Role of Microorganisms in the Remediation of Wastewater in Floating Treatment Wetlands: A Review
Abstract
1. Introduction
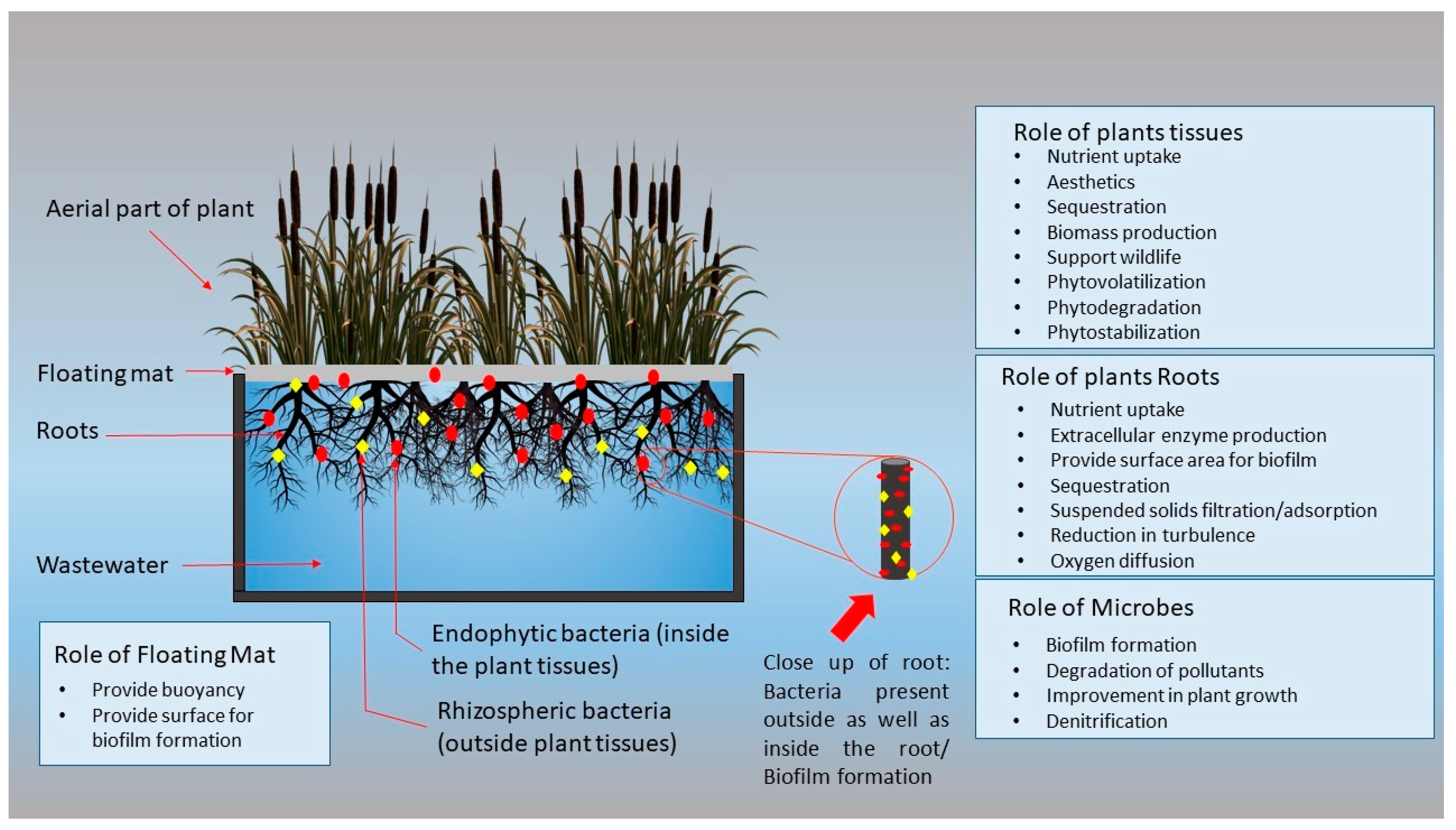
2. Mechanism of FTWs
3. Important Components of FTWs
3.1. Growth Media
3.2. Buoyancy
3.3. Plants
3.4. Bacterial Biofilm
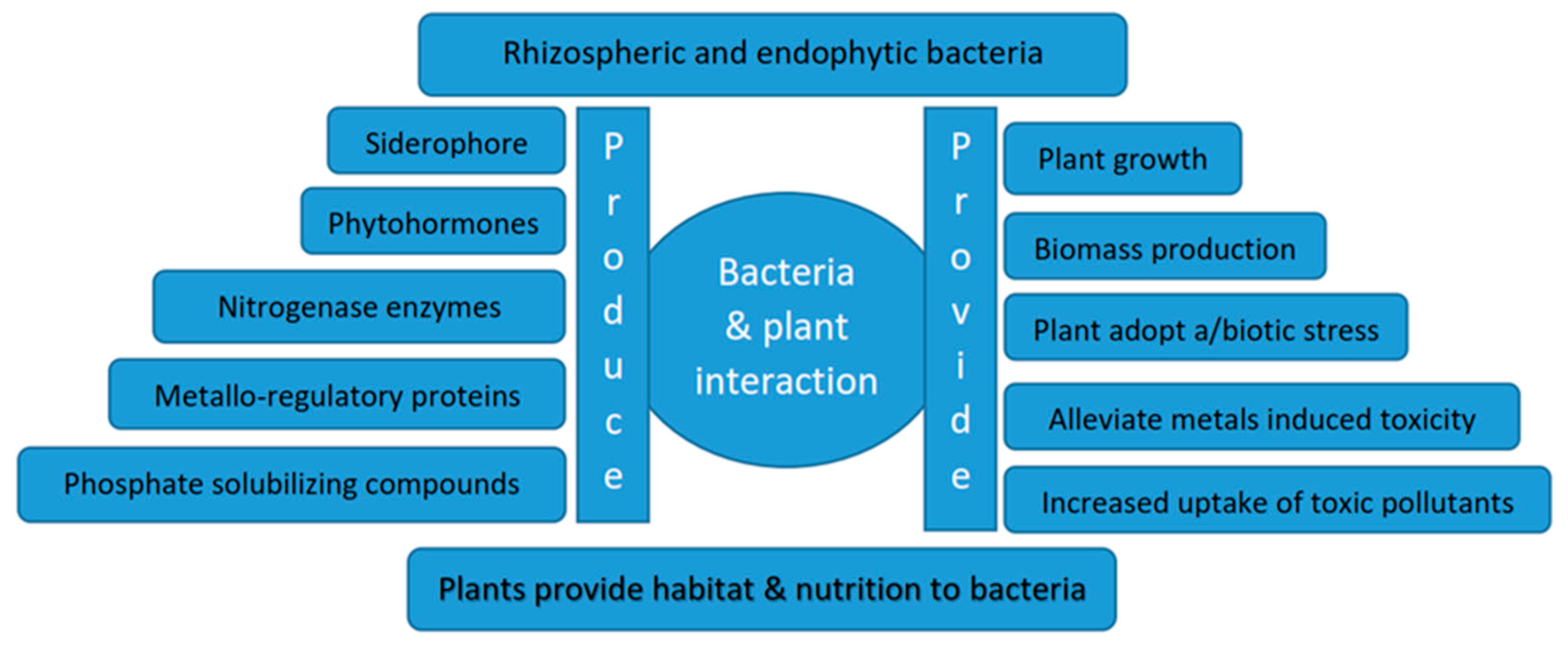
4. Microorganisms
4.1. Role of Endophytes
4.2. Role of Rhizospheric Bacteria
5. Role of Bacteria in Pollutant Removal Process
5.1. Nitrogen Fixation
5.2. Degradation of Organic Pollutants
5.3. Removal of Heavy Metals
5.4. Metal Biosorption and Bioaccumulation
6. Role of Fungi
7. Role of Inoculated Bacteria
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vymazal, J. Constructed wetlands for treatment of industrial wastewaters: A review. Ecol. Eng. 2014, 73, 724–751. [Google Scholar] [CrossRef]
- Stefanakis, A.I. The role of constructed wetlands as green infrastructure for sustainable urban water management. Sustainability 2019, 11, 6981. [Google Scholar] [CrossRef]
- Calheiros, C.S.; Castro, P.M.; Gavina, A.; Pereira, R. Toxicity abatement of wastewaters from tourism units by constructed wetlands. Water 2019, 11, 2623. [Google Scholar] [CrossRef]
- Riva, V.; Mapelli, F.; Syranidou, E.; Crotti, E.; Choukrallah, R.; Kalogerakis, N.; Borin, S. Root bacteria recruited by Phragmites australis in constructed wetlands have the potential to enhance azo-dye phytodepuration. Microorganisms 2019, 7, 384. [Google Scholar] [CrossRef]
- Donoso, N.; van Oirschot, D.; Kumar Biswas, J.; Michels, E.; Meers, E. Impact of aeration on the removal of organic matter and nitrogen compounds in constructed wetlands treating the liquid fraction of piggery manure. Appl. Sci. 2019, 9, 4310. [Google Scholar] [CrossRef]
- Kujala, K.; Karlsson, T.; Nieminen, S.; Ronkanen, A.-K. Design parameters for nitrogen removal by constructed wetlands treating mine waters and municipal wastewater under Nordic conditions. Sci. Total Environ. 2019, 662, 559–570. [Google Scholar] [CrossRef]
- Anawar, H.M.; Ahmed, G.; Strezov, V. Long-term Performance and Feasibility of Using Constructed Wetlands for Treatment of Emerging and Nanomaterial Contaminants in Municipal and Industrial Wastewater. In Emerging and Nanomaterial Contaminants in Wastewater; Elsevier: Amsterdam, The Netherlands, 2019; pp. 63–81. [Google Scholar]
- Smith, E.L.; Kellman, L.; Brenton, P. Restoration of on-farm constructed wetland systems used to treat agricultural wastewater. J. Agric. Sci. 2019, 11, 1–12. [Google Scholar] [CrossRef]
- Jones, T.G.; Willis, N.; Gough, R.; Freeman, C. An experimental use of floating treatment wetlands (FTWs) to reduce phytoplankton growth in freshwaters. Ecol. Eng. 2017, 99, 316–323. [Google Scholar] [CrossRef]
- Headley, T.; Tanner, C. Floating Treatment Wetlands: An Innovative Option for Stormwater Quality Applications. In Proceedings of the 11th International Conference on Wetland Systems for Water Pollution Control, Indore, India, 1–7 November 2008. [Google Scholar]
- Tanner, C.C.; Headley, T.R. Components of floating emergent macrophyte treatment wetlands influencing removal of stormwater pollutants. Ecol. Eng. 2011, 37, 474–486. [Google Scholar] [CrossRef]
- Hubbard, R.; Anderson, W.; Newton, G.; Ruter, J.; Wilson, J. Plant growth and elemental uptake by floating vegetation on a single-stage swine wastewater lagoon. Trans. ASABE 2011, 54, 837–845. [Google Scholar] [CrossRef]
- Shahid, M.J.; Arslan, M.; Ali, S.; Siddique, M.; Afzal, M. Floating wetlands: A sustainable tool for wastewater treatment. CLEAN–Soil Air Water 2018, 46, 1800120. [Google Scholar] [CrossRef]
- Ladislas, S.; Gerente, C.; Chazarenc, F.; Brisson, J.; Andres, Y. Floating treatment wetlands for heavy metal removal in highway stormwater ponds. Ecol. Eng. 2015, 80, 85–91. [Google Scholar] [CrossRef]
- Chen, Z.; Cuervo, D.P.; Müller, J.A.; Wiessner, A.; Köser, H.; Vymazal, J.; Kästner, M.; Kuschk, P. Hydroponic root mats for wastewater treatment—a review. Environ. Sci. Pollut. Res. 2016, 23, 15911–15928. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Tripathi, B. Effect of aeration and mixed culture of Eichhornia crassipes and Salvinia natans on removal of wastewater pollutants. Ecol. Eng. 2014, 62, 48–53. [Google Scholar] [CrossRef]
- Li, W.; Li, Z. In situ nutrient removal from aquaculture wastewater by aquatic vegetable Ipomoea aquatica on floating beds. Water Sci. Technol. 2009, 59, 1937–1943. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, J.; Cui, N.; Dai, Y.; Kong, L.; Wu, J.; Cheng, S. Enhancing the water purification efficiency of a floating treatment wetland using a biofilm carrier. Environ. Sci. Pollut. Res. 2016, 23, 7437–7443. [Google Scholar] [CrossRef]
- Pavlineri, N.; Skoulikidis, N.T.; Tsihrintzis, V.A. Constructed floating wetlands: A review of research, design, operation and management aspects, and data meta-analysis. Chem. Eng. J. 2017, 308, 1120–1132. [Google Scholar] [CrossRef]
- Shahid, M.J.; Arslan, M.; Siddique, M.; Ali, S.; Tahseen, R.; Afzal, M. Potentialities of floating wetlands for the treatment of polluted water of river Ravi, Pakistan. Ecol. Eng. 2019, 133, 167–176. [Google Scholar] [CrossRef]
- Borne, K.E.; Fassman, E.A.; Tanner, C.C. Floating treatment wetland retrofit to improve stormwater pond performance for suspended solids, copper and zinc. Ecol. Eng. 2013, 54, 173–182. [Google Scholar] [CrossRef]
- Faulwetter, J.; Burr, M.D.; Cunningham, A.B.; Stewart, F.M.; Camper, A.K.; Stein, O.R. Floating treatment wetlands for domestic wastewater treatment. Water Sci. Technol. 2011, 64, 2089–2095. [Google Scholar] [CrossRef]
- Chen, Z.; Kuschk, P.; Paschke, H.; Kästner, M.; Müller, J.A.; Köser, H. Treatment of a sulfate-rich groundwater contaminated with perchloroethene in a hydroponic plant root mat filter and a horizontal subsurface flow constructed wetland at pilot-scale. Chemosphere 2014, 117, 178–184. [Google Scholar] [CrossRef]
- Borne, K.E.; Fassman-Beck, E.A.; Tanner, C.C. Floating treatment wetland influences on the fate of metals in road runoff retention ponds. Water Res. 2014, 48, 430–442. [Google Scholar] [CrossRef]
- Afzal, M.; Rehman, K.; Shabir, G.; Tahseen, R.; Ijaz, A.; Hashmat, A.J.; Brix, H. Large-scale remediation of oil-contaminated water using floating treatment wetlands. npj Clean Water 2019, 2, 3. [Google Scholar] [CrossRef]
- Ijaz, A.; Iqbal, Z.; Afzal, M. Remediation of sewage and industrial effluent using bacterially assisted floating treatment wetlands vegetated with Typha domingensis. Water Sci. Technol. 2016, 74, 2192–2201. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guo, R. Comparison of nitrogen removal in floating treatment wetlands constructed with Phragmites australis and acorus calamus in a cold temperate zone. Water Air Soil Pollut. 2017, 228, 132. [Google Scholar] [CrossRef]
- Nawaz, N.; Ali, S.; Shabir, G.; Rizwan, M.; Shakoor, M.B.; Shahid, M.J.; Afzal, M.; Arslan, M.; Hashem, A.; Abd_Allah, E.F. Bacterial augmented floating treatment wetlands for efficient treatment of synthetic textile dye wastewater. Sustainability 2020, 12, 3731. [Google Scholar] [CrossRef]
- Fahid, M.; Ali, S.; Shabir, G.; Rashid Ahmad, S.; Yasmeen, T.; Afzal, M.; Arslan, M.; Hussain, A.; Hashem, A.; Abd Allah, E.F. Cyperus laevigatus L. enhances diesel oil remediation in synergism with bacterial inoculation in floating treatment wetlands. Sustainability 2020, 12, 2353. [Google Scholar] [CrossRef]
- Zhang, C.-B.; Liu, W.-L.; Pan, X.-C.; Guan, M.; Liu, S.-Y.; Ge, Y.; Chang, J. Comparison of effects of plant and biofilm bacterial community parameters on removal performances of pollutants in floating island systems. Ecol. Eng. 2014, 73, 58–63. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Sample, D.J.; Bell, C. Vegetation effects on floating treatment wetland nutrient removal and harvesting strategies in urban stormwater ponds. Sci. Total Environ. 2014, 499, 384–393. [Google Scholar] [CrossRef]
- Afzal, M.; Shabir, G.; Tahseen, R.; Islam, E.U.; Iqbal, S.; Khan, Q.M.; Khalid, Z.M. Endophytic Burkholderia sp. strain PsJN improves plant growth and phytoremediation of soil irrigated with textile effluent. CLEAN–Soil Air Water 2014, 42, 1304–1310. [Google Scholar] [CrossRef]
- Arslan, M.; Imran, A.; Khan, Q.M.; Afzal, M. Plant–bacteria partnerships for the remediation of persistent organic pollutants. Environ. Sci. Pollut. Res. 2017, 24, 4322–4336. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.-B.; Islam, K.; Marimon, Z.; Wanielista, M.P. Assessing biological and chemical signatures related to nutrient removal by floating islands in stormwater mesocosms. Chemosphere 2012, 88, 736–743. [Google Scholar] [CrossRef]
- Khan, S.; Afzal, M.; Iqbal, S.; Khan, Q.M. Plant–bacteria partnerships for the remediation of hydrocarbon contaminated soils. Chemosphere 2013, 90, 1317–1332. [Google Scholar] [CrossRef]
- Hussain, I.; Aleti, G.; Naidu, R.; Puschenreiter, M.; Mahmood, Q.; Rahman, M.M.; Wang, F.; Shaheen, S.; Syed, J.H.; Reichenauer, T.G. Microbe and plant assisted-remediation of organic xenobiotics and its enhancement by genetically modified organisms and recombinant technology: A review. Sci. Total Environ. 2018, 628, 1582–1599. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Ren, L.; Wu, Q. Epiphytic bacterial communities on two common submerged macrophytes in Taihu Lake: Diversity and host-specificity. Chin. J. Oceanol. Limnol. 2012, 30, 237–247. [Google Scholar] [CrossRef]
