The Impact of Metolachlor Applications and Phytoremediation Processes on Soil Microorganisms: Insights from Functional Metagenomics Analysis
Abstract
1. Introduction
2. Materials and Methods
- Microbial community metagenome
- UHPLC MS/MS analysis residue of the herbicide
3. Results
- Soil Bioremediation
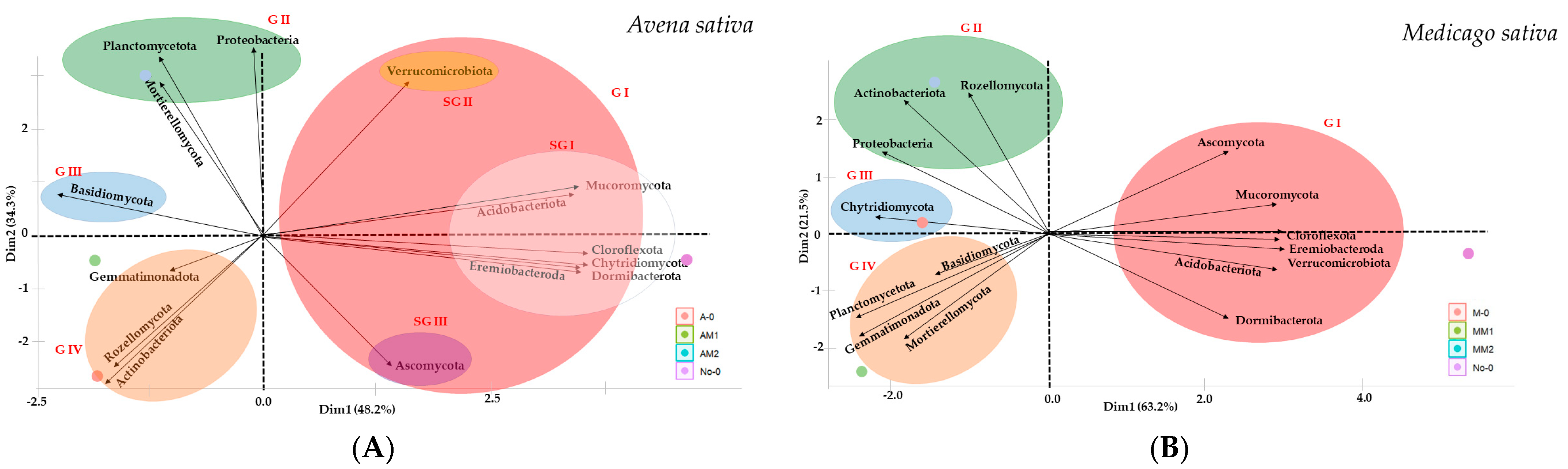
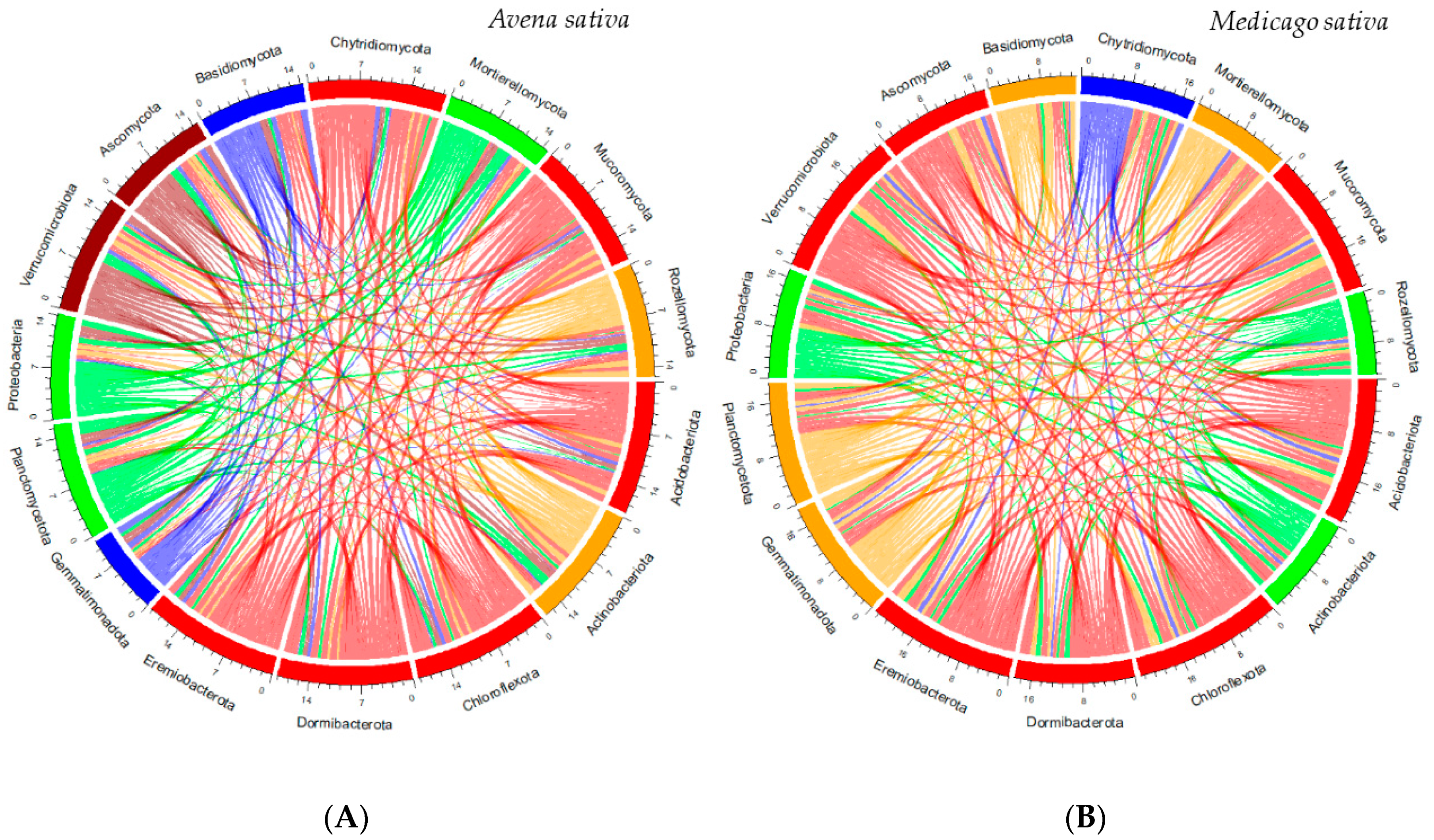
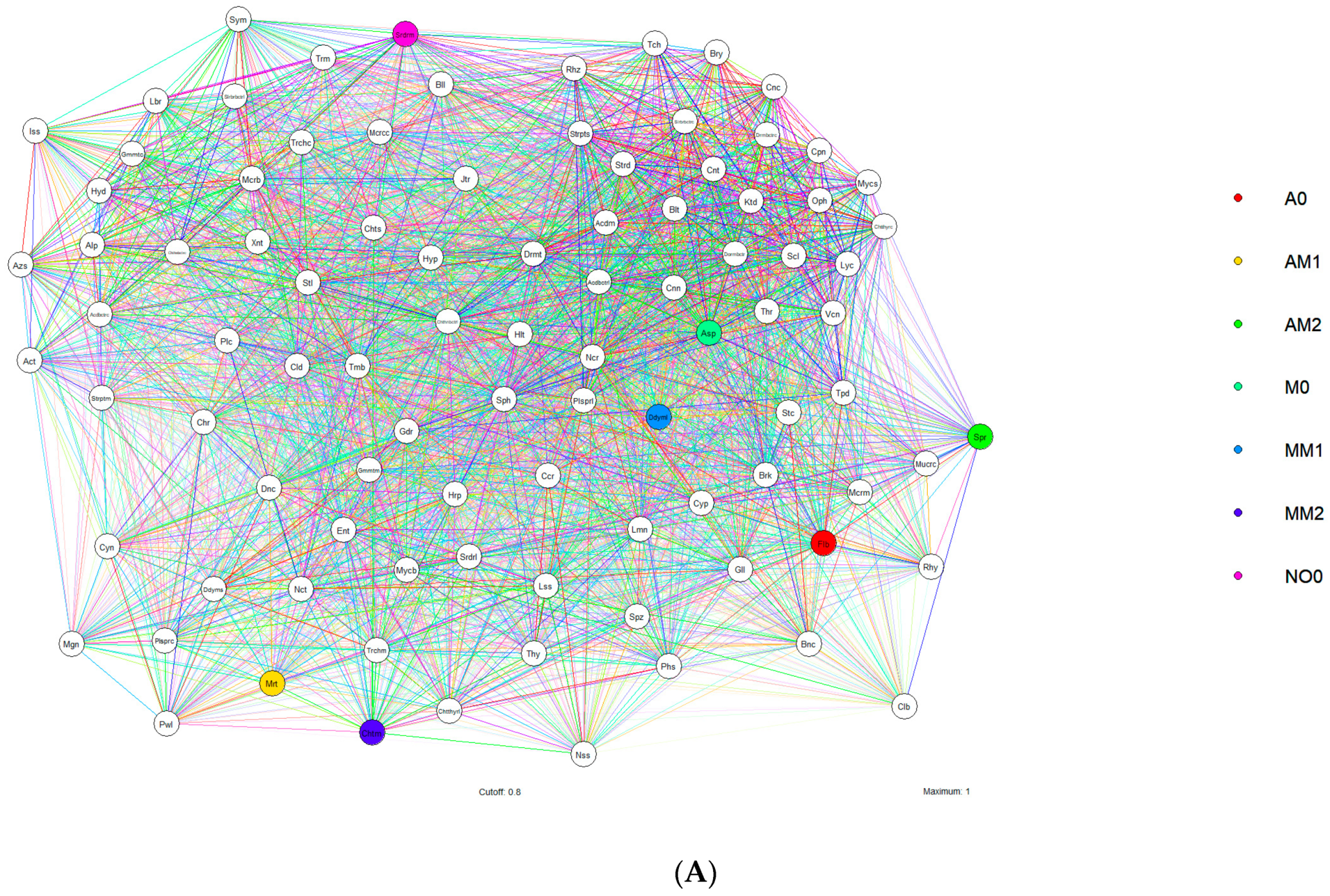
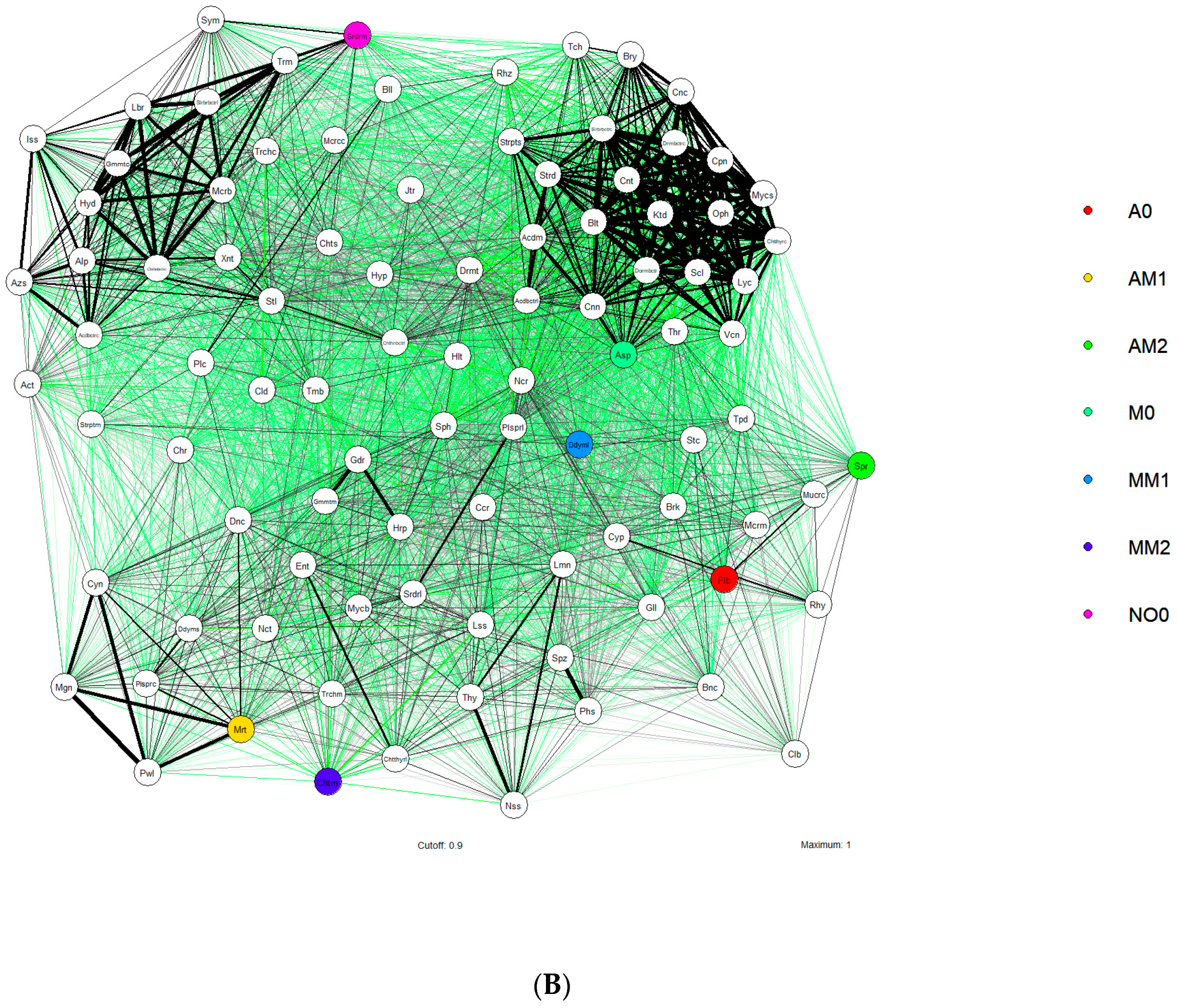
- Global Biodiversity and Microbiome Composition
- Ecological indices of bacterial population in soil
- Ecological indices of the fungal population in the soil
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nath, C.P.; Singh, R.G.; Choudhary, V.K.; Datta, D.; Nandan, R.; Singh, S.S. Challenges and Alternatives of Herbicide-Based Weed Management. Agronomy 2024, 14, 126. [Google Scholar] [CrossRef]
- Lima, A.d.C.; Mendes, K.F. Variable Rate Application of Herbicides for Weed Management in Pre- and Postemergence; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- Bhardwaj, L.; Reddy, B.; Nath, A.J.; Dubey, S.K. Influence of herbicide on rhizospheric microbial communities and soil properties in irrigated tropical rice field. Ecol. Indic. 