Abstract
A remarkable portion of crop genetic diversity is represented by landraces and obsolete cultivars, that have not yet been integrated into the collections of genetic resources in genebanks. Fortunately, they are still maintained by small farmers in rural areas. Their acquisition is an important goal for collecting expeditions, followed by the evaluation of their genetic background. Molecular markers are powerful tools to evaluate the effectiveness of acquisition of new genetic variants. This approach was used for common bean landraces, that were collected through expeditions in the Slovak and Ukrainian Carpathians. In this study, microsatellite markers, developed from expressed sequence tags, were used for genotyping the collected materials. The sub-population of collected landraces contained higher total and average number of different alleles in comparison to equally large sub-populations of already-maintained cultivars. The same was true for the Shannon’s information index, observed heterozygosity, and expected heterozygosity. Both sub-populations showed the presence of private alleles: Average values of 0.500 and 0.833 private alleles per genotype were detected in landraces, and cultivars, respectively. Obtained results emphasized the importance of collecting expeditions to specific regions where landraces are cultivated, even at the present time. The effectiveness of the acquisition of new genetic variability can be determined by molecular tools, as confirmed by microsatellite markers in this study.
1. Introduction
The common bean (Phaseolus vulgaris L.) is a worldwide cultivated crop and important source of foods rich in proteins. Common bean has undergone two independent domestication events: One from each gene pool, the Mesoamerican and Andean, that diverged from a common ancestral wild population [1,2]. The ways of the common bean to Europe have been directed from both the Mesoamerican and Andean centre of origins, through the Iberian Peninsula in the early 16th century. Reposition across continents has had consequences such as reduced genetic diversity, because only a part of the original gene pools has been transferred into Europe [3]. The effect, related to the bottleneck of introduction, was mitigated in Europe by frequent inter-gene pool (Mesoamerican and Andean) hybridization, that was extensive and greater than in the Americas [4,5]. During more than five centuries after introduction into Europe specific common bean variants have been created. This development was the consequence of the adaptation to specific and variable soils, climatic conditions, new pathogens and pests, later divergent and extensive breeding process, as well as new cultivation practices. Many studies have supported the emergence of new genetic diversity in the secondary centers of diversification [4,5,6,7,8,9,10]. The entire European continent can be regarded as one of the secondary diversification centers for Phaseolus vulgaris L. and European common bean landraces are of great value for bean breeding [4,5]. Different developments occurred in Africa and China, where the distribution of the Mesoamerican and Andean common bean gene pools was more uneven [11,12]. Genetic variation is currently maintained in the European collections of bean genetic resources, that has increased to such an extent, that it is possible to create core collections also in common bean [13].
However, modern crop production prefers improved cultivars that replaced traditional landraces and obsolete cultivars over time. This also happened in the common bean. Less yielding landraces disappeared from agricultural production and from agro-ecosystems. Fortunately, they are still maintained by small farmers in the countryside. Even if the common bean is a predominantly autogamous species, genetic diversity within-population (also within-landraces) can be rather high. This diversity can help buffering stresses but, more importantly, can drive adaptation to specific, local environmental conditions.
The responsibility of curators managing collections of genetic resources in genebanks is to enrich already existing collections by the acquisition of new plant accessions, that represent genetic diversity as widely as possible, but with a minimal level of redundancy. Curators should find, collect, characterize, and preserve new materials, especially landraces and obsolete cultivars as sources of new genes, genotypes, and phenotypes. One approach to enrich the collection with new germplasm is to organize collecting expeditions. Such missions, which are organized into relatively small and specific regions, can reveal autochthonous landraces that have a variation in plant habit, seed characteristics, and other traits [14,15,16,17]. Plant accessions, that are collected in the same area, are commonly expected to be mutually more similar than those from distant regions. Nevertheless, distinct regional conditions can objectively create variations within species. Therefore, collecting expeditions over there can be very effective and new germplasm can be acquired [5]. Landraces and obsolete cultivars, that are maintained by local growers, are prospective sources of genetic variation and should be collected, saved, and used in breeding programs. This was confirmed in germplasm collected in marginal areas in the Southern European countries [14,18] and in the Eastern and Central Europe [19,20]. The lines selected from landraces possessed tolerance or resistance against anthracnose, bacterial blights, bean common mosaic virus, and other superior agronomical characteristics [21]. On the other side, collected accessions can enhance a level of redundancy in the species gene pool if they are not characterized efficiently. An appropriate sampling strategy determines the efficiency of new germplasm acquisition. The value of new-acquired germplasm can be increased by the identification of genotype and characterization of phenotypic traits useful for cultivation and breeding. Evaluating approaches are based on tens of morphological and agronomical characteristics, according to species-specific descriptors. Molecular markers, such as seed storage proteins, isoenzymes, and DNA [1,22,23,24,25,26,27] are used intensively for the characterization of genotypes. Microsatellite sequences, located in non-coding and regulatory DNA regions [28,29,30], generate valuable variation for the characterization of genetic diversity in plants, including the common bean [31,32,33]. Large sets of expressed sequence tags single sequence repeats (EST-SSR), i.e., microsatellites within coding DNA sequences, have already been developed for common bean [34,35] and are currently available in public databases. These markers can be used for genetic diversity evaluation of different genepools, functional diversity analysis, selection of breeding lines with specific traits [35,36].
The goal of this study was based on the assumption that: i) There is still a common bean germplasm maintained by rural growers in the countryside that was not yet included in the genetic diversity conservation program, ii) the efficiency of new genetic variation acquisition, by collecting expeditions, can be determined by easily accessible microsatellite markers. Therefore, the aim of this study was to evaluate the significance of already realized collection missions, in terms of the effective acquisition of new common bean germplasm, possessing new genetic variants that could enrich the collection of common bean genetic resources maintained in the genebank.