- Headley, T.; Tanner, C. Constructed wetlands with floating emergent macrophytes: An innovative stormwater treatment technology. Crit. Rev. Environ. Sci. Technol. 2012, 42, 2261–2310. [Google Scholar] [CrossRef]
- Gao, L.; Zhou, W.; Huang, J.; He, S.; Yan, Y.; Zhu, W.; Wu, S.; Zhang, X. Nitrogen removal by the enhanced floating treatment wetlands from the secondary effluent. Bioresour. Technol. 2017, 234, 243–252. [Google Scholar] [CrossRef]
- Jun-Xing, Y.; Yong, L.; Zhi-Hong, Y. Root-induced changes of pH, Eh, Fe (II) and fractions of Pb and Zn in rhizosphere soils of four wetland plants with different radial oxygen losses. Pedosphere 2012, 22, 518–527. [Google Scholar]
- Hussain, F.; Tahseen, R.; Arslan, M.; Iqbal, S.; Afzal, M. Removal of hexadecane by hydroponic root mats in partnership with alkane-degrading bacteria: Bacterial augmentation enhances system’s performance. Int. J. Environ. Sci. Technol. 2019, 16, 4611–4620. [Google Scholar] [CrossRef]
- Dutta, D.; Puzari, K.C.; Gogoi, R.; Dutta, P. Endophytes: Exploitation as a tool in plant protection. Braz. Arch. Biol Technol. 2014, 57, 621–629. [Google Scholar] [CrossRef]
- White, S.A.; Cousins, M.M. Floating treatment wetland aided remediation of nitrogen and phosphorus from simulated stormwater runoff. Ecol. Eng. 2013, 61, 207–215. [Google Scholar] [CrossRef]
- Wang, P.; Jeelani, N.; Zuo, J.; Zhang, H.; Zhao, D.; Zhu, Z.; Leng, X.; An, S. Nitrogen removal during the cold season by constructed floating wetlands planted with Oenanthe javanica. Mar. Freshw. Res. 2018, 69, 635–647. [Google Scholar] [CrossRef]
- Govarthanan, M.; Mythili, R.; Selvankumar, T.; Kamala-Kannan, S.; Rajasekar, A.; Chang, Y.-C. Bioremediation of heavy metals using an endophytic bacterium Paenibacillus sp. RM isolated from the roots of Tridax procumbens. 3 Biotech 2016, 6, 242. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Kang, S.H.; Mulchandani, A.; Chen, W. Bioremediation: Environmental clean-up through pathway engineering. Curr. Opin. Biotechnol. 2008, 19, 437–444. [Google Scholar] [CrossRef]
- Borne, K.E. Floating treatment wetland influences on the fate and removal performance of phosphorus in stormwater retention ponds. Ecol. Eng. 2014, 69, 76–82. [Google Scholar] [CrossRef]
- Urakawa, H.; Dettmar, D.L.; Thomas, S. The uniqueness and biogeochemical cycling of plant root microbial communities in a floating treatment wetland. Ecol. Eng. 2017, 108, 573–580. [Google Scholar] [CrossRef]
- Ashraf, S.; Afzal, M.; Naveed, M.; Shahid, M.; Ahmad Zahir, Z. Endophytic bacteria enhance remediation of tannery effluent in constructed wetlands vegetated with Leptochloa fusca. Int. J. Phytorem. 2018, 20, 121–128. [Google Scholar] [CrossRef]
- Ahsan, M.T.; Najam-ul-Haq, M.; Idrees, M.; Ullah, I.; Afzal, M. Bacterial endophytes enhance phytostabilization in soils contaminated with uranium and lead. Int. J. Phytorem. 2017, 19, 937–946. [Google Scholar] [CrossRef]
- Afzal, M.; Yousaf, S.; Reichenauer, T.G.; Sessitsch, A. Ecology of alkane-degrading bacteria and their interaction with the plant. Mol. Microb. Ecol. Rhizosphere 2013, 2, 975–989. [Google Scholar]
- Billore, S.; Sharma, J. Treatment performance of artificial floating reed beds in an experimental mesocosm to improve the water quality of river Kshipra. Water Sci. Technol. 2009, 60, 2851–2859. [Google Scholar] [CrossRef]
- Van de Moortel, A.M.; Meers, E.; De Pauw, N.; Tack, F.M. Effects of vegetation, season and temperature on the removal of pollutants in experimental floating treatment wetlands. Water Air Soil Pollut. 2010, 212, 281–297. [Google Scholar] [CrossRef]
- Tranvik, L.J. Degradation of Dissolved Organic Matter in Humic Waters by Bacteria. In Aquatic Humic Substances; Springer: Berlin/Heidelberg, Germany, 1998; pp. 259–283. [Google Scholar]
- Zhao, F.; Xi, S.; Yang, X.; Yang, W.; Li, J.; Gu, B.; He, Z. Purifying eutrophic river waters with integrated floating island systems. Ecol. Eng. 2012, 40, 53–60. [Google Scholar] [CrossRef]
- Cao, W.; Zhang, Y. Removal of nitrogen (N) from hypereutrophic waters by ecological floating beds (EFBs) with various substrates. Ecol. Eng. 2014, 62, 148–152. [Google Scholar] [CrossRef]
- Curt, M.; Aguado, P.; Fernandez, J. Nitrogen Absorption by Sparganium Erectum L. and Typha Domingensis (Pers.) Steudel Grown as Floaters. In Proceedings of the International Meeting on Phytodepuration, Lorca, Spain, 20–22 July 2005. [Google Scholar]
- Hu, G.-J.; Zhou, M.; Hou, H.-B.; Zhu, X.; Zhang, W.-H. An ecological floating-bed made from dredged lake sludge for purification of eutrophic water. Ecol. Eng. 2010, 36, 1448–1458. [Google Scholar] [CrossRef]
- Shin, C.J.; Nam, J.M.; Kim, J.G. Floating mat as a habitat of Cicuta virosa, a vulnerable hydrophyte. Landsc. Ecol. Eng. 2015, 11, 111–117. [Google Scholar] [CrossRef]
- Seo, E.-Y.; Kwon, O.-B.; Choi, S.-I.; Kim, J.-H.; Ahn, T.-S. Installation of an artificial vegetating island in oligomesotrophic Lake Paro, Korea. Sci. World J. 2013, 2013, 857670. [Google Scholar] [CrossRef]
- Sun, L.; Liu, Y.; Jin, H. Nitrogen removal from polluted river by enhanced floating bed grown canna. Ecol. Eng. 2009, 35, 135–140. [Google Scholar] [CrossRef]
- Boonsong, K.; Chansiri, M. Domestic wastewater treatment using vetiver grass cultivated with floating platform technique. AU J. Technol. 2008, 12, 73–80. [Google Scholar]
- Li, M.; Wu, Y.-J.; Yu, Z.-L.; Sheng, G.-P.; Yu, H.-Q. Nitrogen removal from eutrophic water by floating-bed-grown water spinach (Ipomoea aquatica Forsk.) with ion implantation. Water Res. 2007, 41, 3152–3158. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, J.; Li, P.; Zhang, J.; Xie, H.; Zhang, B. Nutrient removal in constructed microcosm wetlands for treating polluted river water in northern China. Ecol. Eng. 2011, 37, 560–568. [Google Scholar] [CrossRef]
- Zhao, F.; Yang, W.; Zeng, Z.; Li, H.; Yang, X.; He, Z.; Gu, B.; Rafiq, M.T.; Peng, H. Nutrient removal efficiency and biomass production of different bioenergy plants in hypereutrophic water. Biomass Bioenergy 2012, 42, 212–218. [Google Scholar] [CrossRef]
- Wu, Q.-T.; Gao, T.; Zeng, S.; Chua, H. Plant-biofilm oxidation ditch for in situ treatment of polluted waters. Ecol. Eng. 2006, 28, 124–130. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Sample, D.J. Assessment of the nutrient removal effectiveness of floating treatment wetlands applied to urban retention ponds. J. Environ. Manag. 2014, 137, 23–35. [Google Scholar] [CrossRef]
- Kadlec, R.H.; Wallace, S. Treatment Wetlands; CRC press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Li, H.; Zhao, H.-P.; Hao, H.-L.; Liang, J.; Zhao, F.-L.; Xiang, L.-C.; Yang, X.-E.; He, Z.-L.; Stoffella, P.J. Enhancement of nutrient removal from eutrophic water by a plant–microorganisms combined system. Environ. Eng. Sci. 2011, 28, 543–554. [Google Scholar] [CrossRef]
- Lamers, L.P.; Govers, L.L.; Janssen, I.C.; Geurts, J.J.; Van der Welle, M.E.; Van Katwijk, M.M.; Van der Heide, T.; Roelofs, J.G.; Smolders, A.J. Sulfide as a soil phytotoxin—A review. Front. Plant Sci. 2013, 4, 268. [Google Scholar] [CrossRef] [PubMed]
- Pavan, F.; Breschigliaro, S.; Borin, M. Screening of 18 species for digestate phytodepuration. Environ. Sci. Pollut. Res. 2015, 22, 2455–2466. [Google Scholar] [CrossRef]
- Battin, T.J.; Kaplan, L.A.; Newbold, J.D.; Cheng, X.; Hansen, C. Effects of current velocity on the nascent architecture of stream microbial biofilms. Appl. Environ. Microbiol. 2003, 69, 5443–5452. [Google Scholar] [CrossRef]
- Thorén, A.-K. Urea transformation of wetland microbial communities. Microb. Ecol. 2007, 53, 221–232. [Google Scholar] [CrossRef]
- Pang, S.; Zhang, S.; Lv, X.; Han, B.; Liu, K.; Qiu, C.; Wang, C.; Wang, P.; Toland, H.; He, Z. Characterization of bacterial community in biofilm and sediments of wetlands dominated by aquatic macrophytes. Ecol. Eng. 2016, 97, 242–250. [Google Scholar] [CrossRef]
- Branda, S.S.; Vik, Å.; Friedman, L.; Kolter, R. Biofilms: The matrix revisited. Trends Microbiol. 2005, 13, 20–26. [Google Scholar] [CrossRef]
- Latasa, C.; Solano, C.; Penadés, J.R.; Lasa, I. Biofilm-associated proteins. C. R. Biol. 2006, 329, 849–857. [Google Scholar] [CrossRef]
- Lasa, I.; Penadés, J.R. Bap: A family of surface proteins involved in biofilm formation. Res. Microbiol. 2006, 157, 99–107. [Google Scholar] [CrossRef] [PubMed]
- López, D.; Vlamakis, H.; Kolter, R. Biofilms. Cold Spring Harbor Perspect. Biol. 2010, 2, a000398. [Google Scholar] [CrossRef] [PubMed]
- Rice, K.C.; Mann, E.E.; Endres, J.L.; Weiss, E.C.; Cassat, J.E.; Smeltzer, M.S.; Bayles, K.W. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2007, 104, 8113–8118. [Google Scholar] [CrossRef] [PubMed]
- Branda, S.S.; Chu, F.; Kearns, D.B.; Losick, R.; Kolter, R. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 2006, 59, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, M.; Saunders, A.M.; Schramm, A. Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora. Appl. Environ. Microbiol. 2008, 74, 3279–3283. [Google Scholar] [CrossRef]