2024, 158, 111534. [Google Scholar] [CrossRef]
- Singh, A.K.; Pandey, A.K. Chapter 9—Exploitation of mycometabolites in weed management: Global scenario and future application. In Entrepreneurship with Microorganisms; Academic Press: Cambridge, MA, USA, 2024; pp. 179–188. [Google Scholar] [CrossRef]
- Li, X.; Zhang, H.; Sun, M.; Xu, N.; Zhao, M. Land use change from upland to paddy field in Mollisols drives soil aggregation and associated microbial communities. Appl. Soil Ecol. 2020, 146, 103351. [Google Scholar] [CrossRef]
- Thiour-Mauprivez, C.; Martin-Laurent, F.; Calvayrac, C. Effects of herbicide on non-target microorganisms: Towards a new class of biomarkers? Sci. Total Environ. 2019, 684, 314–325. [Google Scholar] [CrossRef]
- Hachisu, S. Strategies for discovering resistance-breaking, safe, and sustainable commercial herbicides with novel modes of action and chemotypes. Soc. Chem. Ind. 2021, 77, 3042–3048. [Google Scholar] [CrossRef] [PubMed]
- Adomako, M. Effect of Some Commonly Used Herbicides on Soil Microbial Population. J. Environ. Earth Sci. 2016, 6, 30–38. [Google Scholar]
- Hussain Lone, A.; Raverkar, K.P.; Pareek, N. In-vitro effects of herbicides on soil microbial communities. Bioscan 2014, 9, 11–16. [Google Scholar]
- Wilfred, K.; Kwame Dzomeku, I.; Xorse, K.J. Efficacy of pre-emergence and post-emergence herbicides for weed management in groundnut (Arachis hypogaea L.) production in Guinea Savannah. Int. J. Sci. Res. Manag. (IJSRM) 2020, 8, 263–276. [Google Scholar] [CrossRef]
- Yilmaz, G.; Feruzan, D.A.N. Phytotoxic effects of herbicide Attribut and surfactant BioPower on the root, stem, and leaf anatomy of Triticum aestivum ‘Pehlivan’. Turk. J. Bot. 2013, 37, 10. [Google Scholar] [CrossRef]
- Atwood, D.; Paisley, J.C. Pesticides Industry Sales and Usage: Market Estimates; US Environmental Protection Agency: Washington, DC, USA, 2017. Available online: https://www.epa.gov/pesticides/pesticides-industry-sales-and-usage-2008-2012-market-estimates (accessed on 25 May 2024).