2. Materials and Methods
2.1. Plant Material
The set of 173 common bean (Phaseolus vulgaris L.) accessions, maintained within the collection of bean genetic resources, were obtained from the Genebank of the Slovak Republic (Research Institute of Plant Production, Piešťany). Eighty-seven of them were cultivars originated and released in Slovakia (SVK), Czech Republic (CZE), Germany (DEU), France (FRA), Hungary (HUN), Netherlands (NLD), and the USA (Table S1). The set of landraces included 86 accessions collected by collecting expeditions in Carpathian Mountains in Slovakia and Ukraine (UKR) (Table S2) in the period 1992–1996. Most of landraces collected during the expeditions were mixtures containing seeds differing in size, shape, color, drawing, and other parameters. Seeds with identical characteristics, within each individual mixture, were sorted according to basic seed characteristics, pooled, and subsequently multiplicated to produce pure lines [37].
2.2. Genetic Characterization
The genomic DNA was extracted from young leaves [38]. DNA from each common bean accession was bulk DNA prepared from equivalent amounts of DNA, extracted from ten individual plants. Altogether, 18 microsatellite-derived primer pairs were used for analyses. Eight pairs (PHVPVPK, PVGLND5, PVGSR1, PVME1, YU-1, YU-2, YU-3, YU-4) were developed previously [39,40]. Another ten pairs of primers (BNG1, BNG2, BNG3, BNG4, BNG5, BNG6, BNG91, BNG95, BNG191, PDX1) were designed within our study after searching for tandem repeats in DNA sequences of common bean within the GenBank database [41]. Primers used in polymerase chain reactions (PCR) were designed using the software Primer3 [42]. All amplification reactions were done in 15 μl volumes containing 25 ng of DNA, 1 x PCR buffer (50 mmol. L−1 KCl, 10 mmol. L−1 TRIS-HCl, pH 8.3), 1.5 mmol L−1 MgCl2, 0.1 μmol·L−1 both of primers, 0.1 μmol L−1 each of dNTPs, and 0.6 U of Taq-DNA polymerase. The amplification conditions included an initial denaturation 3 min at 94 °C, followed by 30 cycles of denaturation 1 min at 94 °C, annealing 1 min at 48 °C, extension 1 min at 72 °C; and final extension 8 min at 72 °C. All PCR reactions were performed in the PTC-200 Thermal Cycler (MJ Research, Inc.). The equivalent volume of loading buffer (95% formamide, 10 mmol L−1 NaOH, 0.05% bromphenol blue) was added to each sample. The samples were denatured for 3 min at 99 °C and 5 μl of each sample was loaded into 6% polyacrylamide gel containing 7 mol L−1 of urea. Gels in electrophoretic unit (SEQ 3341, Scie-Plas) were run in 0.5 x TBE buffer at constant power of 45 W for 3.5–5 h, depending on the size of amplified fragments. Microsatellite DNA was stained by silver staining method [43]. The 100 bp, 50 bp, 25 bp, and 10 bp DNA ladders (Invitrogen) were used as microsatellite allele size markers. DNA data were documented by the software Kodak Digital Science 1D v. 3.0.0 (Eastman Kodak Co., Rochester, NY, USA).
2.3. Data Analysis
Polymorphic DNA microsatellite markers were considered as different alleles. The number of effective alleles, Shannon’s information index, observed and expected heterozygosity, and the fixation index were calculated using the software GenAIEx v. 6.5 [44]. The polymorphism information content (PIC) was calculated by the software Cervus 3.0.7 [45]. The analysis of molecular variance (AMOVA) was performed using the software Arlequin v. 3.5 [46]. Population structure was assessed by the STRUCTURE v. 2.3.4 software [47] using the default setting of the admixture model for the ancestry of individuals and correlated allele frequencies. The models were tested for K-values ranging from one to fifteen, with ten independent runs each. Burn-in and Markov Chain Monte Carlo iterations were set to 100,000. The most likely number of clusters was chosen by plotting the LnP(D) values against ΔK values with the best K value selected according to the Evanno test [48]. The tree in STRUCTURE was estimated using the program NEIGHBOR by Mary Kuhner and John Yamato, implementing Saitou and Nei’s “Neighbor Joining Method” [49]. The plot was produced using DRAWTREE by Joe Felsenstein [50]. Both programs are distributed by Joe Felsenstein as part of his PHYLIP phylogeny package [50].
3. Results
All 18 used primer pairs originated from the genic regions of common bean DNA and reflected genetic information of specific genes. Amplified microsatellite markers were located over nine out of the 11 common bean chromosomes, with the exception of chromosomes Pv08 and Pv10 (Table S3). The position of the markers was verified using on-line tools implemented on Phytozome v 12.1.6. [51]. Six EST-SSR markers were excluded from analyses, either due to the absence of the amplification product (YU-4) or the generation of monomorphic patterns (BNG1, BNG2, BNG3, BNG95, and BNG191). The remaining twelve markers generated polymorphic patterns, producing 69 different alleles. The number of alleles per locus ranged from 2 to 18 with a mean of 5.08 per locus (Table 1). The highest allelic variation (18 different alleles) was in the microsatellite containing di-nucleotide motif (AT)n located within the 5'-untransribed region of the gene PvME1 encoding the NADP-dependent malic enzyme (Table S3).

Table 1.
Parameters of genetic diversity of sub-populations of cultivars (87 accessions) and landraces (86 accessions) of common bean (Phaseolus vulgaris L.).