- Crump, B.C.; Koch, E.W. Attached bacterial populations shared by four species of aquatic angiosperms. Appl. Environ. Microbiol. 2008, 74, 5948–5957. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, G. Nutrient concentration variations during Oenanthe javanica growth and decay in the ecological floating bed system. J. Environ. Sci. 2010, 22, 1710–1717. [Google Scholar] [CrossRef]
- Stewart, F.M.; Muholland, T.; Cunningham, A.B.; Kania, B.G.; Osterlund, M.T. Floating islands as an alternative to constructed wetlands for treatment of excess nutrients from agricultural and municipal wastes–results of laboratory-scale tests. Land Contam. Reclam. 2008, 16, 25–33. [Google Scholar] [CrossRef]
- Nihorimbere, V.; Ongena, M.; Smargiassi, M.; Thonart, P. Beneficial effect of the rhizosphere microbial community for plant growth and health. Biotechnologie Agronomie Société et Environ. 2011, 15, 327–337. [Google Scholar]
- Acosta-Martínez, V.; Dowd, S.; Sun, Y.; Allen, V. Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem. 2008, 40, 2762–2770. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- el Zahar Haichar, F.; Marol, C.; Berge, O.; Rangel-Castro, J.I.; Prosser, J.I.; Balesdent, J.; Heulin, T.; Achouak, W. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2008, 2, 1221. [Google Scholar] [CrossRef] [PubMed]
- Achá, D.; Iniguez, V.; Roulet, M.; Guimaraes, J.R.D.; Luna, R.; Alanoca, L.; Sanchez, S. Sulfate-reducing bacteria in floating macrophyte rhizospheres from an Amazonian floodplain lake in Bolivia and their association with Hg methylation. Appl. Environ. Microbiol. 2005, 71, 7531–7535. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, X.; Liu, G.; Chen, T.; Zhang, G.; Dong, Z.; Yang, X.; Hu, P. Pyrosequencing reveals bacterial diversity in the rhizosphere of three Phragmites australis ecotypes. Geomicrobiol. J. 2013, 30, 593–599. [Google Scholar] [CrossRef]
- Van Bodegom, P.; Stams, F.; Mollema, L.; Boeke, S.; Leffelaar, P. Methane oxidation and the competition for oxygen in the rice rhizosphere. Appl. Environ. Microbiol. 2001, 67, 3586–3597. [Google Scholar] [CrossRef]
- Tanaka, Y.; Tamaki, H.; Matsuzawa, H.; Nigaya, M.; Mori, K.; Kamagata, Y. Microbial community analysis in the roots of aquatic plants and isolation of novel microbes including an organism of the candidate phylum OP10. Microbes Environ. 2012, 27, 149–157. [Google Scholar] [CrossRef]
- Unno, Y.; Shinano, T. Metagenomic analysis of the rhizosphere soil microbiome with respect to phytic acid utilization. Microbes Environ. 2013, 28, 120–127. [Google Scholar] [CrossRef]
- Zhang, L.; Shao, H. Heavy metal pollution in sediments from aquatic ecosystems in China. Clean–Soil Air Water 2013, 41, 878–882. [Google Scholar] [CrossRef]
- Uroz, S.; Buée, M.; Murat, C.; Frey-Klett, P.; Martin, F. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ. Microbiol. Rep. 2010, 2, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Tribedi, P.; Sil, A.K. Low-density polyethylene degradation by Pseudomonas sp. AKS2 biofilm. Environ. Sci. Pollut. Res. 2013, 20, 4146–4153. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Yu, X.; Zhang, S.; Gu, L. Comparison of the community structures of ammonia-oxidizing bacteria and archaea in rhizoplanes of floating aquatic macrophytes. Microbiol. Res. 2011, 166, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Avrahami, S.; Conrad, R.; Braker, G. Effect of ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl. Environ. Microbiol. 2003, 69, 3027. [Google Scholar] [CrossRef]
- Hempel, M.; Botté, S.E.; Negrin, V.L.; Chiarello, M.N.; Marcovecchio, J.E. The role of the smooth cordgrass Spartina alterniflora and associated sediments in the heavy metal biogeochemical cycle within Bahía Blanca estuary salt marshes. J. Soils Sed. 2008, 8, 289. [Google Scholar] [CrossRef]
- Rastogi, G.; Sbodio, A.; Tech, J.J.; Suslow, T.V.; Coaker, G.L.; Leveau, J.H. Leaf microbiota in an agroecosystem: Spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012, 6, 1812. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.I.; Kühl, M.; Priemé, A. Different bacterial communities associated with the roots and bulk sediment of the seagrass Zostera marina. FEMS Microbiol. Ecol. 2007, 62, 108–117. [Google Scholar] [CrossRef]
- Faulwetter, J.L.; Burr, M.D.; Parker, A.E.; Stein, O.R.; Camper, A.K. Influence of season and plant species on the abundance and diversity of sulfate reducing bacteria and ammonia oxidizing bacteria in constructed wetland microcosms. Microb. Ecol. 2013, 65, 111–127. [Google Scholar] [CrossRef]
- Chen, N.; Yang, J.-S.; Qu, J.-H.; Li, H.-F.; Liu, W.-J.; Li, B.-Z.; Wang, E.T.; Yuan, H.-L. Sediment prokaryote communities in different sites of eutrophic Lake Taihu and their interactions with environmental factors. World J. Microbiol. Biotechnol. 2015, 31, 883–896. [Google Scholar] [CrossRef]
- Glick, B.R. Using soil bacteria to facilitate phytoremediation. Biotechnol. Adv. 2010, 28, 367–374. [Google Scholar] [CrossRef]
- Ma, Y.; Prasad, M.; Rajkumar, M.; Freitas, H. Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol. Adv. 2011, 29, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Hou, H.; ShangGuan, Y.; Li, J.; Li, F. Genetic diversity of endophytic bacteria of the manganese-hyperaccumulating plant Phytolacca americana growing at a manganese mine. Eur. J. Soil Biol. 2014, 62, 15–21. [Google Scholar] [CrossRef]
- Erdei, L.; Mezôsi, G.; Mécs, I.; Vass, I.; Fôglein, F.; Bulik, L. Phytoremediation as a program for decontamination of heavy-metal polluted environment. Acta Biol. Szeged. 2005, 49, 75–76. [Google Scholar]
- Dharni, S.; Srivastava, A.K.; Samad, A.; Patra, D.D. Impact of plant growth promoting Pseudomonas monteilii PsF84 and Pseudomonas plecoglossicida PsF610 on metal uptake and production of secondary metabolite (monoterpenes) by rose-scented geranium (Pelargonium graveolens cv. bourbon) grown on tannery sludge amended soil. Chemosphere 2014, 117, 433–439. [Google Scholar]
- Ma, Y.; Rajkumar, M.; Rocha, I.; Oliveira, R.S.; Freitas, H. Serpentine bacteria influence metal translocation and bioconcentration of Brassica juncea and Ricinus communis grown in multi-metal polluted soils. Front. Plant Sci. 2015, 5, 757. [Google Scholar] [CrossRef] [PubMed]
- Chadha, N.; Mishra, M.; Rajpal, K.; Bajaj, R.; Choudhary, D.K.; Varma, A. An ecological role of fungal endophytes to ameliorate plants under biotic stress. Arch. Microbiol. 2015, 197, 869–881. [Google Scholar] [CrossRef]
- Marella, S. Bacterial endophytes in sustainable crop production: Applications, recent developments and challenges ahead. Int. J. Life Sci. Res. 2014, 2, 46–56. [Google Scholar]
- Ryan, R.P.; Germaine, K.; Franks, A.; Ryan, D.J.; Dowling, D.N. Bacterial endophytes: Recent developments and applications. FEMS Microbiol. Lett. 2008, 278, 1–9. [Google Scholar] [CrossRef]
- Bacon, C.W.; White, J.F. Functions, mechanisms and regulation of endophytic and epiphytic microbial communities of plants. Symbiosis 2016, 68, 87–98. [Google Scholar] [CrossRef]
- Berg, G.; Hallmann, J. Control of Plant Pathogenic Fungi with Bacterial Endophytes. In Microbial Root Endophytes; Springer: Berlin/Heidelberg, Germany, 2006; pp. 53–69. [Google Scholar]
- White, J.F.; Kingsley, K.I.; Kowalski, K.P.; Irizarry, I.; Micci, A.; Soares, M.A.; Bergen, M.S. Disease protection and allelopathic interactions of seed-transmitted endophytic pseudomonads of invasive reed grass (Phragmites australis). Plant Soil 2018, 422, 195–208. [Google Scholar] [CrossRef]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, E.A.; Carvalho, J.M.; dos Santos, D.C.; Feitosa, A.O.; Marinho, P.S.; Guilhon, G.M.S.; Santos, L.S.; de Souza, A.L.; Marinho, A.M. Chemical constituents of Aspergillus sp EJC08 isolated as endophyte from Bauhinia guianensis and their antimicrobial activity. An. Acad. Brasi. Ciênc. 2013, 85, 1247–1253. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, H.; Akiyama, M.; Kobayashi, K.; Yamaji, K. Fe and P solubilization under limiting conditions by bacteria isolated from Carex kobomugi roots at the Hasaki coast. Curr. Microbiol. 2013, 66, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Knoth, J.L.; Kim, S.H.; Ettl, G.J.; Doty, S.L. Effects of cross host species inoculation of nitrogen-fixing endophytes on growth and leaf physiology of maize. Gcb Bioenergy 2013, 5, 408–418. [Google Scholar] [CrossRef]
- Jasim, B.; Jimtha John, C.; Shimil, V.; Jyothis, M.; Radhakrishnan, E. Studies on the factors modulating indole-3-acetic acid production in endophytic bacterial isolates from P iper nigrum and molecular analysis of ipdc gene. J. Appl. Microbiol. 2014, 117, 786–799. [Google Scholar] [CrossRef]
- Khan, A.L.; Waqas, M.; Kang, S.-M.; Al-Harrasi, A.; Hussain, J.; Al-Rawahi, A.; Al-Khiziri, S.; Ullah, I.; Ali, L.; Jung, H.-Y. Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 2014, 52, 689–695. [Google Scholar] [CrossRef]
- Chaturvedi, H.; Singh, V.; Gupta, G. Potential of bacterial endophytes as plant growth promoting factors. J. Plant Pathol. Microbiol. 2016, 7, 2. [Google Scholar] [CrossRef]
- Van Loon, L.; Bakker, P.; Pieterse, C. Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 1998, 36, 453–483. [Google Scholar] [CrossRef]
- Rajkumar, M.; Prasad, M.N.V.; Swaminathan, S.; Freitas, H. Climate change driven plant–metal–microbe interactions. Environ. Int. 2013, 53, 74–86. [Google Scholar] [CrossRef]