- Kouame, K.B.J.; Bertucci, M.B.; Savin, M.C.; Bararpour, T.; Steckel, L.E.; Butts, T.R.; Willett, C.D.; Machado, F.G.; Roma-Burgos, N. Resistance of Palmer amaranth (Amaranthus palmeri) to Metolachlor in the midsouthern United States. Weed Sci. 2022, 70, 380–389. [Google Scholar] [CrossRef]
- Silva, Q.M.; Palmieri, M.J.; Andrade-Vieira, L.F. Effects of a S-Metolachlor Based Herbicide on Two Plant Models: Zea mays L. and Lactuca sativa L. Preprint, 2022; Version 1; [CrossRef] [PubMed]
- Da Silva, M.S.; Furtado, J.A.L.; Castro, J.Q.; dos Santos, L.L.; Almeida, E.I.B.; de Oliveira, L.B.T.; Sousa, W.d.S.; Araújo, R.C.d.A. Weed control and selectivity of different pre-emergence active ingredients in a soybean crop. Agron. Colomb. 2022, 39, 392–404. [Google Scholar] [CrossRef]
- MAPA; Ministério da Agricultura e Pecuária. Plano Setorial de Mitigação e de Adaptação às Mudanças Climáticas para a Consolidação de uma Economia de Baixa Emissão de Carbono na Agricultura; MAPA/ACS; Governo Federal: Brasília, Brazil, 2023; p. 173. Available online: https://www.gov.br/agricultura/pt-br/assuntos/sustentabilidade/plano-abc (accessed on 5 February 2024).
- Grădilă, M.; Jalobă, D.; Ciontu, V.; Erban, M.; Petcu, V. Research regarding weed control in grain legumes crops. Sci. Pap. Ser. A Agron. 2021, 64, 344–349. [Google Scholar]
- Chauhan, B.S.; Gill, G.S.; Preston, C. Timing and Dose of Metolachlor Affect Rigid Ryegrass (Lolium rigidum) Control in Wheat. Weed Technol. 2007, 21, 225–229. [Google Scholar] [CrossRef]
- Mesnage, R.; Panzacchi, S.; Bourne, E.; Mein, C.A.; Perry, M.J.; Hu, J.; Chen, J.; Mandrioli, D.; Belpoggi, F.; Antoniou, M.N. Glyphosate and its formulations Roundup Bioflow and RangerPro alter bacterial and fungal community composition in the rat caecum microbiome. Front. Microbiol. 2022, 13, 888853. [Google Scholar] [CrossRef] [PubMed]
- Curran, W.S. Persistence of herbicides in soil. Crops Soils Mag. 2016, 49, 16–21. [Google Scholar] [CrossRef]
- Sanyal, D.; Yaduraju, N.T.; Kulshestha, G. Metolachlor persistence in laboratory and field soils under Indian tropical conditions. J. Environ. Sci. Health Part B Pestic. Food Contam. Agric. Wastes 2008, 43, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Dinelli, G.; Accinelli, C.; Vicari, A.; Catizone, P. Comparison of the Persistence of Atrazine and Metolachlor under Field and Laboratory Conditions. J. Agric. Food Chem. 2000, 48, 3239–3245. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, L.M.; de Freitas Souza, M.; de Laia, M.L.; de Oliveira Melo, J.; da Costa, M.R.; Gonçalves, J.F.; Dos Santos, J.B. Metagenomic analysis reveals mechanisms of atrazine biodegradation promoted by tree species. Environ. Pollut. 2020, 267, 115636. [Google Scholar] [CrossRef]
- Barroso, G.M.; Dos Santos, E.A.; Ribeiro Pires, F.; Galon, L.; Cabral, G.M.; Dos Santos, J.B. Phytoremediation: A green and low-cost technology to remediate herbicides in the environment. Chemosphere 2023, 334, 138943. [Google Scholar] [CrossRef]
- Oliveira, M.C.; Osipitan, O.A.; Begcy, K.; Werle, R. Cover crops, hormones and herbicides: Priming an integrated weed management strategy. Plant Sci. 2020, 301, 110550. [Google Scholar] [CrossRef]
- Scavo, A.; Mauromicale, G. Integrated Weed Management in Herbaceous Field Crops. Agronomy 2020, 10, 466. [Google Scholar] [CrossRef]
- Silva, T.S.; Arneson, N.J.; DeWerff, R.P.; Smith, D.H.; Silva, D.V.; Werle, R. Preemergence herbicide premixes reduce the risk of soil residual weed control failure in corn. Weed Technol. 2023, 37, 410–421. [Google Scholar] [CrossRef]
- Vasileiou, M.; Vasileiou, M.; Kyrgiakos, S.; Kleisiari, L.; Kleftodimos, C.; Vlontzos, G.; Belhouchette, G.; Pardalos, P.M. Transforming weed management in sustainable agriculture with artificial intelligence: A systematic literature review towards weed identification and deep learning. Crop Prot. 2024, 176, 106522. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Barroso, G.M.; dos Santos, J.B.; de Oliveira, I.T.; Nunes, T.K.M.R.; Ferreira, E.A.; Pereira, I.M.; Silva, D.V.; Souza, M.F. Tolerance of Bradyrhizobium sp. BR 3901 to herbicides and their ability to use these pesticides as a nutritional source. Ecol. Indic. 2020, 119, 106783. [Google Scholar] [CrossRef]
- Mauser, K.M.; Brühl, C.A.; Zaller, J.G. Herbicide Effects on Nontarget Organisms, Biodiversity and Ecosystem Functions. Encycl. Biodivers. 2024, 4, 239–257. [Google Scholar] [CrossRef]
- Scheepmaker, J.W.A.; van de Kassteele, J. Effects of chemical control agents and microbial biocontrol agents on numbers of non-target microbial soil organisms: A meta-analysis. Biocontrol Sci. Technol. 2011, 21, 1225–1242. [Google Scholar] [CrossRef]
- Liao, H.; Li, X.; Yang, Q.; Bai, Y.; Wen, C.; Liu, C.; Chen, Z.; Tang, J.; Yu, Z.; Che, J.; et al. Herbicide selection promotes antibiotic resistance in soil microbiomes. Mol. Biol. Evol. 2021, 38, 2337–2350. [Google Scholar] [CrossRef]
- Rose, M.T.; Cavagnaro, T.R.; Scanlan, C.A.; Rose, T.J.; Vancov, T.; Kimber, S.; Kennedy, I.R.; Kookana, R.S.; Zwieten, L.V. Impact of herbicides on soil biology and function. Adv. Agron. 2016, 136, 133–220. [Google Scholar] [CrossRef]
- Gonçalves dos Santos, H.; Jacomine, P.K.T.; Cunha dos Anjos, L.H.; de Oliveira, V.A.; de Oliveira, J.B.; Coelho, M.R.; Lumbreras, J.F.; Ferreira Cunha, T.J. Sistema Brasileiro de Classificação de Solos, 2nd ed.; Ministério da Agricultura, Pecuária e Abastecimento: Brasília, Brazil, 2006; pp. 1–306.