The 173 analyzed common beans accessions belong to two sub-populations. One represented 87 registered (released) cultivars, the other 86 landraces, acquired by collecting expeditions, respectively. The registered cultivars possessed EST-SSR loci altogether 59 alleles, whereas landraces possessed 63 alleles. The analysis of molecular diversity, performed by the AMOVA associated genetic variance among sub-populations (cultivars, and landraces, respectively), among individuals, and within individuals. Proportion of molecular variation among sub-populations accounted 6.5% of the total variation and was significant (p < 0.001), as indicated by the randomization test. Variation caused by differences among individuals of sub-populations was 75.8% and variation within individuals caused by their heterozygosity was 17.7% (Figure 1).
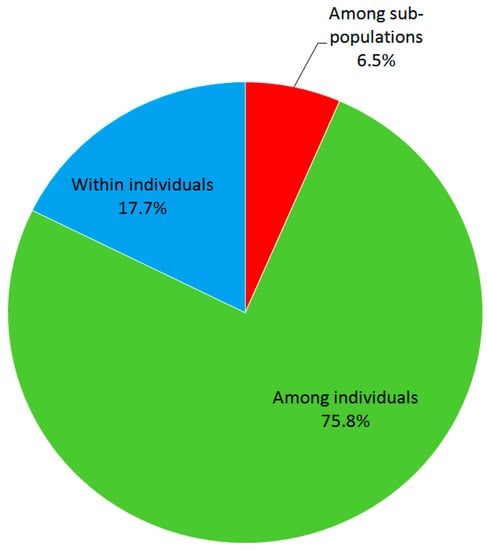
Figure 1.
The AMOVA analysis presenting the percentage of molecular variance among, and within, individuals, and among sub-populations (cultivars versus landraces).
The mean number of different alleles (Na) was 4.917 and 5.250, the mean number of effective alleles (Ne) was 2.357 and 3.147 within sub-populations of registered cultivars, and landraces, respectively (Table 1, Figure 2). The Ne values indicated that allele frequencies were different in both sub-populations and only around one half of alleles had significant frequencies. Higher degrees of genetic diversity, presented by the number of alleles, was in the sub-population of landraces, compared to sub-population of registered cultivars.

Figure 2.
Mean values of number of alleles (Na), different alleles found in 25% or fewer populations (NaFreq.), number of effective alleles (Ne), Shannon’s information index (I), number of private alleles, and heterozygosity (He) in the common bean sub-populations of registered cultivars and landraces, respectively.
The Shannon’s information index (I) indicated efficiency of microsatellite loci to reveal genetic variation. The highest values were for the locus PvME1 (2.547 and 1.999), the lowest for BNG5 locus (0.188 and 0.186) in sub-populations of landraces and cultivars, respectively (Table 1). The mean values of Shannon’s information index indicated that higher genetic diversity has been in sub-population of landraces (I = 1.012), lower in sub-population of cultivars (I = 0.889), respectively. It also means that this parameter indicated slightly higher genetic variation contained in landraces in comparison with cultivars.
The observed heterozygosity (Ho) at individual EST-SSR loci ranged from zero to 0.779. The higher mean value of Ho was in sub-population of landraces indicating that landraces were more diverse population than registered cultivars. Observed heterozygosity in all loci, as well as the mean values of this parameter was distinctly different from expected heterozygosity (He) under the Hardy-Weinberg equilibrium in both sub-populations (Table 1, Figure 2). The mean He was 0.449 in registered cultivars and 0.521 in landraces (Table 1, Figure 2) and was much higher than the mean Ho in both sub-populations (0.074 in cultivars and 0.109 in landraces). This suggests higher genetic diversity contained in landraces, and at the same time, reveals a tendency towards increasing homozygosity and deficiency of heterozygotes in registered cultivars.
The polymorphic information content (PIC) of individual EST-SSR markers ranged from 0.084 to 0.891. The lowest discrimination power for both sub-populations was in the least variable locus BNG5, the opposite was the locus PvME1 (Table 1). Nevertheless, an average PIC value in both sub-populations were relatively high (0.405–0.457) and markers BNG-4, PHPVPK, PVGLND5, PvME1 had PIC value higher than 0.5.
The mean fixation indices (F) within both sub-populations were very high, 0.876, and 0.844, respectively (Table 1). This may indicate that crossing between accessions of sub-populations of registered cultivars, as well as landraces, was limited. High estimates of the fixation index (0.860) between sub-populations of cultivar and landraces was observed.
Both sub-populations also contained private alleles [52,53], i.e., alleles found only in one, or only in other sub-populations. Released cultivars possessed six, and landraces ten of the private alleles, respectively. On average, it was 0.833 of private allele per genotype in sub-population of landraces and 0.500 in sub-population of cultivars (Figure 2). Five from all 16 private alleles detected in this study were at the locus PvME1, four in landraces, one in cultivars. A high total number of private alleles both, in cultivars and landraces, showed that both sub-populations are abundant in genetic variation. The pooling of both germplasms into a common collection is beneficial for expanding the conserved bean genetic diversity.
The modeled population structure of common beans constructed by the STRUCTURE software using the program STRUCTURE HARVESTER [54] at the best selected value K = 3 (Figure S1) organized 173 accessions into three groups (Figure 3). All analyzed common beans were for graphical presentations, hierarchically organized according to affiliation either, to cultivars, or to landraces, respectively. The percentage of genotypes with unambiguous membership (coefficient of membership ≥ 90%) to one of the three groups was 74.57%. This indicates that the vast majority of cultivars, and landraces, respectively, were strongly assigned to relevant sub-population. The first group (Figure 3, blue bars) contained predominantly registered cultivars. Of the 87 registered cultivars, up to 60, i.e., 68.97% were clearly assigned to this group. The second group consists mainly of landraces collected during missions (green bars in Figure 3). Of the 86 landraces, 70 (81.40%) were included in this group. The STRUCTURE analysis revealed also the third group (Figure 3 and Figure 4) whose 43 members (red bars) were scattered in both other groups (Figure 3). One accession, the Slovak cultivar Žltostruková (No. 85 in Figure 3) was not unambiguously assigned to any group. Its coefficients of membership were 0.292 to the first group, 0.321 to the second group, and 0.387 to the third group.