- Yihui, B.; Zhouying, X.; Yurong, Y.; ZHANG, H.; Hui, C.; Ming, T. Effect of dark septate endophytic fungus Gaeumannomyces cylindrosporus on plant growth, photosynthesis and Pb tolerance of maize (Zea mays L.). Pedosphere 2017, 27, 283–292. [Google Scholar]
- Shi, P.; Zhu, K.; Zhang, Y.; Chai, T. Growth and cadmium accumulation of Solanum nigrum L. seedling were enhanced by heavy metal-tolerant strains of Pseudomonas aeruginosa. Water Air Soil Pollut. 2016, 227, 459. [Google Scholar] [CrossRef]
- Babu, A.G.; Shea, P.J.; Sudhakar, D.; Jung, I.-B.; Oh, B.-T. Potential use of Pseudomonas koreensis AGB-1 in association with Miscanthus sinensis to remediate heavy metal (loid)-contaminated mining site soil. J. Environ. Manag. 2015, 151, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Visioli, G.; Vamerali, T.; Mattarozzi, M.; Dramis, L.; Sanangelantoni, A.M. Combined endophytic inoculants enhance nickel phytoextraction from serpentine soil in the hyperaccumulator Noccaea caerulescens. Front. Plant Sci. 2015, 6, 638. [Google Scholar] [CrossRef] [PubMed]
- Afzal, M.; Shabir, G.; Hussain, I.; Khalid, Z.M. Paper and board mill effluent treatment with the combined biological–coagulation–filtration pilot scale reactor. Bioresour. Technol. 2008, 99, 7383–7387. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.J.; Tahseen, R.; Siddique, M.; Ali, S.; Iqbal, S.; Afzal, M. Remediation of polluted river water by floating treatment wetlands. Water Supply 2019, 19, 967–977. [Google Scholar] [CrossRef]
- Newman, L.A.; Reynolds, C.M. Bacteria and phytoremediation: New uses for endophytic bacteria in plants. Trends Biotechnol. 2005, 23, 6–8. [Google Scholar] [CrossRef]
- Shehzadi, M.; Fatima, K.; Imran, A.; Mirza, M.S.; Khan, Q.M.; Afzal, M. Ecology of bacterial endophytes associated with wetland plants growing in textile effluent for pollutant-degradation and plant growth-promotion potentials. Plant Biosyst. 2016, 150, 1261–1270. [Google Scholar] [CrossRef]
- Faußer, A.C.; Hoppert, M.; Walther, P.; Kazda, M. Roots of the wetland plants Typha latifolia and Phragmites australis are inhabited by methanotrophic bacteria in biofilms. Flora-Morphol. Distrib. Funct. Ecol. Plants 2012, 207, 775–782. [Google Scholar] [CrossRef]
- Görres, C.-M.; Conrad, R.; Petersen, S.O. Effect of soil properties and hydrology on Archaeal community composition in three temperate grasslands on peat. FEMS Microbiol. Ecol. 2013, 85, 227–240. [Google Scholar] [CrossRef]
- Shpigel, M.; Ben-Ezra, D.; Shauli, L.; Sagi, M.; Ventura, Y.; Samocha, T.; Lee, J. Constructed wetland with Salicornia as a biofilter for mariculture effluents. Aquaculture 2013, 412, 52–63. [Google Scholar] [CrossRef]
- Vymazal, J. Removal of nutrients in various types of constructed wetlands. Sci. Total Environ. 2007, 380, 48–65. [Google Scholar] [CrossRef] [PubMed]
- García-Martínez, M.; López-López, A.; Calleja, M.L.; Marbà, N.; Duarte, C.M. Bacterial community dynamics in a seagrass (Posidonia oceanica) meadow sediment. Estuar. Coasts. 2009, 32, 276–286. [Google Scholar] [CrossRef]
- Vymazal, J. Plants used in constructed wetlands with horizontal subsurface flow: A review. Hydrobiologia 2011, 674, 133–156. [Google Scholar] [CrossRef]
- Xu, X.; Thornton, P.E.; Post, W.M. A global analysis of soil microbial biomass carbon, nitrogen and phosphorus in terrestrial ecosystems. Global Ecol. Biogeogr. 2013, 22, 737–749. [Google Scholar] [CrossRef]
- Nelson, E.B.; Karp, M.A. Soil pathogen communities associated with native and non-native P hragmites australis populations in freshwater wetlands. Ecol. Evol. 2013, 3, 5254–5267. [Google Scholar] [CrossRef]
- Ehrenfeld, J.G.; Ravit, B.; Elgersma, K. Feedback in the plant-soil system. Annu. Rev. Environ. Resour. 2005, 30, 75–115. [Google Scholar] [CrossRef]
- Wang, L.; Wu, J.; Ma, F.; Yang, J.; Li, S.; Li, Z.; Zhang, X. Response of arbuscular mycorrhizal fungi to hydrologic gradients in the rhizosphere of Phragmites australis (Cav.) Trin ex. Steudel growing in the Sun Island Wetland. BioMed Res. Int. 2015, 2015, 810124. [Google Scholar]
- Curl, E.; Harper, J. Rhizosphere. In Fauna-microflora Interactions; John Wiley and Sons Ltd.: Chichester, UK, 1990; pp. 369–388. [Google Scholar]
- Petersen, N.R.; Jensen, K. Nitrification and denitrification in the rhizosphere of the aquatic macrophyte Lobelia dortmanna L. Limnol. Oceanogr. 1997, 42, 529–537. [Google Scholar] [CrossRef]
- Bodelier, P.; Dedysh, S.N. Microbiology of wetlands. Front. Microbiol. 2013, 4, 79. [Google Scholar] [CrossRef]
- Han, G.; Luo, Y.; Li, D.; Xia, J.; Xing, Q.; Yu, J. Ecosystem photosynthesis regulates soil respiration on a diurnal scale with a short-term time lag in a coastal wetland. Soil Biol. Biochem. 2014, 68, 85–94. [Google Scholar] [CrossRef]
- Koelbener, A.; Ström, L.; Edwards, P.J.; Venterink, H.O. Plant species from mesotrophic wetlands cause relatively high methane emissions from peat soil. Plant Soil 2010, 326, 147–158. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789. [Google Scholar] [CrossRef] [PubMed]
- Weston, D.J.; Timm, C.M.; Walker, A.P.; Gu, L.; Muchero, W.; Schmutz, J.; Shaw, A.J.; Tuskan, G.A.; Warren, J.M.; Wullschleger, S.D. S phagnum physiology in the context of changing climate: Emergent influences of genomics, modelling and host–microbiome interactions on understanding ecosystem function. Plant, Cell Environ. 2015, 38, 1737–1751. [Google Scholar] [CrossRef] [PubMed]
- Husson, O. Redox potential (Eh) and pH as drivers of soil/plant/microorganism systems: A transdisciplinary overview pointing to integrative opportunities for agronomy. Plant Soil 2013, 362, 389–417. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Cassman, K.; Cleveland, C.; Crews, T.; Field, C.B.; Grimm, N.B.; Howarth, R.W.; Marino, R.; Martinelli, L.; Rastetter, E.B. Towards an Ecological Understanding of Biological Nitrogen Fixation. In The Nitrogen Cycle at Regional to Global Scales; Springer: Berlin/Heidelberg, Germany, 2002; pp. 1–45. [Google Scholar]
- Lamers, L.P.; Van Diggelen, J.M.; Op Den Camp, H.J.; Visser, E.J.; Lucassen, E.C.; Vile, M.A.; Jetten, M.S.; Smolders, A.J.; Roelofs, J.G. Microbial transformations of nitrogen, sulfur, and iron dictate vegetation composition in wetlands: A review. Front Microbiol. 2012, 3, 156. [Google Scholar] [CrossRef]
- Bañeras, L.; Ruiz-Rueda, O.; López-Flores, R.; Quintana, X.D.; Hallin, S. The role of plant type and salinity in the selection for the denitrifying community structure in the rhizosphere of wetland vegetation. Int. Microbiol. 2012, 15, 89–99. [Google Scholar]
- Trias, R.; Ruiz-Rueda, O.; García-Lledó, A.; Vilar-Sanz, A.; López-Flores, R.; Quintana, X.D.; Hallin, S.; Bañeras, L. Emergent macrophytes act selectively on ammonia-oxidizing bacteria and archaea. Appl. Environ. Microbiol. 2012, 78, 6352–6356. [Google Scholar] [CrossRef]
- He, J.-Z.; Shen, J.-P.; Zhang, L.-M.; Di, H.J. A review of ammonia-oxidizing bacteria and archaea in Chinese soils. Front. Microbiol. 2012, 3, 296. [Google Scholar]
- Reddy, K.; Patrick, W.; Lindau, C. Nitrification-denitrification at the plant root-sediment interface in wetlands. Limnol. Oceanogr. 1989, 34, 1004–1013. [Google Scholar] [CrossRef]
- Galloway, J.N.; Schlesinger, W.H.; Levy, H.; Michaels, A.; Schnoor, J.L. Nitrogen fixation: Anthropogenic enhancement-environmental response. Global Biogeochem. Cycles 1995, 9, 235–252. [Google Scholar] [CrossRef]
- López-Guerrero, M.G.; Ormeño-Orrillo, E.; Acosta, J.L.; Mendoza-Vargas, A.; Rogel, M.A.; Ramírez, M.A.; Rosenblueth, M.; Martínez-Romero, J.; Martínez-Romero, E. Rhizobial extrachromosomal replicon variability, stability and expression in natural niches. Plasmid 2012, 68, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Ormeño-Orrillo, E.; Hungria, M.; Martinez-Romero, E. Dinitrogen-fixing prokaryotes. Prokaryotes Prokaryotic Physiol. Biochem. 2013, 427–451. [Google Scholar]
- Oyewole, O. Microbial communities and their activities in paddy fields: A review. J. Vet. Adv. 2012, 2, 74–80. [Google Scholar]
- Arima, Y.; Yoshida, T. Nitrogen fixation and denitrification in the roots of flooded crops. Soil Sci. Plant Nutr. 1982, 28, 483–489. [Google Scholar] [CrossRef]
- Knapp, A. The sensitivity of marine N2 fixation to dissolved inorganic nitrogen. Front. Microbiol. 2012, 3, 374. [Google Scholar] [CrossRef] [PubMed]
- Waisel, Y.; Agami, M. Ecophysiology of Roots of Submerged Aquatic Plants. In Plant Roots: The Hidden Half, 2nd ed.; Marcel Dekker, Inc.: New York, NY, USA, 1996; pp. 895–909. [Google Scholar]
- Neori, A.; Agami, M. The functioning of rhizosphere biota in wetlands–a review. Wetlands 2017, 37, 615–633. [Google Scholar] [CrossRef]
- Hiraishi, A. Biodiversity of dehalorespiring bacteria with special emphasis on polychlorinated biphenyl/dioxin dechlorinators. Microbes Environ. 2008, 23, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fennell, D.; Du, S.; Liu, F.; Liu, H.; Haggblom, M. Dehalogenation of Polychlorinated Dibenzo-p-Dioxins and Dibenzofurans, Polychlorinated Biphenyls, and Brominated Flame Retardants, and Potential as a Bioremediation Strategy. In Environmental Biotechnology and Safety; Elsevier Inc.: Amsterdam, The Netherlands, 2011; pp. 135–149. [Google Scholar]