- Primavesi, A.C.; De Andrade Rodrigues, A.; Godoy, R. Recomendações Técnicas para o Cultivo de Aveia; Publisher: São Carlos, Brazil, 2000; pp. 1–100. [Google Scholar]
- Rassini, J.B. Alfafa (Medicago sativa L.): Estabelecimento e Cultivo no Estado de São Paulo; Publisher: São Carlos, Brazil, 1999; pp. 1–50. [Google Scholar]
- Ghosh, P.K.; Bandyopadhyay, K.K.; Wanjari, R.H.; Manna, M.C.; Misra, A.K.; Mohanty, M.; Subba Rao, A. Legume Effect for Enhancing Productivity and Nutrient Use-Efficiency in Major Cropping Systems—An Indian Perspective: A Review. J. Sustain. Agric. 2014, 30, 59–86. [Google Scholar] [CrossRef]
- Prasad, R. Efficient fertilizer use: The key to food security and better environment. J. Trop. Agric. 2009, 47, 1–17. [Google Scholar]
- Geronime, E.; Aparicio, V. Changes in soil pH and addition of inorganic phosphate affect glyphosate adsorption in agricultural soil. Eur. J. Soil Sci. 2022, 73, c13188. [Google Scholar] [CrossRef]
- Guiasu, R.C.; Guiasu, S. The Rich-Gini-Simpson quadratic index of biodiversity. Nat. Sci. 2010, 2, 1130–1137. [Google Scholar] [CrossRef]
- Mouillot, D.; Lepretre, A. A comparison of species diversity estimators. Rest Popul. Ecol. 1999, 41, 203–215. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, R.; Song, P.; Liu, S.; Li, Y.; Li, H. Diversity and Distribution of 18 Cephalopod Species, and Their Link with Some Environmental Factors in the NW Pacific. Diversity 2023, 15, 694. [Google Scholar] [CrossRef]
- Brondi, S.H.; Macedo, A.N.; Vicente, G.H.; Nogueira, A.R. Evaluation of the QuEChERS Method and Gas Chromatography–Mass Spectrometry for the Analysis of Pesticide Residues in Water and Sediment. Bull. Environ. Contam. Toxicol. 2011, 86, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Kemmerich, M. Resíduos de Agrotóxicos em Ameixa, Maçã, pera e Pêssego: Desenvolvimento de Métodos de Análise e Monitoramento. Ph.D. Thesis, Universidade Federal de Santa Maria, Santa Maria, Brazil, 2017. Available online: https://repositorio.ufsm.br/handle/1/12732 (accessed on 10 March 2024).
- Yu, L.; Van Eerd, L.L.; O’Halloran, I.; Sikkema, P.H.; Robinson, D.E. Response of four fall-seeded cover crops to residues of selected herbicides. Crop Prot. 2015, 75, 11–17. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, J.; Xie, P.; Tan, L.; Han, L. Discontinuous low temperature stress and plant growth regulators during the germination period promote roots growth in alfalfa (Medicago sativa L.). Plant Physiol. Biochem. 2023, 197, 107624. [Google Scholar] [CrossRef]
- Paul, S.K.; Mazumder, S.; Naidu, R. Herbicidal weed management practices: History and prospects of nanotechnology in an eco-friendly crop production system. Heliyon 2024, 10, e26527. [Google Scholar] [CrossRef]
- Mackay, J.E.; Bernhardt, L.T.; Smith, R.G.; Ernakovich, J.G. Seedage and pesticide seed treatments have distinct effects on soil microbial diversity and function. Soil Biol. Biochem. 2023, 176, 108860. [Google Scholar] [CrossRef]
- Pathak, H.K.; Seth, C.S.; Chauhan, P.K.; Dubey, G.; Singh, G.; Jain, D.; Upadhyay, S.K.; Dwivedi, P.; Khoo, K.S. Recent advancement of nano-biochar for the remediation of heavy metals and emerging contaminants: Mechanism, adsorption kinetic model, plant growth and development. Environ. Res. 2024, 255, 119136. [Google Scholar] [CrossRef] [PubMed]
- Wołejko, E.; Kaczyński, P.; Łozowicka, B.; Wydro, U.; Borusiewicz, A.; Hrynko, I.; Konecki, R.; Snarska, K.; Dec, D.; Malinowski, P. Dissipation of Metolachlor in plant and soil and effect on enzymatic activities. Environ. Monit. Assess. 2017, 189, 355. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Kucharski, J.; Bacmaga, M.; Tomkiel, M. Response of microorganisms and enzymes to soil contamination with a mixture of terbuthylazine, mesotrione, and Metolachlor. Environ. Sci. Pollut. Res. 2017, 24, 1910–1925. [Google Scholar] [CrossRef] [PubMed]