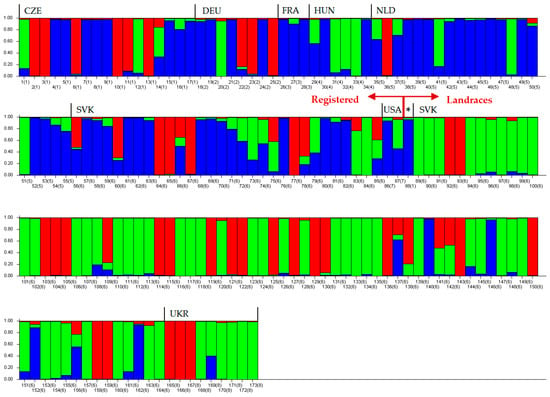
Figure 3.
Assignment of 173 common beans accessions to three populations by the STRUCTURE analysis (ΔK = 3). Each bar represents individual accession, numbers of bars are the same as in supplementary Tables S1 and S2. Numbers 1–87 = registered cultivars, 88–173 = landraces (* only one landrace—No. 88 Maslová královná, from Czech Republic).
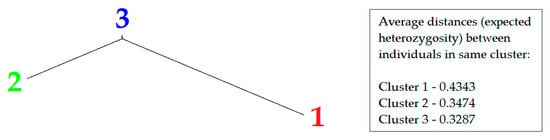
Figure 4.
Genetic tree of 173 common beans grouped into three clusters according to differences in expected heterozygosity within them (colors of numbers correlate with colors of accessions in Figure 3).
Expected heterozygosity (He) is an estimator of gene diversity present in subdivided populations [55]. Based on the average distances of the He parameter landraces (mainly belonging to group 1 and 2) are more diverse, on average, than cultivars (mainly belonging to group 3).
4. Discussion
The collecting expeditions, organized into the Inner Western and Outer Eastern Carpathians, in the territories of Slovakia and Ukraine, were aimed at rural areas with the goal of acquiring new common bean germplasm. The seed samples were obtained from locals who grow beans usually only for their own consumption, and for their income, as part of the productions sold in local markets. After multiplication of obtained seeds, an extensive evaluation of morphological and agronomical traits was done in field trials and the phenotype variation, described [37]. During former bean breeding program supported in Slovakia a lot of new cultivars were released for farmers and gardeners; later, many foreign cultivars were also made available for them. This could suggest that bean landraces, acquired during the collection expeditions around the country, could have been cultivars already developed and released in the past. If it was so, the genebank collection of bean genetic resource would have been supplemented only by redundant accessions. However, the purpose of the collecting expeditions is to find and acquire new genetic diversity. Such diversity, represented by landraces and obsolete cultivars, can still exist in situ, and its collection can increase the genetic diversity kept in ex situ collections [11].
The molecular analysis in our study confirmed differences between analyzed cultivars and landraces collected during expeditions, third group of beans accessions was also separated according to allelic variants at analyzed microsatellite loci. The beans of this group could be brought by small farmers and gardeners from other, neighboring countries. However, the existence of this group can also reflect the effect of the gene flow and gene exchange by hybridization between obsolete cultivars, landraces, and released cultivars. Hybridization between common bean genotypes is common, present everywhere in Europe, distributed unevenly, but highly frequent in central Europe [4]. This study of a large collection of European landraces revealed that different accession possibly derived from hybridization between Andean and Mesoamerican gene pools [4]. Multiple natural hybridizations certainly occurred within, and between, registered cultivars and landraces, especially when these materials are grown at the same time and location. The pressure of varying microenvironmental conditions also contributes shaping the genetic diversity and structure of common bean landrace populations [56]. The experiments with historical common bean varieties (some mentioned already in the year 1855) revealed that their genetic composition, as well as phenotypic traits, can change during multiplication in different environments within as little as three growing seasons [57].
The percentage of molecular variation among individuals in our study was 75.8% that could indicate to the existence of heterogeneity in compared common bean cultivars and landraces. High heterozygosity within individuals also contributed to molecular variation largely (17.7%). This parameter should be affected by the high proportion (one half) of landraces from the total analyzed beans. It has been reported that in progenies of some common bean lines the outcrossing rate was extreme, up to 85% [58]. However, the average rates are usually much lower [57,59]. The reasons for the gene flow include, contrasting environmental conditions, short distances between grown adjacent landraces, and higher frequency of pollination by insects. Another reason for the high heterozygosity within landraces is the fact that seeds of individual landraces could be mixtures of different genotypes with similar seed characteristics. Nevertheless, molecular diversity, that has been observed between sub-populations of cultivars and landraces, was present in our study (6.5%) and statistically significant (p < 0.001).
The parameters of the genetic diversity Na, Ne, Ho, and He, determined within landraces collected in the Inner West and Outer Eastern Carpathians, were almost the same or very similar as were determined for landraces from Italy [5], China [32], and Turkey [60]. These parameters were also significantly higher than in the landraces collected in Portugal [61]. However, it should be borne in mind that other genotypes and other markers have been used in these studies. Nevertheless, this may indicate that the Carpathian countryside in Slovakia and Ukraine is also a unique reservoir of European common bean diversity.