- Fenchel, T.; Blackburn, H.; King, G.M.; Blackburn, T.H. Bacterial Biogeochemistry: The Ecophysiology of Mineral Cycling; Academic press: Cambridge, MA, USA, 2012. [Google Scholar]
- Stottmeister, U.; Wießner, A.; Kuschk, P.; Kappelmeyer, U.; Kästner, M.; Bederski, O.; Müller, R.; Moormann, H. Effects of plants and microorganisms in constructed wetlands for wastewater treatment. Biotechnol. Adv. 2003, 22, 93–117. [Google Scholar] [CrossRef]
- Paris, D.F.; Steen, W.C.; Baughman, G.L.; Barnett, J.T. Second-order model to predict microbial degradation of organic compounds in natural waters. Appl. Environ. Microbiol. 1981, 41, 603–609. [Google Scholar] [CrossRef]
- Calheiros, C.S.; Duque, A.F.; Moura, A.; Henriques, I.S.; Correia, A.; Rangel, A.O.; Castro, P.M. Changes in the bacterial community structure in two-stage constructed wetlands with different plants for industrial wastewater treatment. Bioresour. Technol. 2009, 100, 3228–3235. [Google Scholar] [CrossRef]
- Mori, K.; Toyama, T.; Sei, K. Surfactants degrading activities in the rhizosphere of giant duckweed (Spirodela polyrrhiza). Jpn. J. Water Treat. Biol. 2005, 41, 129–140. [Google Scholar] [CrossRef][Green Version]
- Mordukhova, E.A.; Sokolov, S.L.; Kochetkov, V.V.; Kosheleva, I.A.; Zelenkova, N.F.; Boronin, A.M. Involvement of naphthalene dioxygenase in indole-3-acetic acid biosynthesis by Pseudomonas putida. FEMS Microbiol. Lett. 2000, 190, 279–285. [Google Scholar] [CrossRef]
- Jouanneau, Y.; Willison, J.C.; Meyer, C.; Krivobok, S.; Chevron, N.; Besombes, J.-L.; Blake, G. Stimulation of pyrene mineralization in freshwater sediments by bacterial and plant bioaugmentation. Environ. Sci. Technol. 2005, 39, 5729–5735. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Golubev, S.N.; Schelud’ko, A.V.; Muratova, A.Y.; Makarov, O.E.; Turkovskaya, O.V. Assessing the potential of rhizobacteria to survive under phenanthrene pollution. Water Air Soil Pollut. 2009, 198, 5–16. [Google Scholar] [CrossRef]
- Huang, X.-D.; El-Alawi, Y.; Penrose, D.M.; Glick, B.R.; Greenberg, B.M. A multi-process phytoremediation system for removal of polycyclic aromatic hydrocarbons from contaminated soils. Environ. Pollut. 2004, 130, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Escalante-Espinosa, E.; Gallegos-Martínez, M.; Favela-Torres, E.; Gutiérrez-Rojas, M. Improvement of the hydrocarbon phytoremediation rate by Cyperus laxus Lam. inoculated with a microbial consortium in a model system. Chemosphere 2005, 59, 405–413. [Google Scholar] [CrossRef]
- Simpson, D.R. Biofilm processes in biologically active carbon water purification. Water Res. 2008, 42, 2839–2848. [Google Scholar] [CrossRef]
- Ghosh, U.; Weber, A.S.; Jensen, J.N.; Smith, J.R. Granular activated carbon and biological activated carbon treatment of dissolved and sorbed polychlorinated biphenyls. Water Environ. Res 1999, 71, 232–240. [Google Scholar] [CrossRef]
- Guasch, H.; Lehmann, V.; Van Beusekom, B.; Sabater, S.; Admiraal, W. Influence of phosphate on the response of periphyton to atrazine exposure. Arch. Environ. Contam. Toxicol. 2007, 52, 32–37. [Google Scholar] [CrossRef]
- Semrau, J.D.; DiSpirito, A.A.; Yoon, S. Methanotrophs and copper. FEMS Microbiol. Rev. 2010, 34, 496–531. [Google Scholar] [CrossRef]
- Yoon, S. Towards Practical Application of Methanotrophic Metabolism in Chlorinated Hydrocarbon Degradation, Greenhouse Gas Removal, and Immobilization of Heavy Metals. Ph.D. Thesis, University of Michigan, Ann Arbor, MI, USA, 2010. [Google Scholar]
- Pandey, V.C.; Singh, J.; Singh, D.; Singh, R.P. Methanotrophs: Promising bacteria for environmental remediation. Int. J. Environ Sci. Technol. 2014, 11, 241–250. [Google Scholar] [CrossRef]
- Han, B.; Zhang, S.; Zhang, L.; Liu, K.; Yan, L.; Wang, P.; Wang, C.; Pang, S. Characterization of microbes and denitrifiers attached to two species of floating plants in the wetlands of Lake Taihu. PLoS ONE 2018, 13, e0207443. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yi, N.; Wang, Y.; Ma, T.; Zhou, Q.; Zhang, Z.; Yan, S. Effect of Eichhornia crassipes on production of N2 by denitrification in eutrophic water. Ecol. Eng. 2014, 68, 14–24. [Google Scholar] [CrossRef]
- Mullen, M.; Wolf, D.; Ferris, F.; Beveridge, T.; Flemming, C.; Bailey, G. Bacterial sorption of heavy metals. Appl. Environ. Microbiol. 1989, 55, 3143–3149. [Google Scholar] [CrossRef] [PubMed]
- Sessitsch, A.; Kuffner, M.; Kidd, P.; Vangronsveld, J.; Wenzel, W.W.; Fallmann, K.; Puschenreiter, M. The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 2013, 60, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.U.; Sessitsch, A.; Harris, M.; Fatima, K.; Imran, A.; Arslan, M.; Shabir, G.; Khan, Q.M.; Afzal, M. Cr-resistant rhizo-and endophytic bacteria associated with Prosopis juliflora and their potential as phytoremediation enhancing agents in metal-degraded soils. Front. Plant Sci. 2015, 5, 755. [Google Scholar] [CrossRef] [PubMed]
- Cobbett, C.; Goldsbrough, P. Phytochelatins and metallothioneins: Roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 2002, 53, 159–182. [Google Scholar] [CrossRef]
- Kaegi, J.H.; Schaeffer, A. Biochemistry of metallothionein. Biochemistry 1988, 27, 8509–8515. [Google Scholar] [CrossRef]
- Huckle, J.W.; Morby, A.P.; Turner, J.S.; Robinson, N.J. Isolation of a prokaryotic metallothionein locus and analysis of transcriptional control by trace metal ions. Mol. Microbiol. 1993, 7, 177–187. [Google Scholar] [CrossRef]
- Valls, M.; Atrian, S.; de Lorenzo, V.; Fernández, L.A. Engineering a mouse metallothionein on the cell surface of Ralstonia eutropha CH34 for immobilization of heavy metals in soil. Nat. Biotechnol. 2000, 18, 661. [Google Scholar] [CrossRef]
- Mejáre, M.; Bülow, L. Metal-binding proteins and peptides in bioremediation and phytoremediation of heavy metals. Trends Biotechnol. 2001, 19, 67–73. [Google Scholar] [CrossRef]
- Shin, M.-N.; Shim, J.; You, Y.; Myung, H.; Bang, K.-S.; Cho, M.; Kamala-Kannan, S.; Oh, B.-T. Characterization of lead resistant endophytic Bacillus sp. MN3-4 and its potential for promoting lead accumulation in metal hyperaccumulator Alnus firma. J. Hazard. Mater. 2012, 199, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Mindlin, S.Z.; Bass, I.A.; Bogdanova, E.S.; Gorlenko, Z.M.; Kalyaeva, E.S.; Petrova, M.A.; Nikiforov, V.G. Horizontal transfer of mercury resistance genes in environmental bacterial populations. Mol. Biol. 2002, 36, 160–170. [Google Scholar] [CrossRef]
- Li, T.; Liu, M.; Zhang, X.; Zhang, H.; Sha, T.; Zhao, Z. Improved tolerance of maize (Zea mays L.) to heavy metals by colonization of a dark septate endophyte (DSE) Exophiala pisciphila. Sci. Total Environ. 2011, 409, 1069–1074. [Google Scholar] [CrossRef]
- Zhang, X.; Li, C.; Nan, Z. Effects of cadmium stress on growth and anti-oxidative systems in Achnatherum inebrians symbiotic with Neotyphodium gansuense. J. Hazard. Mater. 2010, 175, 703–709. [Google Scholar] [CrossRef]
- Wan, Y.; Luo, S.; Chen, J.; Xiao, X.; Chen, L.; Zeng, G.; Liu, C.; He, Y. Effect of endophyte-infection on growth parameters and Cd-induced phytotoxicity of Cd-hyperaccumulator Solanum nigrum L. Chemosphere 2012, 89, 743–750. [Google Scholar] [CrossRef]
- Brown, N.L.; Stoyanov, J.V.; Kidd, S.P.; Hobman, J.L. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 2003, 27, 145–163. [Google Scholar] [CrossRef]
- Sofu, A.; Sayilgan, E.; Guney, G. Experimental design for removal of Fe (II) and Zn (II) ions by different lactic acid bacteria biomasses. Int. J. Environ. Res. 2015, 9, 93–100. [Google Scholar]
- Li, C.; Wang, S.; Du, X.; Cheng, X.; Fu, M.; Hou, N.; Li, D. Immobilization of iron-and manganese-oxidizing bacteria with a biofilm-forming bacterium for the effective removal of iron and manganese from groundwater. Bioresour. Technol. 2016, 220, 76–84. [Google Scholar] [CrossRef]
- Franzblau, R.E.; Daughney, C.J.; Swedlund, P.J.; Weisener, C.G.; Moreau, M.; Johannessen, B.; Harmer, S.L. Cu (II) removal by Anoxybacillus flavithermus–iron oxide composites during the addition of Fe (II) aq. Geochim. Cosmochim. Acta 2016, 172, 139–158. [Google Scholar] [CrossRef]
- Guo, Y.; Huang, T.; Wen, G.; Cao, X. The simultaneous removal of ammonium and manganese from groundwater by iron-manganese co-oxide filter film: The role of chemical catalytic oxidation for ammonium removal. Chem. Eng. J. 2017, 308, 322–329. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Kim, J.-H.; Kim, C.-J.; Oh, D.-K. Metal adsorption of the polysaccharide produced from Methylobacterium organophilum. Biotechnol. Lett 1996, 18, 1161–1164. [Google Scholar] [CrossRef]
- Marchal, M.; Briandet, R.; Koechler, S.; Kammerer, B.; Bertin, P. Effect of arsenite on swimming motility delays surface colonization in Herminiimonas arsenicoxydans. Microbiology 2010, 156, 2336–2342. [Google Scholar] [CrossRef]
- Iyer, A.; Mody, K.; Jha, B. Biosorption of heavy metals by a marine bacterium. Mar. Pollut. Bull. 2005, 50, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Oshima, T.; Kondo, K.; Ohto, K.; Inoue, K.; Baba, Y. Preparation of phosphorylated bacterial cellulose as an adsorbent for metal ions. React. Funct. Polym. 2008, 68, 376–383. [Google Scholar] [CrossRef]