- Munoz, A.; Koskinen, W.C.; Cox, L.; Sadowsky, M.J. Biodegradation and Mineralization of Metolachlor and Alachlor by Candida xestobii. J. Agric. Food Chem. 2011, 59, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Morales-Becerra, C.E.; Ortiz-Rojas, L.Y.; Chaves-Bedoya, G. Assessment of Burkholderia glumae control in rice (Oryza sativa) FEDEARROZ 67, using silver nanoparticles (AgNPs) under greenhouse conditions. Revista Colombiana de Ciencias Hortícolas 2023, 17, e16358. [Google Scholar] [CrossRef]
- Qin, P.; Hu, L.; Liu, Y.; Hu, X.; Zhang, X.; Rosado, A.S.; Wei, G.; Chen, C. Responses of soil microbial communities and nutrient dynamics under continuous alfalfa (Medicago sativa L.) cultivation. Soil Ecol. 2024, 197, 105356. [Google Scholar] [CrossRef]
- Lyra, W.d.S.; da Silva, E.C.; de Araújo, M.C.U.; Fragoso, W.D.; Veras, G. Classificação periódica: Um exemplo didático para ensinar análise de componentes principais. Química Nova 2010, 33, 1594–1597. [Google Scholar] [CrossRef]
- El Jaouhari, M.; Damour, G.; Tixier, P.; Coulis, M. Glyphosate reduces the biodiversity of soil macrofauna and benefits exotic over native species in a tropical agroecosystem. Basic Appl. Ecol. 2023, 73, 18–26. [Google Scholar] [CrossRef]
- Jousset, A.; Bienhold, C.; Chatzinotas, A.; Gallien, L.; Gobet, A.; Kurm, V.; Hol, W.H.G. Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 2017, 11, 853–862. [Google Scholar] [CrossRef]
- Vryzas, Z.; Papadakis, E.N.; Oriakli, K.; Moysiadis, T.P.; Papadopoulou-Mourkidou, E. Biotransformation of atrazine and metolachlor within soil profile and changes in microbial communities. Chemosphere 2012, 89, 1330–1338. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Song, S.; Chen, S.; Du, Z.; Kong, J. Adaptive evaluation of green manure rotation for a low fertility farmland system: Impacts on crop yield, soil nutrients, and soil microbial community. Catena 2022, 213, 106873. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, L.; Fan, J.; Wen, Y. Impacts of Rac- and S-metolachlor on cyanobacterial cell integrity and release of microcystins at different nitrogen levels. Chemosphere 2017, 191, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Bernat, P.; Jasinska, A.; Niedziałkowska, K.; Słaba, M.; Ro’zalska, S.; Paraszkiewicz, K.; Sas-Paszt, L.; Heipieper, H.J. Adaptation of the metolachlor-degrading fungus Trichoderma harzianum to the simultaneous presence of low-density polyethylene (LDPE) microplastics. Ecotoxicol. Environ. Saf. 2023, 267, 115656. [Google Scholar] [CrossRef] [PubMed]
- Bickel, S.; Or, D. The chosen few variations in common and rare soil bacteria across biomes. ISME J. 2021, 15, 3315–3325. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, O.F.; Maillard, E.; Vuilleumier, S.; Imfeld, G. Bacterial communities in batch and continuous-flow wetlands treating the herbicide S-metolachlor. Sci. Total Environ. 2014, 499, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Emamifar, S.; Abolmaali, S.; Sohrabi, S.M.; Mohammadi, M.; Shahmohammadi, M. Molecular characterization and evaluation of the antibacterial activity of a plant defensin peptide derived from a gene of oat (Avena sativa L.). Phytochemistry 2020, 181, 112586. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, I.; Sadanov, A.; Baimakhanova, G.; Faizulina, E.; Tatarkina, L. Metabolic interaction at the level of extracellular amino acids between plant growth-promoting rhizobacteria and plants of alfalfa (Medicago sativa L.). Rhizosphere 2022, 21, 100477. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Z.; Solanki, M.K.; Yu, Z.-X.; Anas, M.; Dong, D.-F.; Xing, Y.-X.; Malviya, M.K.; Pang, F.; Li, Y.-R. Genome Characteristics Reveal the Biocontrol Potential of Actinobacteria Isolated From Sugarcane Rhizosphere. Front. Microbiol. 2021, 12, 797889. [Google Scholar] [CrossRef] [PubMed]
- Viegas, C.A.; Costa, C.; André, S.; Viana, P.; Ribeiro, R.; Moreira-Santos, M. Does S-Metolachlor affect the performance of Pseudomonas sp. strain ADP as a bioaugmentation bacterium for atrazine-contaminated soils? PLoS ONE 2012, 7, e37140. [Google Scholar] [CrossRef]