The average value of PIC (0.431) confirmed the usefulness and good discrimination capacity of used genic microsatellite marker system in common bean. Similar PIC values (0.47 and 0.44, respectively) were declared for EST-SSRs also in other studies in common beans [33,62]. The most interesting among analyzed loci was the locus PVME1 encoding cytosolic NADP-dependent malic enzyme. In this case, the amplified region contains a (AT)n tandem repeats located within the 5'-untranslated leader sequence of the gene [63]. This EST-SSR marker showed a very high variation (Table 1).
The most important intent of expeditions, collecting plant genetic resources, is to discover and sample new genetic variants, i.e. new allelic variation. Such genetic variants, but also other parameters, can be identified and characterized in collected samples by using different approaches, including DNA analyses. The basic parameter at the level of genes is the number of distinct alleles (allelic richness) present at single loci within the population of collected genotypes [64]. The successful collection of new germplasm depends generally on the presence and identification of unique or rare alleles, called private alleles [52], in sampled accessions. Monitoring mission-specific alleles is also convenient for studies focused on gene flow and genetic diversity shift over time. In our study, landraces contained more private alleles than registered cultivars. This is interesting in comparison with results of similar study from Portugal [61].
New plant accessions can be obtained also by material exchange between genebanks. It is an effective and easy way of getting new germplasm usable in research and developmental programs facilitated also by international initiatives [65]. Molecular marker-based approaches can effectively assist in the acquisition of new accessions, formation of core collections, and for reducing the redundancy in genebank collections [66,67]. Genomics and molecular markers help to identify and characterize newly acquired specific and unique alleles, thereby enriching the collections of genetic resources [68], but it can also help in reducing the issues related to bio-piracy arising from international exchange of plant germplasm [69]. The assessment of molecular diversity within germplasm collections is also helpful in the characterization of accessions that lack passport data [70], which is common in landraces, collected during early missions. Microsatellite markers, also those derived from coding DNA sequences, can efficiently assist the acquisition of new genetic variants (alleles) potentially useful in future breeding programs. The benefits of including landraces in plant breeding programs is the introgression of genes that might ensure, for example, improvement in absorption and utilization of nutrients, adaptation to environmental stresses, increasing productivity, and stability of yield in environments. Many common bean landraces were used as donors of important traits, such as resistance to biotic and abiotic stresses, seeds characteristics, nutritional properties (high content of proteins, fatty acids, minerals, etc.) [71,72,73,74]. In this regard, it has also been shown that common bean lines, developed from landraces, can outperform commercial cultivars under organic conditions [75]. The sampling of landraces and their introduction into already existing collections of common bean has been beneficial for the extension of already conserved genetic diversity. Moreover, genetic diversity of common bean landraces could increase nutritional composition and taste of foods prepared from the beans [14]. This can increase the demand by consumers for specific and local products with high quality and peculiar taste.
In conclusion, our results showed that rural areas still offer new genetic variants of common bean. Landraces can be collected and after specific characterization included into the genetic diversity conservation programs. Unselected germplasm still maintained in situ (on-farm) harbors a number of new allelic variants, including alleles useful in classical as well as molecular breeding approaches. Based on the results obtained in our study, organizing collecting expeditions to rigorously selected geographical areas, is still of great importance even at the present time.
Supplementary Materials
The following are available online at https://www.mdpi.com/2071-1050/11/19/5270/s1, Table S1: List of 87 registered and released common bean (Phaseolus vulgaris L.) cultivars and their origin; Table S2: List of 86 landraces of common bean (Phaseolus vulgaris L.) acquired during collecting missions in the Slovak and Ukrainian Carpathians; Table S3: List of 18 microsatellite markers used in study of common beans; Figure S1: Comparison of ln(K) and ∆K values for microsatellite dataset calculated from program STRUCTURE using the web-based program STRUCTURE HARVESTER [54], where the hypothesized number of populations ranged from 1 to 15.
Author Contributions
M.S.: molecular analyses; K.O.: statistical analyses; P.H.: collecting expeditions; D.M.: methodology and laboratory analyses; M.G.: methodology and formal analysis; J.K.: supervision, project administration, methodology, writing—draft preparation, review, and editing.