- Ozdemir, G.; Ceyhan, N.; Manav, E. Utilization of an exopolysaccharide produced by Chryseomonas luteola TEM05 in alginate beads for adsorption of cadmium and cobalt ions. Bioresour. Technol. 2005, 96, 1677–1682. [Google Scholar] [CrossRef]
- Ozdemir, G.; Ozturk, T.; Ceyhan, N.; Isler, R.; Cosar, T. Heavy metal biosorption by biomass of Ochrobactrum anthropi producing exopolysaccharide in activated sludge. Bioresour. Technol. 2003, 90, 71–74. [Google Scholar] [CrossRef]
- Freire-Nordi, C.S.; Vieira, A.A.H.; Nascimento, O.R. The metal binding capacity of Anabaena spiroides extracellular polysaccharide: An EPR study. Process Biochem. 2005, 40, 2215–2224. [Google Scholar] [CrossRef]
- Pugazhendhi, A.; Boovaragamoorthy, G.M.; Ranganathan, K.; Naushad, M.; Kaliannan, T. New insight into effective biosorption of lead from aqueous solution using Ralstonia solanacearum: Characterization and mechanism studies. J. Clean. Prod. 2018, 174, 1234–1239. [Google Scholar] [CrossRef]
- Wu, Y.; Zhao, X.; Jin, M.; Li, Y.; Li, S.; Kong, F.; Nan, J.; Wang, A. Copper removal and microbial community analysis in single-chamber microbial fuel cell. Bioresour. Technol. 2018, 253, 372–377. [Google Scholar] [CrossRef]
- Pugazhendhi, A.; Ranganathan, K.; Kaliannan, T. Biosorptive removal of copper (II) by Bacillus cereus isolated from contaminated soil of electroplating industry in India. Water Air Soil Pollut. 2018, 229, 76. [Google Scholar] [CrossRef]
- Wen, X.; Du, C.; Zeng, G.; Huang, D.; Zhang, J.; Yin, L.; Tan, S.; Huang, L.; Chen, H.; Yu, G. A novel biosorbent prepared by immobilized Bacillus licheniformis for lead removal from wastewater. Chemosphere 2018, 200, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, K.; Mitra, S.; Sarkar, A.; Maiti, T.K. Alleviation of phytotoxic effects of cadmium on rice seedlings by cadmium resistant PGPR strain Enterobacter aerogenes MCC 3092. J. Hazard. Mater. 2018, 351, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Choińska-Pulit, A.; Sobolczyk-Bednarek, J.; Łaba, W. Optimization of copper, lead and cadmium biosorption onto newly isolated bacterium using a Box-Behnken design. Ecotoxicol. Environ. Saf. 2018, 149, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.-J.; Zhang, Z.-M.; Yang, G.-E.; Li, B.-Z. Bioremediation of cadmium by growing Rhodobacter sphaeroides: Kinetic characteristic and mechanism studies. Bioresour. Technol. 2008, 99, 7716–7722. [Google Scholar] [CrossRef]
- Vijayaraghavan, K.; Yun, Y.-S. Bacterial biosorbents and biosorption. Biotechnol. Adv. 2008, 26, 266–291. [Google Scholar] [CrossRef]
- Volesky, B. Advances in biosorption of metals: Selection of biomass types. FEMS Microbiol. Rev. 1994, 14, 291–302. [Google Scholar] [CrossRef]
- Ma, Y.; Rajkumar, M.; Luo, Y.; Freitas, H. Inoculation of endophytic bacteria on host and non-host plants—effects on plant growth and Ni uptake. J. Hazard. Mater. 2011, 195, 230–237. [Google Scholar] [CrossRef]
- Guo, H.; Luo, S.; Chen, L.; Xiao, X.; Xi, Q.; Wei, W.; Zeng, G.; Liu, C.; Wan, Y.; Chen, J. Bioremediation of heavy metals by growing hyperaccumulaor endophytic bacterium Bacillus sp. L14. Bioresour. Technol. 2010, 101, 8599–8605. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Luo, S.; Li, X.; Wan, Y.; Chen, J.; Liu, C. Interaction of Cd-hyperaccumulator Solanum nigrum L. and functional endophyte Pseudomonas sp. Lk9 on soil heavy metals uptake. Soil Biol. Biochem. 2014, 68, 300–308. [Google Scholar] [CrossRef]
- Sheng, X.-F.; Xia, J.-J.; Jiang, C.-Y.; He, L.-Y.; Qian, M. Characterization of heavy metal-resistant endophytic bacteria from rape (Brassica napus) roots and their potential in promoting the growth and lead accumulation of rape. Environ. Pollut. 2008, 156, 1164–1170. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, J.-L. Characteristics of Zn2+ biosorption by Saccharomyces cerevisiae. Biomed. Environ. Sci. BES 2007, 20, 478–482. [Google Scholar] [PubMed]
- Talos, K.; Pager, C.; Tonk, S.; Majdik, C.; Kocsis, B.; Kilar, F.; Pernyeszi, T. Cadmium biosorption on native Saccharomyces cerevisiae cells in aqueous suspension. Acta Univ. Sapientiae Agric. Environ 2009, 1, 20–30. [Google Scholar]
- Tigini, V.; Prigione, V.; Giansanti, P.; Mangiavillano, A.; Pannocchia, A.; Varese, G.C. Fungal biosorption, an innovative treatment for the decolourisation and detoxification of textile effluents. Water 2010, 2, 550–565. [Google Scholar] [CrossRef]
- Rajkumar, M.; Ae, N.; Freitas, H. Endophytic bacteria and their potential to enhance heavy metal phytoextraction. Chemosphere 2009, 77, 153–160. [Google Scholar] [CrossRef]
- Joshi, P.M.; Juwarkar, A.A. In vivo studies to elucidate the role of extracellular polymeric substances from azotobacter in immobilization of heavy metals. Environ. Sci. Technol. 2009, 43, 5884–5889. [Google Scholar] [CrossRef]
- Visioli, G.; D’Egidio, S.; Vamerali, T.; Mattarozzi, M.; Sanangelantoni, A.M. Culturable endophytic bacteria enhance Ni translocation in the hyperaccumulator Noccaea caerulescens. Chemosphere 2014, 117, 538–544. [Google Scholar] [CrossRef]
- Luo, Y.; Guo, W.; Ngo, H.H.; Nghiem, L.D.; Hai, F.I.; Zhang, J.; Liang, S.; Wang, X.C. A review on the occurrence of micropollutants in the aquatic environment and their fate and removal during wastewater treatment. Sci. Total Environ. 2014, 473, 619–641. [Google Scholar] [CrossRef]
- Hempel, M.; Blume, M.; Blindow, I.; Gross, E.M. Epiphytic bacterial community composition on two common submerged macrophytes in brackish water and freshwater. BMC Microbiol. 2008, 8, 58. [Google Scholar] [CrossRef]
- Cai, X.; Gao, G.; Tang, X.; Dong, B.; Dai, J.; Chen, D.; Song, Y. The response of epiphytic microbes to habitat and growth status of Potamogeton malaianus Miq. in Lake Taihu. J. Basic Microbiol. 2013, 53, 828–837. [Google Scholar]
- Aung, K.; Jiang, Y.; He, S.Y. The role of water in plant–microbe interactions. Plant J. 2018, 93, 771–780. [Google Scholar] [CrossRef] [PubMed]
- D’Annibale, A.; Leonardi, V.; Federici, E.; Baldi, F.; Zecchini, F.; Petruccioli, M. Leaching and microbial treatment of a soil contaminated by sulphide ore ashes and aromatic hydrocarbons. Appl. Microbiol. Biotechnol. 2007, 74, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Tunali, S.; Akar, T.; Özcan, A.S.; Kiran, I.; Özcan, A. Equilibrium and kinetics of biosorption of lead (II) from aqueous solutions by Cephalosporium aphidicola. Sep. Purif. Technol. 2006, 47, 105–112. [Google Scholar] [CrossRef]
- Akar, T.; Tunali, S.; Çabuk, A. Study on the characterization of lead (II) biosorption by fungus Aspergillus parasiticus. Appl. Biochem. Biotechnol. 2007, 136, 389–405. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.J.; Budd, K.; Lefebvre, D.D. The biotransformation of mercury in pH-stat cultures of microfungi. Botany 2006, 84, 254–260. [Google Scholar] [CrossRef]
- Ahalya, N.; Ramachandra, T.; Kanamadi, R. Biosorption of heavy metals. Res. J. Chem. Environ 2003, 7, 71–79. [Google Scholar]
- Chihpin, H. Application of Aspergillus oryzae and rhizopus oryzae for Cu (II) removal. Water Res. 1996, 9, 1985–1990. [Google Scholar]
- Shah, M.A. Mycorrhizas in Aquatic Plants. In Mycorrhizas: Novel Dimensions in the Changing World; Springer: Berlin/Heidelberg, Germany, 2014; pp. 63–68. [Google Scholar]
- Twanabasu, B.R.; Smith, C.M.; Stevens, K.J.; Venables, B.J.; Sears, W.C. Triclosan inhibits arbuscular mycorrhizal colonization in three wetland plants. Sci. Total Environ. 2013, 447, 450–457. [Google Scholar] [CrossRef]
- Wang, S.; Ye, J.; Perez, P.G.; Huang, D.-F. Abundance and diversity of ammonia-oxidizing bacteria in rhizosphere and bulk paddy soil under different duration of organic management. Afr. J. Microbiol. Res. 2011, 5, 5560–5568. [Google Scholar]
- Burke, D.J.; Smemo, K.A.; López-Gutiérrez, J.C.; DeForest, J.L. Soil fungi influence the distribution of microbial functional groups that mediate forest greenhouse gas emissions. Soil Biol. Biochem. 2012, 53, 112–119. [Google Scholar] [CrossRef]
- Mohamed, D.J.; Martiny, J.B. Patterns of fungal diversity and composition along a salinity gradient. ISME J. 2011, 5, 379. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-L.; Guan, M.; Liu, S.-Y.; Wang, J.; Chang, J.; Ge, Y.; Zhang, C.-B. Fungal denitrification potential in vertical flow microcosm wetlands as impacted by depth stratification and plant species. Ecol. Eng. 2015, 77, 163–171. [Google Scholar] [CrossRef]
- Saha, M.; Sarkar, S.; Sarkar, B.; Sharma, B.K.; Bhattacharjee, S.; Tribedi, P. Microbial siderophores and their potential applications: A review. Environ. Sci. Pollut. Res. 2016, 23, 3984–3999. [Google Scholar] [CrossRef] [PubMed]
- Persson, B. On the mechanism of adhesion in biological systems. J. Chem. Phy. 2003, 118, 7614–7621. [Google Scholar] [CrossRef]
- Nakai, S.; Inoue, Y.; Hosomi, M.; Murakami, A. Myriophyllum spicatum-released allelopathic polyphenols inhibiting growth of blue-green algae Microcystis aeruginosa. Water Res. 2000, 34, 3026–3032. [Google Scholar] [CrossRef]
- De Stefani, G.; Tocchetto, D.; Salvato, M.; Borin, M. Performance of a floating treatment wetland for in-stream water amelioration in NE Italy. Hydrobiologia 2011, 674, 157–167. [Google Scholar] [CrossRef]