- Feng, G.; Xie, T.; Wang, X.; Bai, J.; Tang, L.; Zhao, H.; Wei, W.; Wang, M.; Zhao, Y. Metagenomic analysis of microbial community and function involved in cd-contaminated soil. BMC Microbiol. 2018, 18, 11. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Bahram, M.; Polme, S.; Koljalg, U.; Yorou, N.S.; Wijesundera, R.; Suija, A. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [PubMed]
- Henn, C.; Monteiro, D.A.; Boscolo, M.; da Silva, R.; Gomes, E. Biodegradation of atrazine and ligninolytic enzyme production by basidiomycete strains. BMC Microbiol. 2020, 20, 266. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Zhao, X.; Zhang, X.; Zhao, Q.; Wang, X.; Li, Y. Restructured fungal community diversity and biological interactions promote metolachlor biodegradation in soil microbial fuel cells. Chemosphere 2019, 221, 735–749. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Omolehin, O.; Hemmings, G.; Tseng, H.T.; Taylor, A.; Taylor, C.; Kong, P.; Daughtrey, M.; Luster, D.; Gouker, F.; et al. Boxwood phyllosphere fungal and bacterial communities and their differential responses to film-forming anti-desiccants. BMC Microbiol. 2023, 23, 219. [Google Scholar] [CrossRef] [PubMed]
- Ghareeb, Y.; Belal, R.; El-Khateeb, E.B.; El-Khateeb NM, M. Utilização da biossíntese de nanomateriais como agentes biológicos para o controle de doenças transmitidas pelo solo em plantas de pimenta: Nematóides das galhas e fungos da podridão radicular. BMC Plant Biol. 2024, 24, 110. [Google Scholar] [CrossRef]
- Bhatt, P.; Verma, A.; Gangola, S.; Bhandari, G.; Chen, S. Microbial glycoconjugates in organic pollutant bioremediation: Recent advances and applications. Microb. Cell Fact. 2021, 20, 72. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

| Treatments | Phytoremediation Species | Herbicide | Dose (g/ha) |
|---|---|---|---|
| AM1 | Avena sativa | Metolachlor | 530.70 |
| AM2 | Avena sativa | Metolachlor | 1061.40 |
| MM1 | Medicago sativa | Metolachlor | 530.70 |
| MM2 | Medicago sativa | Metolachlor | 1061.40 |
| A-0 | Avena sativa | - | - |
| M-0 | Medicago sativa | - | - |
| NoM1 | - | Metolachlor | 530.70 |
| NoM2 | - | Metolachlor | 1061.40 |
| No-0 | - | - | - |
| Avena sativa | Medicago sativa | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Unif 1 | Richness | Dist. 2 | Dom 3 | Unif 1 | Richness | Dist. 2 | Dom 3 | ||
| A0 | 0.71 | 104.0 | 3.3 | 0.92 | M0 | 0.90 | 120.0 | 4.2 | 1.0 |
| AM1 | 0.79 | 121.0 | 3.8 | 0.96 | MM1 | 0.90 | 122.0 | 4.2 | 1.0 |
| AM2 | 0.86 | 116.0 | 4.1 | 0.98 | MM2 | 0.90 | 117.0 | 4.2 | 1.0 |
| No-0 | 0.85 | 109.0 | 4.0 | 0.97 | No-0 | 0.90 | 109.0 | 4.0 | 1.0 |
| Avena sativa | Medicago sativa | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Unif 1 | Richness | Dist. 2 | Dom 3 | Unif 1 | Richness | Dist. 2 | Dom 3 | ||
| A0 | 0.71 | 28.0 | 2.38 | 0.77 | M0 | 0.94 | 44.0 | 3.57 | 0.96 |
| AM1 | 0.90 | 44.0 | 2.40 | 0.94 | MM1 | 0.94 | 41.0 | 3.51 | 0.95 |
| AM2 | 0.93 | 40.0 | 3.43 | 0.95 | MM2 | 0.90 | 39.0 | 3.32 | 0.94 |
| No-0 | 0.95 | 42.0 | 3.30 | 0.96 | No-0 | 0.94 | 42.0 | 3.53 | 0.96 |
| A-0 | AM1 | AM2 | M-0 | MM1 | MM2 | No-0 | |
|---|---|---|---|---|---|---|---|
| ------------------------------------- Family ------------------------------------- | |||||||
| Uncultivated | 8.69 | 9.59 | 8.27 | 7.91 | 9.49 | 14.66 | 17.97 |
| Beijerinckiaceae | 4.42 | 4.09 | 4.16 | 1.84 | 3.4 | 3.25 | 1.65 |
| Bryobacteraceae | 1.84 | 1.75 | 1.28 | * | * | * | 3.60 |
| Burkholderiaceae | 10.63 | 7.80 | 5.12 | 4.75 | 5.20 | 6.46 | 1.89 |
| Undetermined | 6.51 | 7.91 | 9.04 | 4.90 | 6.97 | 7.27 | 9.63 |
| Ktedonobacteraceae | * | * | * | 0.54 | 0.81 | 0.72 | 5.68 |
| Micrococcaceae | 11.01 | 10.20 | 14.27 | 36.60 | 17.50 | 3.58 | 2.78 |
| Mycobacteriaceae | 3.82 | 2.58 | 3.26 | 1.91 | 2.82 | 1.83 | 2.76 |
| Solirubrobacteraceae | 4.89 | 4.33 | 5.47 | 3.20 | 4.57 | 4.74 | 11.91 |
| Sphingomonadaceae | 3.60 | 4.20 | 3.92 | 2.64 | 3.25 | 1.58 | 1.05 |
| Xanthobacteraceae | 8.17 | 7.90 | 12.56 | 7.68 | 7.32 | 17.74 | 12.85 |
| ------------------------------------- Genus ------------------------------------- | |||||||