Funding
This research was funded by the Slovak Research and Development Agency, grants number APVV-15-0156, APVV-16-0026, APVV-16-0051, APVV-14-0055, and by the Operational Programme Research and Development co-financed by the European Regional Development Fund, grant number ITMS 26210120039.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gepts, P.; Osborn, T.C.; Rashka, T.; Bliss, F.A. Phaseolin protein variability in wild forms and landraces of the common bean (Phaseolus vulgaris L.): Evidence for multiple centers of domestication. Econ. Bot. 1986, 40, 451–468. [Google Scholar] [CrossRef]
- Schmutz, J.; McClean, P.E.; Jackson, S.A. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Gepts, P.; Bliss, F.A. Dissemination pathways of common bean (Phaseolus vulgaris; Fabaceae) deduced from phaseolin electrophoretic variability. II Europe and Africa. Econ. Bot. 1988, 42, 86–104. [Google Scholar] [CrossRef]
- Angioi, S.A.; Rau, D.; Attene, G.; Nanni, L.; Bellucci, E.; Logozzo, G.; Negri, V.; Spagnoletti Zeuli, P.L.; Papa, R. Beans in Europe: Origin and structure of the European landraces of Phaseolus vulgaris L. Theor. Appl. Genet. 2010, 121, 829–843. [Google Scholar] [CrossRef] [PubMed]
- Gioia, T.; Logozzo, G.; Attene, G.; Bellucci, E.; Benedettelli, S.; Negri, V.; Papa, R.; Spagnoletti Zeuli, P. Evidence for introduction bottleneck and extensive inter-gene pool (Mesoamerica x Andes) hybridization in the European common bean (Phaseolus vulgaris L.) germplasm. PLoS ONE 2013, 8, 75974. [Google Scholar] [CrossRef] [PubMed]
- Rodino, A.P.; Santalla, M.; Montero, I.; Casquero, P.A.; De Ron, A.M. Diversity of common bean (Phaseolus vulgaris L.) germplasm from Portugal. Genet. Resour. Crop Evol. 2001, 48, 409–417. [Google Scholar] [CrossRef]
- Santalla, M.; Rodino, A.; de Ron, A.M. Allozyme evidence supporting southwestern Europe as a secondary center of genetic diversity for common bean. Theor. Appl. Genet. 2002, 104, 934–944. [Google Scholar] [CrossRef] [PubMed]
- Sicard, D.; Nanni, L.; Porfiri, O.; Bulfon, D.; Papa, R. Genetic diversity of Phaseolus vulgaris L. and P. coccineus L. landraces in central Italy. Plant Breed. 2005, 124, 464–473. [Google Scholar] [CrossRef]
- Raggi, L.; Tiranti, B.; Negri, V. Italian common bean landraces: Diversity and population structure. Genet. Resour. Crop Evol. 2013, 60, 1515–1530. [Google Scholar] [CrossRef]
- Bitocchi, E.; Rau, D.; Bellucci, E.; Rodriguez, M.; Murgia, M.L.; Gioia, T.; Santo, D.; Nanni, L.; Attene, G.; Papa, R. Beans (Phaseolus vulgaris L.) as a model for understanding crop evolution. Front. Plant Sci. 2017, 8, 722. [Google Scholar] [CrossRef]
- Blair, M.W.; González, L.F.; Kimani, M.; Butare, L. Genetic diversity, inter-gene pool introgression and nutritional quality of common beans (Phaseolus vulgaris L.) from Central Africa. Theor. Appl. Genet. 2010, 121, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Blair, M.W.; Wang, S. Genetic diversity in Chinese common bean (Phaseolus vulgaris L.) landraces assessed with simple sequence repeats markers. Theor. Appl. Genet. 2008, 117, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Logozzo, G.; Donnoli, R.; Macaluso, L.; Papa, R.; Knüpffer, H.; Spagnoletti Zeuli, P. Analysis of the contribution of Mesoamerican and Andean gene pools to European common bean (Phaseolus vulgaris L.) germplasm and strategies to establish a core collection. Genet. Resour. Crop Evol. 2007, 54, 1763–1779. [Google Scholar] [CrossRef]
- Negri, V.; Tosti, N. Phaseolus genetic diversity maintained on-farm in central Italy. Genet. Resour. Crop Evol. 2002, 49, 511–520. [Google Scholar] [CrossRef]
- Negri, V. Landraces in central Italy: Where and why they are conserved and perspectives for their on-farm conservation. Genet. Resour. Crop Evol. 2003, 50, 871–885. [Google Scholar] [CrossRef]
- Piergiovanni, A.R.; Taranto, G.; Losavio, F.P.; Pignone, D. Common bean (Phaseolus vulgaris L.) landraces from Abruzzo and Lazio regions (Central Italy). Genet. Resour. Crop Evol. 2006, 53, 313–322. [Google Scholar] [CrossRef]
- Casals, J.; Casañas, F.; Simó, J. Is it still necessary to continue to collect crop genetic resources in the Mediterranean area? A case study in Catalonia. Econ. Bot. 2017, 71, 330–341. [Google Scholar] [CrossRef]
- Rodino, A.P.; Santalla, M.; de Ron, A.M.; Singh, S.P. A core collection of common bean from the Iberian peninsula. Euphytica 2003, 131, 165–175. [Google Scholar] [CrossRef]
- Zeven, A.C.; Waninge, J.; Van Hintum, T.; Singh, S.P. Phenotypic variation in a core collection of common bean (Phaseolus vulgaris L.) in the Netherlands. Euphytica 1999, 109, 93–106. [Google Scholar] [CrossRef]
- Eichenberger, K.; Gugerli, F.; Schneller, J.J. Morphological and molecular diversity of Swiss common bean cultivars (Phaseolus vulgaris L., Fabaceae) and their origin. Bot. Helv. 2000, 110, 61–77. [Google Scholar]
- Rodino, P.A.; Monteagudo, A.B.; De Ron, A.M.; Santalla, M. Ancestral landraces of common bean from the south of Europe and their agronomical value for breeding programs. Crop Sci. 2009, 49, 2087–2099. [Google Scholar] [CrossRef]
- Singh, S.P.; Nodari, R.; Gepts, P. Genetic diversity in cultivated common bean: I. Allozymes. Crop Sci. 1991, 31, 19–23. [Google Scholar] [CrossRef]