- Afzal, M.; Khan, Q.M.; Sessitsch, A. Endophytic bacteria: Prospects and applications for the phytoremediation of organic pollutants. Chemosphere 2014, 117, 232–242. [Google Scholar] [CrossRef] [PubMed]
- De Meyer, S.E.; De Beuf, K.; Vekeman, B.; Willems, A. A large diversity of non-rhizobial endophytes found in legume root nodules in Flanders (Belgium). Soil Biol. Biochem. 2015, 83, 1–11. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, W.; Lang, D.; Zhang, X.; Cui, G.; Zhang, X. Interactions between endophytes and plants: Beneficial effect of endophytes to ameliorate biotic and abiotic stresses in plants. J. Plant Biol. 2019, 62, 1–13. [Google Scholar]
- Momose, A.; Hiyama, T.; Nishimura, K.; Ishizaki, N.; Ishikawa, S.; Yamamoto, M.; Hung, N.V.P.; Ohtake, N.; Sueyoshi, K.; Ohyama, T. Characteristics of nitrogen fixation and nitrogen release from diazotrophic endophytes isolated from sugarcane (Saccharum officinarum L.) stems. Bull. Facul. Agric. Niigata Univ. 2013, 66, 66. [Google Scholar]
- Afzal, M.; Yousaf, S.; Reichenauer, T.G.; Sessitsch, A. The inoculation method affects colonization and performance of bacterial inoculant strains in the phytoremediation of soil contaminated with diesel oil. Int. J. Phytorem. 2012, 14, 35–47. [Google Scholar] [CrossRef]
- Zhao, S.; Zhou, N.; Zhao, Z.-Y.; Zhang, K.; Wu, G.-H.; Tian, C.-Y. Isolation of endophytic plant growth-promoting bacteria associated with the halophyte Salicornia europaea and evaluation of their promoting activity under salt stress. Curr. Microbiol. 2016, 73, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.R.; Dubey, G.; Kollah, B. Endophytes of Jatropha curcas promote growth of maize. Rhizosphere 2017, 3, 20–28. [Google Scholar] [CrossRef]
- Ma, Y.; Rajkumar, M.; Freitas, H. Inoculation of plant growth promoting bacterium Achromobacter xylosoxidans strain Ax10 for the improvement of copper phytoextraction by Brassica juncea. J. Environ. Manag. 2009, 90, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.-J.; Guan, D.-X.; Luo, J.; Rathinasabapathi, B.; Ma, L.Q. Characterization of arsenic-resistant endophytic bacteria from hyperaccumulators Pteris vittata and Pteris multifida. Chemosphere 2014, 113, 9–16. [Google Scholar] [CrossRef]
- Ijaz, A.; Shabir, G.; Khan, Q.M.; Afzal, M. Enhanced remediation of sewage effluent by endophyte-assisted floating treatment wetlands. Ecol. Eng. 2015, 84, 58–66. [Google Scholar] [CrossRef]
- Rehman, K.; Imran, A.; Amin, I.; Afzal, M. Inoculation with bacteria in floating treatment wetlands positively modulates the phytoremediation of oil field wastewater. J. Hazard. Mater. 2018, 349, 242–251. [Google Scholar] [CrossRef]
- Li, X.-N.; Song, H.-L.; Li, W.; Lu, X.-W.; Nishimura, O. An integrated ecological floating-bed employing plant, freshwater clam and biofilm carrier for purification of eutrophic water. Ecol. Eng. 2010, 36, 382–390. [Google Scholar] [CrossRef]
- Wu, Q.; Hu, Y.; Li, S.; Peng, S.; Zhao, H. Microbial mechanisms of using enhanced ecological floating beds for eutrophic water improvement. Bioresour. Technol. 2016, 211, 451–456. [Google Scholar] [CrossRef]
- Zhao, T.; Fan, P.; Yao, L.; Yan, G.; Li, D.; Zhang, W. Ammonifying bacteria in plant floating island of constructed wetland for strengthening decomposition of organic nitrogen. Trans. Chin. Soc. Agric. Eng. 2011, 27, 223–226. [Google Scholar]
- Feng, Y.; Zhang, H. Experimental study on nitrogen and phosphorus removal efficiency of eutrophical lake by floating combination biological technology. J. Kunming Univ. Sci. Technol. (Nat. Sci. Ed.) 2012, 1. [Google Scholar]
- Cao, W.; Zhang, H.; Wang, Y.; Pan, J. Bioremediation of polluted surface water by using biofilms on filamentous bamboo. Ecol. Eng. 2012, 42, 146–149. [Google Scholar] [CrossRef]
- Bhaskaran, K.; Nadaraja, A.V.; Tumbath, S.; Shah, L.B.; Veetil, P.G.P. Phytoremediation of perchlorate by free floating macrophytes. J. Hazard. Mater. 2013, 260, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zhang, S.; Ding, Z.; Aziz, R.; Rafiq, M.T.; Li, H.; He, Z.; Stoffella, P.J.; Yang, X. Enhanced purification of eutrophic water by microbe-inoculated stereo floating beds. Pol. J. Environ. Stud. 2013, 22, 957–964. [Google Scholar]
- Liu, J.; Wang, F.; Liu, W.; Tang, C.; Wu, C.; Wu, Y. Nutrient removal by up-scaling a hybrid floating treatment bed (HFTB) using plant and periphyton: From laboratory tank to polluted river. Bioresour. Technol. 2016, 207, 142–149. [Google Scholar] [CrossRef]
- Saleem, H.; Rehman, K.; Arslan, M.; Afzal, M. Enhanced degradation of phenol in floating treatment wetlands by plant-bacterial synergism. Int. J. Phytorem. 2018, 20, 692–698. [Google Scholar] [CrossRef]
- Saleem, H.; Arslan, M.; Rehman, K.; Tahseen, R.; Afzal, M. Phragmites australis—A helophytic grass—Can establish successful partnership with phenol-degrading bacteria in a floating treatment wetland. Saudi J. Biol. Sci. 2018, 26, 1179–1186. [Google Scholar] [CrossRef]
- Sun, Z.; Xie, D.; Jiang, X.; Fu, G.; Xiao, D.; Zheng, L. Effect of eco-remediation and microbial community using multilayer solar planted floating island (MS-PFI) in the drainage channel. bioRxiv 2018, 1, 327965. [Google Scholar]
- Rehman, K.; Imran, A.; Amin, I.; Afzal, M. Enhancement of oil field-produced wastewater remediation by bacterially-augmented floating treatment wetlands. Chemosphere 2019, 217, 576–583. [Google Scholar] [CrossRef]
- Tara, N.; Iqbal, M.; Mahmood Khan, Q.; Afzal, M. Bioaugmentation of floating treatment wetlands for the remediation of textile effluent. Water Environ. J. 2019, 33, 124–134. [Google Scholar] [CrossRef]
- Tara, N.; Arslan, M.; Hussain, Z.; Iqbal, M.; Khan, Q.M.; Afzal, M. On-site performance of floating treatment wetland macrocosms augmented with dye-degrading bacteria for the remediation of textile industry wastewater. J. Clean. Prod. 2019, 217, 541–548. [Google Scholar] [CrossRef]
| Bacteria | Metal | Reference |
|---|---|---|
| Lactobacillus delbrueckii and Streptococcus thermophillus | Fe, Zn | [200] |
| Acinetobacter sp., Bacillus megaterium and Sphingobacterium sp. | Fe, Mn | [201] |
| Anoxybacillus flavithermus | Fe, Cu | [202] |
| Leptothrix, Pseudomonas, Hyphomicrobium and Planctomyces | Mn | [203] |
| Methylobacterium organophilum | Cu, Pb | [204] |
| Herminiimonas arsenicoxydans | As | [205] |
| Enterobacter cloaceae | Cd, Cu, Cr | [206] |
| Acetobacter | Pb, Cu, Mn, Zn, Co | [207] |
| Chryseomonas luteola | Cd, Co, Cu, Ni | [208] |
| Ochrobactrum anthropi | Cr, Cu | [209] |
| Anabaena spiroides | Mn | [210] |
| Ralstonia solanacearum | Pb | [211] |
| Proteobacteria and Bacteroidetes | Cu | [212] |
| Bacillus cereus | Cu | [213] |
| Bacillus licheniformis | Pb | [214] |
| Ralstonia solanacearum | Pb | [211] |
| Enterobacter aerogenes | Cd | [215] |
| SPseudomonas azotoformans | Cd, Cu, Pb | [216] |
| Bacteria/Bacterial Biofilm | Nature of Bacteria | Plant | Plant–Bacteria Interaction | Summary | Reference |
|---|---|---|---|---|---|
| Bacterial Biofilm | __ | Ipomoea aquatic and Corbicula fluminea | __ | The removal efficiencies of TN, NH4+-N, TP, total organic carbon (TOC), Chl-a, total microcystin-LR and extracellular microcystin-LR were 52.7%, 33.7%, 54.5%, 49.2%, 80.2%, 77.4% and 68.0%, respectively. | [261] |
| Proteobacteria | Nitrosomonadaceae | Canna Indica and Iris pseudacorus | Bacteria were mainly attached on the fiber filling of floating mat and plant roots | The average removal efficiencies of chemical oxygen demand (COD), TN, NH3-N and TP for Canna indica set-up were 23.1%, 15.3%, 18.1% and 19.4% higher, respectively, than that of the setup with only substrate, and 14.2%, 12.8%, 7.9% and 11.9% higher than Iris pseudacorus. FTWs. | [262] |
| Nitrifying and Denitrifying | Carrying nirS, nirK and amoA genes | Unplanted | Specific microbial communities were visualized with denaturing gradient gel electrophores (DGGE) | COD was efficiently removed in all systems examined (>90% removal). Ammonia was efficiently removed by nitrification. Removal of total dissolved nitrogen was ∼50% by day 28 | [22] |
| Biofilms | __ | Carex virgate, Cyperus ustulatus, Juncus edgariae, and Schoenoplectus tabemaemontani | Biofilm performed a key role in the removal of Cu, P and FSS. Plant roots and biofilm interaction enhanced metal speciation | The presence of a planted floating mat with biofilms improved removal of copper (>six-fold), fine suspended particles (∼threefold reduction in turbidity) and dissolved reactive P compared to the control. | [11] |
| Ammonifying bacterial strains | Engineering bacterial strain | Cymbidium faberi | The ammonifying bacteria adhered to plants roots enhanced oxygen supply to microorganism involved in nitrification process and increased capacity of plants roots to absorb ammonia nitrogen. | The organic nitrogen decomposition rate was up to 86.50% by adding the strain agent while it was 75.66% without them in the control test group in FTWs | [263] |
| Adsorptive biofilm | Natural | Thalia dealbata | Combined action of plant and biofilms | The average removal rates for TN, NH4+-N, NO3−-N NO2−-N, TP and chlorophyll-a in summer–autumn season were 36.9%, 44.8%, 25.6%, 53.2%, 43.3% and 64.5%, respectively, effectively reduced the concentrations of total suspended solids (TSS), Escherichia coli and heavy metals. | [55] |
