickpeas and Beans in a Climate Change Scenario: The Mediterranean Basin. Sustainability. 2020; 12(4):1315. https://doi.org/10.3390/su12041315
Chicago/Turabian StyleMartinelli, Federico, Francisco Javier Ollero, Antonio Giovino, Anna Perrone, Abdelkader Bekki, Sanja Sikora, Rania El Nabbout, Mariem Bouhadida, Derya Yucel, Marco Bazzicalupo, and et al. 2020. "Proposed Research for Innovative Solutions for Chickpeas and Beans in a Climate Change Scenario: The Mediterranean Basin" Sustainability 12, no. 4: 1315. https://doi.org/10.3390/su12041315
APA StyleMartinelli, F., Ollero, F. J., Giovino, A., Perrone, A., Bekki, A., Sikora, S., El Nabbout, R., Bouhadida, M., Yucel, D., Bazzicalupo, M., Mengoni, A., & Pérez-Montaño, F. (2020). Proposed Research for Innovative Solutions for Chickpeas and Beans in a Climate Change Scenario: The Mediterranean Basin. Sustainability, 12(4), 1315. https://doi.org/10.3390/su12041315