- Gill-Langarica, H.R.; Muruaga-Martínez, J.S.; Vargas-Vázquez, M.L.P.; Rosales-Serna, R.; Mayek-Pérez, N.; Rosales-Serna, R.; Mayek-Pérez, N. Genetic diversity analysis of common beans based on molecular markers. Genet. Mol. Biol. 2011, 34, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Müller, B.S.F.; Pappas, G.J., Jr.; Valdisser, P.A.M.R.; Coelho, G.R.C.; de Menezes, I.P.P.; Abreu, A.G.; Borba, T.O.C.; Sakamoto, T.; Brondani, C.; Barros, E.G.; et al. An operational SNP panel integrated to SSR marker for the assessment of genetic diversity and population structure of the common bean. Plant Mol. Biol. Rep. 2015, 33, 1697–1711. [Google Scholar] [CrossRef]
- Valdisser, P.A.M.R.; Pereira, W.J.; Filho, J.E.A.; Müller, B.S.F.; Coelho, G.R.C.; de Menezes, J.P.; Vianna, J.P.G.; Zucchi, M.I.; Lanna, A.C.; Coelho, A.S.G.; et al. In-depth genome characterization of a Brazilian common bean core collection using DArTseq high-density SNP genotyping. BMC Genomics 2017, 18, 423. [Google Scholar] [CrossRef] [PubMed]
- Zargar, S.M.; Mahajan, R.; Nazir, M.; Nagar, P.; Kim, S.T.; Rai, V.; Masi, A.; Ahmad, S.M.; Shah, R.A.; Ganai, N.A.; et al. Common bean proteomics: Present status and future strategies. J. Proteomics 2017, 169, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Raatz, B.; Mukankusi, C.; Lobaton, J.D.; Male, A.; Chisale, V.; Amsalu, B.; Fourie, D.; Mukamuhirwa, F.; Muimui, K.; Mutari, B.; et al. Analyses of African common bean (Phaseolus vulgaris L.) germplasm using a SNP fingerprinting platform: Diversity, quality control and molecular breeding. Genet. Resour. Crop Evol. 2019, 66, 707–722. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Korol, A.B.; Fahima, T.; Beiles, A.; Nevo, E. Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Mol. Ecol. 2002, 11, 2453–2465. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Korol, A.B.; Fahima, T.; Nevo, E. Microsatellites within genes: Structure, function and evolution. Mol. Biol. Evol. 2004, 21, 991–1007. [Google Scholar] [CrossRef]
- Martin, P.; Makepeace, K.; Hill, S.A.; Hood, D.W.; Moxon, E.R. Microsatellite instability regulates transcription factor binding and gene expression. Proc. Natl. Acad. Sci. USA 2005, 102, 3800–3804. [Google Scholar] [CrossRef]
- Blair, M.W.; Pedraza, F.; Buendia, H.F.; Gaitán-Solís, E.; Beebe, S.E.; Gepts, P.; Tohme, J. Development of genome-wide anchored microsatellite map for common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2003, 107, 1362–1374. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Wang, G.; Mao, W.; Hu, Q.; Liu, N.; Ye, L.; Gong, Y. Genetic diversity and population structure of common bean (Phaseolus vulgaris) landraces from China revealed by a new set of EST-SSR markers. Biochem. Syst. Ecol. 2014, 57, 250–256. [Google Scholar] [CrossRef]
- Blair, M.W.; Lorigados, S.M. Diversity of common bean landraces, breeding lines, and varieties from Cuba. Crop Sci. 2016, 56, 1–9. [Google Scholar] [CrossRef]
- Garcia, R.A.V.; Rangel, P.N.; Brondani, C.; Martins, W.S.; Melo, L.C.; Carniero, M.S.; Borba, T.C.O.; Brondani, R.P.V. The characterization of a new set of EST-derived simple sequence repeat (SSR) markers as a resource for the genetic analysis of Phaseolus vulgaris. BMC Genet. 2011, 12, 41. [Google Scholar] [CrossRef] [PubMed]
- Blair, M.W.; Hurtado, N. EST-SSR markers from five sequenced cDNA libraries of common bean (Phaseolus vulgaris L.) comparing three bioinformatic alghoritms. Mol. Ecol. Resour. 2013, 13, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Blair, M.W.; Hurtado, N.; Chavarro, C.M.; Muñoz-Torres, M.C.; Giraldo, M.C.; Pedraya, F.; Tomkins, J.; Wing, R. Gene-based SSR markers for common bean (Phaseolus vulgaris L.) derived from root and leaf tissue ESTs: An integration of the BMc series. BMC Plant Biol. 2011, 11, 50. [Google Scholar] [CrossRef] [PubMed]
- Horňáková, O.; Závodná, M.; Žáková, M.; Kraic, J.; Debre, F. Diversity of common bean landraces collected in the western and eastern Carpatien. Czech J. Genet. Plant Breed. 2003, 39, 73–83. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Woods, J.; Hicks, J.B. A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Yu, K.; Park, S.J.; Poysa, V. Abundance and variation of microsatellite DNA sequences in beans (Phaseolus and Vigna). Genome 1999, 42, 27–34. [Google Scholar] [CrossRef]
- Yu, K.; Park, S.J.; Poysa, V.; Gepts, P. Integration of simple sequence repeat (SSR) markers into molecular linkage map of common bean (Phaseolus vulgaris L.). J. Hered. 2000, 91, 429–434. [Google Scholar] [CrossRef]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank. Nucleic Acid Res. 2005, 33, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the www for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [PubMed]
- Bassam, B.J.; Caetano-Anolles, G.; Gresshoff, P.M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 1991, 196, 80–83. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. PHYLIP (Phylogeny Inference Package), 3.6 ed.; Distributed by the author; Department of Genome Sciences, University of Washington: Seattle, WA, USA, 2005. [Google Scholar]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acid Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Neel, J.V. “Private” genetic variants and the frequency of mutation among South American Indians. Proc. Natl. Acad. Sci. USA 1973, 70, 3311–3315. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M. Gene flow in natural populations. Annu. Rev. Ecol. Syst. 1985, 16, 393–430. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. Structure Harvester: A website and program for visualizing Structure output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [PubMed]