| Photosynthetic bacteria | __ | Vetiveria zizanioids | Combined action of plant and inoculated bacteria improved purifying effect of FTWs | Efficiently removed TN and TP | [264] |
| Biofilm Reactor | Protozoa and Metazoa | Bambusoideae | In the batch reactor, COD was mainly removed by the biofilm on the filamentous bamboo | The removal rate of the COD, NH4+–N, turbidity, and total bacteria were 11.2–74.3%, 2.2–56.1%, 20–100% | [265] |
| Acinetobacter sp. | Perchlorate reducing bacterium | Pistia stratiotes | Phyto-accumulation and rhizo-degradation were key mechanisms involved in perchlorate removal | Pistia showed 63.8 ± 4% (w/v) removal of 5 mg/L level perchlorate in 7 days | [266] |
| Denitrifying polyphosphate accumulating microorganisms | __ | Festuca arundinacea | Improved the growth of plant and biomass | The average removal rates were 86.32%, 93.60%, 90.12%, 72.09%, and 84.29%, respectively, for NH4+-N, NO3¯-N, TN, TP, and ortho-P. | [267] |
| Acinetobacter, Bacillus cereus and Bacillus licheniformis | Endophytic bacteria | Brachiaria mutica | The inoculated bacteria showed persistence in water as well as successfully colonized the root and shoots of the plants | Maximum reduction in COD, biological oxygen demand (BOD5), TN, and PO4 was achieved by the combined use of plants and bacteria. | [259] |
| Biofilms | Natural | Juncus effuses Carex riparia | Metals were found in the root biofilm, probably due to microbial respiration activity | Analysis showed Ni concentration in leaves were between 23 and 31 μg/g dry matter, and between 113 and 131 μg/g in roots. Accumulation of Zn was 45-80 μg/g in leaves and 168–210 μg/g in roots. | [14] |
| Klebsiella sp., Pseudomonas sp. and Acinetobacter sp. | Endophytic Bacteria | Typha domingensis | Possessed pollutant-degrading and plant growth-promoting abilities and successful survival of bacteria was found in plant tissues | The average reduction in COD and BOD5 was 87% and 87.5%, and significantly removed heavy metals. | [26] |
| Biofilm | Nitrifying and denitrifying bacteria | Canna indica | Improved nitrification and denitrification process and overall high removal of total nitrogen | Significantly higher removal rates of ammonia nitrogen (85.2%), total phosphorus (82.7%), and orthophosphate (82.5%) were observed | [18] |
| The community was mainly composed of Cyanobacteria, Proteobacteria, Bacteroidetes, Planctomycetes, Firmicutes, Actinobacteria, Chlorobi and Acidobacteria. | Periphyton | __ | Improved its nutrient removal capacity | Successfully maintained TN and TP concentration in the river water at less than 2.0 and 0.02 mg L−1 respectively | [268] |
| Dechloromonas, Thiobacillus and Nitrospira | Heterotrophic and autotrophic | __ | Mixotrophic denitrification occurred in auto and heterotrophic bacteria | About 89.4% of the TN was removed from autotrophic coupled floating wetlands, and 88.5% from heterotrophic enhanced floating wetlands | [39] |
| Bacillus subtilis, Klebsiella sp., Acinetobacter Junii and Acinetobacter sp. | Hydrocarbon degrading bacteria | Brachiara mutica and Phragmites australis | Alkane-degrading gene (alkB) abundance confirmed microbial growth in plant’s root and shoot and in water. | Reduced oil content (97%), COD (93%), and BOD (97%), in wastewater | [260] |
| Acinetobacter lwofii, Bacillus cereus, and Pseudomonas sp. | Phenol-degrading bacteria | Typha domingensis | The inoculated bacteria showed successful colonization and survival in the rhizosphere, root interior and shoot interior of the plant and enhanced plant growth and biomass | Bacterial augmentation enhanced the removal potential significantly, i.e., 0.146 g/m2/day vs. 0.166 g/m2/day without bacterial inoculation | [269] |
| Acinetobacter lwofii, Bacillus cereus, and Pseudomonas sp. | Phenol degrading bacteria | Phragmite australis | Improved plant biomass and high rate of inoculated bacteria survival observed in plant roots, shoot and water | Plant–bacteria synergism significantly improved the phenol degradation and removal. Highest reduction in COD, BOD, and TOC was achieved by bacterial augmentation | [270] |
| Acinetobacter, Acinetobacter sp., and Bacillus niabensis | Hydrocarbons degrading bacteria | Leptochloa fusca | Achieved successful degradation of Hexadecane The Inoculated bacteria displayed highest persistence in the roots followed by shoots and then in the wastewater and improved plant growth promoting (PGP) activities | Hydrocarbons degradation was recorded up to 92%, COD was reduced up to 95%, BOD up to 84%, and TDS up to 47% and alleviated the toxicity | [41] |
| Archaea, anaerobic ammonium oxidation (Anammox) bacteria | Natural | Oenanthe javanica | High abundance and diversity of bacteria in planted floating wetland | The average removal rates of NH4+-N, NO3–-N and total nitrogen were 78.3, 44.4 and 49.7% respectively | [44] |
| Proteobacteria Actinobacteria Cyanobacteria, and Rhizorhapis | __ | Eichhornia crassipes | Bacteria were involved in pollutant degradation and nutrients removal | Suspended solids, TN, TP, NO3–-N and COD was 86%, 75%, 80%, 95% and 84%, respectively. | [271] |
| Bacillus subtilis, Klebsiella sp., Acinetobacter Junii, and Acinetobacter sp. | Hydrocarbon degrading bacteria | Typha domingensis and Leptochloa fusca | Persistence of bacteria and expression of the alkB gene in the rhizoplane of inoculated plants | Reduction in hydrocarbon (95%), COD (90%), and BOD content (93%) | [272] |
| Acinetobacter junii, Pseudomonas indoloxydans, and Rhodococcus sp. | Rhizospheric and endophytes | Phragmites australis and Typha domingensis | Removal efficiency was further enhanced by augmentation with bacteria and promoted plant growth | Color, COD and BOD after an 8-day period were 97, 87 and 92%, respectively, 87–99% reduction in heavy metals | [273] |
| Consortium of five strains namely Aeromonas salmonicida, Bacillus cerus, Pseudomonas indoloxydans, Pseudomonas gessardii, and Rhodococcus sp. | Rhizospheric and endophytes | Phragmites australis and Brachia mutica | Persistence and survival of inoculated bacteria in roots and shoots, and inoculated bacteria improved the plant growth and biomass production | Reduced COD, BOD5, and TOC up to 85.9%, 83.3%, and 86.6% in 96 h, respectively. TN was reduced from 37.5 to 2.07 mg l−1, N from 33.3 to 1.23 mg l−1, and TP from 2.63 to 0.53 mg l−1. Trace metals were also reduced up to 79.5% for iron, 91.4% for nickel, 91.8% for manganese, 36.14% for lead, and 85.19% for chromium. | [20] |
| Acinetobacter juniistrain, Rhodococcus sp. strain, and Pseudomonas indoloxydans | Dye degrading bacteria | Phragmites australis | The inoculated bacteria showed persistence in water, roots and shoots of inoculated plants of FTWs | The COD was reduced to 92%, BOD to 91%, color to 86%, and trace metals to approximately 87% in the treated wastewater. | [274] |
| Bacillus cerus, Cyperus laevigatus, Aeromonas salmonicida and Pseudomonas gessardii, | Rhizospheric and endophytes | Typha domingensis and Leptochloa fusca | Improved remediation performance of inoculated plants, inoculated bacteria were found in root and shoots of inoculated plants | The TN, NO3−1 and TP contents decreased to 1.77 mg l−1, 0.80 mg l−1 and 0.60 mg l−1, respectively. Additionally, the concentration of iron, nickel, manganese, lead, and chromium in the water lowered to 0.41, 0.16, 0.10, 0.25, and 0.08 mg l−1, | [131] |
| These strains were Ochrobactrum intermedium, Microbacterium oryzae, Pseudomonas, Acinetobacter sp., Klebsiella sp., Acinetobacter sp., P. aeruginosa, Bacillus subtilus, and Acinetobacter junii | Bacteria possessing capabilities of hydrocarbon degradation, rhamnolipid production, and plant growth promotion. | Phragmites australis, Typha domingensis, Leptochloa fusca, and Brachiaria mutica | Produced biosurfactants and promoted plant growth. Bacteria showed persistent in the rhizoplane, roots and shoots of plants | Reduced COD, BOD, TDS, hydrocarbon content, and heavy metals by 97.4%, 98.9%, 82.4%, 99.1%, and 80%, respectively, within 18 months. | [25] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahid, M.J.; AL-surhanee, A.A.; Kouadri, F.; Ali, S.; Nawaz, N.; Afzal, M.; Rizwan, M.; Ali, B.; Soliman, M.H. Role of Microorganisms in the Remediation of Wastewater in Floating Treatment Wetlands: A Review. Sustainability 2020, 12, 5559. https://doi.org/10.3390/su12145559
Shahid MJ, AL-surhanee AA, Kouadri F, Ali S, Nawaz N, Afzal M, Rizwan M, Ali B, Soliman MH. Role of Microorganisms in the Remediation of Wastewater in Floating Treatment Wetlands: A Review. Sustainability. 2020; 12(14):5559. https://doi.org/10.3390/su12145559
Chicago/Turabian StyleShahid, Munazzam Jawad, Ameena A. AL-surhanee, Fayza Kouadri, Shafaqat Ali, Neeha Nawaz, Muhammad Afzal, Muhammad Rizwan, Basharat Ali, and Mona H. Soliman. 2020. "Role of Microorganisms in the Remediation of Wastewater in Floating Treatment Wetlands: A Review" Sustainability 12, no. 14: 5559. https://doi.org/10.3390/su12145559
APA StyleShahid, M. J., AL-surhanee, A. A., Kouadri, F., Ali, S., Nawaz, N., Afzal, M., Rizwan, M., Ali, B., & Soliman, M. H. (2020). Role of Microorganisms in the Remediation of Wastewater in Floating Treatment Wetlands: A Review. Sustainability, 12(14), 5559. https://doi.org/10.3390/su12145559