- Tiranti, B.; Negri, V. Selective microenvironmental effects play a role in shaping genetic diversity and structure in a Phaseolus vulgaris L. landrace: Implications for on-farm conservation. Mol. Ecol. 2007, 16, 4942–4955. [Google Scholar] [CrossRef] [PubMed]
- Klaedtke, S.M.; Caproni, L.; Klauck, J.; de la Grandville, P.; Dutartre, M.; Stassart, P.M.; Chable, V.; Negri, V.; Raggi, L. Short-term local adaptation of historical common bean (Phaseolus vulgaris L.) varieties and implications for in situ management of bean diversity. Int. J. Mol. Sci. 2017, 18, 493. [Google Scholar] [CrossRef] [PubMed]
- Wells, W.C.; Isom, W.H.; Waines, J.G. Outcrossing rates of six common bean lines. Crop Sci. 1998, 28, 177–178. [Google Scholar] [CrossRef]
- Ibarra-Pérez, F.; Ehdale, B.; Waines, J.G. Estimation of outcrossing rate in common bean. Crop Sci. 1997, 37, 60–65. [Google Scholar] [CrossRef]
- Madakbas, S.Y.; Sarikamis, G.; Basak, H.; Karadavut, U.; Özmen, C.Y.; Dasci, M.G.; Cayan, S. Genetic characterization of green bean (Phaseolus vulgaris L.) accessions from Turkey with SCAR and SSR markers. Biochem. Genet. 2016, 54, 495–505. [Google Scholar] [CrossRef]
- Leitão, S.T.; Dinis, M.; Veloso, M.M.; Šatovič, Z.; Vaz Patto, M.C. Establishing the bases for introducing the unexplored Portuguese common bean germplasm into the breeding world. Front. Plant Sci. 2017, 8, 1296. [Google Scholar] [CrossRef]
- Hanai, L.R.; Campos, T.; Camargo, L.E.A.; Benchimol, L.L.; Souza, A.P.; Melotto, M.; Carbonell, A.M.; Chioratto, A.F.; Consoli, L.; Formighieri, E.F.; et al. Development, characterization, and comparative analysis of polymorphism at common bean SSR loci isolated from genic and genomic source. Genome 2007, 50, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.H.; Grima-Pettenati, J.; Feuillet, C. Characterization of a bean (Phaseolus vulgaris L.) malic-enzyme gene. Eur. J. Biochem. 1994, 224, 999–1009. [Google Scholar] [CrossRef] [PubMed]
- Khlestina, E.K.; Huang, X.Q.; Quenum, F.J.-B.; Chebotar, S.; Röder, M.S.; Börner, A. Genetic diversity in cultivated plant-loss or stability? Theor. Appl. Genet. 2004, 108, 1466–1472. [Google Scholar] [CrossRef] [PubMed]
- Khlestina, E.K.; Varshney, R.V.; Röder, M.S.; Graner, A.; Börner, A. A comparative assessment of genetic diversity in cultivated barley collected in different decades of the last century in Austria, Albania and India by using genomic and genic simple sequence repeat (SSR) markers. Plant Genet. Resour. 2006, 4, 125–133. [Google Scholar] [CrossRef]
- Sardos, J.; Christelová, P.; Čížková, J.; Paofa, J.; Sachter-Smith, G.L.; Janssens, S.B.; Rauka, G.; Ruas, M.; Daniells, J.W.; Doležel, J.; et al. Collection of new diversity of wild and cultivated bananas (Musa spp.) in the autochtomous region of Bougainville, Papua New Guinea. Genet. Resour. Crop Evol. 2018, 65, 2267–2286. [Google Scholar] [CrossRef]
- Van Treuren, R.; de Groot, E.C.; Boukema, I.W.; van de Wiel, C.C.M.; van Hintum, T.J.L. Marker-assisted reduction of redundancy in a genbank collection of cultivated lettuce. Plant Genet. Resour. 2010, 8, 95–105. [Google Scholar] [CrossRef]
- Jomová, K.; Benková, M.; Kraic, J. Enrichment of chickpea genetic resources collection monitored by microsatellites. Czech J. Genet. Plant Breed. 2009, 45, 11–17. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Ndjiondjop, M.-N.; Henry, R.J. Role of genomics in promoting the utilization of plant genetic resources in genebanks. Brief. Funct. Genomics 2018, 17, 198–206. [Google Scholar] [CrossRef]
- Cuevas, H.E.; Prom, L.K. Assessment of molecular diversity and population structure of the Ethiopian sorghum [Sorghum bicolour (L.) Moench] germplasm collection maintained by the USDA-ARS National Plant Germplasm System using SSR markers. Genet. Resour. Crop Evol. 2013, 60, 1817–1830. [Google Scholar] [CrossRef]
- Jiménez, O.R.; Korpelainen, H. Microsatellite markers reveal promising genetic diversity and seed trait associations in common bean landraces (Phaseolus vulgaris L.) from Nicaragua. Plant Genet. Resour. 2012, 10, 108–118. [Google Scholar] [CrossRef]
- De Ron, A.M.; Rodiño, A.P.; Santalla, M.; González, A.M.; Lema, M.J.; Martin, I.; Kigel, J. Seedling emergence and phenotypic response of common bean germplasm to different temperatures under controlled conditions and in open field. Front. Plant Sci. 2016, 7, 1087. [Google Scholar] [CrossRef] [PubMed]
- Abenavoli, M.R.; Leone, M.; Sunseri, F.; Bacchi, M.; Sorgonà, A. Root phenotyping for drought tolerance in bean landraces from Calabria (Italy). J. Agron. Crop Sci. 2015, 202, 1–12. [Google Scholar] [CrossRef]
- Celmeli, T.; Sari, H.; Canci, H.; Sari, D.; Adak, A.; Eker, T.; Toker, C. The nutritional content of common bean (Phaseolus vulgaris L.) landraces in comparison to modern varieties. Agronomy 2018, 8, 166. [Google Scholar] [CrossRef]
- Caproni, L.; Raggi, L.; Tissi, C.; Howlett, S.; Torricelli, R.; Negri, V. Multi-environmental evaluation and genetic characterization of common bean breeding lines for organic farming systems. Sustainability 2018, 10, 777. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).