| Undetermined | 47.12 | 40.62 | 32.40 | 31.00 | 32.89 | 38.73 | 34.45 |
| Uncultivated | 11.92 | 16.53 | 19.93 | 16.03 | 17.53 | 14.68 | 31.23 |
| Arthrobacter sp. | 0.71 | 1.19 | 0.53 | * | * | * | 1.78 |
| Baekduia sp. | 0.89 | 0.82 | 0.62 | 1.09 | 0.89 | 1.35 | 1.91 |
| Bradyrhizobium sp. | 2.48 | 2.70 | 4.78 | 3.46 | 2.78 | 3.55 | 2.75 |
| Massilia sp. | 2.13 | 1.56 | 2.06 | 3.49 | 2.22 | 1.95 | 0.30 |
| Methylobacterium sp. | * | * | * | 2.69 | 2.04 | 2.54 | 0.40 |
| Mycobacterium sp. | 1.53 | 2.29 | 1.59 | 3.06 | 2.3 | 2.89 | 2.58 |
| Pseudarthrobacter sp. | 3.47 | 0.80 | 0.46 | 1.78 | 1.65 | 2.46 | 0.06 |
| Sphingomicrobium sp | 2.01 | 2.69 | 1.31 | 2.67 | 2.96 | 2.89 | 0.82 |
| Streptomyces sp. | 2.54 | 1.25 | 0.89 | 1.68 | 1.01 | 2.16 | 1.58 |
| Avena sativa | Medicago sativa | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Unif 1 | Richness | Dist. 2 | Dom 3 | Unif 1 | Richness | Dist. 2 | Dom 3 | ||
| A0 | 0.64 | 76.0 | 2.78 | 0.87 | M0 | 0.81 | 76.0 | 3.52 | 0.95 |
| AM1 | 0.67 | 77.0 | 2.91 | 0.90 | MM1 | 0.79 | 81.0 | 3.48 | 0.94 |
| AM2 | 0.79 | 76.0 | 3.40 | 0.95 | MM2 | 0.82 | 78.0 | 3.58 | 0.96 |
| No-0 | 0.75 | 67.0 | 3.17 | 0.93 | No-0 | 0.75 | 67.0 | 3.17 | 0.93 |
| A-0 | AM1 | AM2 | M-0 | MM1 | MM2 | No-0 | |
|---|---|---|---|---|---|---|---|
| ------------------------------------- Family ------------------------------------- | |||||||
| Undetermined | 7.42 | 17.35 | 18.13 | 3.18 | 2.11 | 2.76 | 11.06 |
| Aspergillaceae | 10.46 | 7.41 | 12.18 | 10.49 | 6.01 | 9.55 | 21.14 |
| Chaetomiaceae | 6.65 | 4.77 | 3.74 | 8.58 | 8.55 | 13.69 | 6.91 |
| Cladosporiaceae | 3.65 | 4.43 | 5.01 | 13.06 | 14.61 | 2.07 | 3.71 |
| Cunninghamellaceae | 1.24 | 0.49 | 2.77 | 1.08 | 0.51 | 2.38 | 7.88 |
| Didymellaceae | 12.38 | 4.57 | 13.19 | 13.84 | 6.06 | 7.06 | 3.25 |
| Filobasidiaceae | * | * | * | 2.73 | 7.01 | 2.93 | 0.01 |
| Hypocreaceae | 9.22 | 1.37 | 1.03 | 0.46 | 2.05 | 7.37 | 8.44 |
| Mortierellaceae | 3.32 | 19.57 | 4.87 | * | * | * | 0.00 |
| Nectriaceae | 12.13 | 4.21 | 6.57 | 28.88 | 11.79 | 12.45 | 18.6 |
| Phaeosphaeriaceae | 1.42 | 1.38 | 2.96 | * | * | * | 2.29 |
| Rhynchogastremataceae | * | * | * | 1.49 | 20.00 | 4.73 | 0.26 |
| ---------------------------------------- Genus ---------------------------------------- | |||||||
| Cladosporium sp. | 13.06 | 14.61 | 2.07 | 19.52 | 24.94 | 31.39 | 3.71 |
| Epicoccum sp. | 3.64 | 4.56 | 3.50 | 5.23 | 1.53 | 8.57 | 1.69 |
| Fusarium sp. | 28.76 | 11.57 | 10.89 | 11.94 | 3.44 | 6.18 | 18.19 |
| Humicola sp. | 5.41 | 6.30 | 10.43 | 4.90 | 2.28 | 1.99 | 1.42 |
| Undetermined | 14.72 | 6.42 | 12.67 | * | * | * | 28.41 |
| Mortierella sp. | * | * | * | 3.32 | 19.57 | 4.87 | 0.00 |
| Papiliotrema sp. | 1.49 | 20.0 | 4.73 | * | * | * | 0.26 |
| Penicillium sp. | 10.06 | 5.82 | 9.08 | 9.32 | 6.40 | 10.89 | 11.08 |
| Pleurophragmium sp. | 1.29 | 2.04 | 7.19 | 1.63 | 3.44 | 2.21 | 0.01 |
| Saitozyma sp. | 1.10 | 2.12 | 6.41 | * | * | * | 0.09 |
| Trichoderma sp. | 0.46 | 1.93 | 7.37 | 9.20 | 1.37 | 1.03 | 8.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hejazirad, S.P.; de Abreu, C.M.; Carneiro, G.H.F.; Gomes, C.R.; Spinola Filho, P.R.d.C.; da Costa, M.R.; Santos, J.B.d. The Impact of Metolachlor Applications and Phytoremediation Processes on Soil Microorganisms: Insights from Functional Metagenomics Analysis. J. Xenobiot. 2024, 14, 970-988. https://doi.org/10.3390/jox14030054
Hejazirad SP, de Abreu CM, Carneiro GHF, Gomes CR, Spinola Filho PRdC, da Costa MR, Santos JBd. The Impact of Metolachlor Applications and Phytoremediation Processes on Soil Microorganisms: Insights from Functional Metagenomics Analysis. Journal of Xenobiotics. 2024; 14(3):970-988. https://doi.org/10.3390/jox14030054
Chicago/Turabian StyleHejazirad, Seyedeh Parvin, Caique Menezes de Abreu, Guilherme Henrique Fernandes Carneiro, Carlos Rodrigues Gomes, Paulo Roberto de Carvalho Spinola Filho, Márcia Regina da Costa, and José Barbosa dos Santos. 2024. "The Impact of Metolachlor Applications and Phytoremediation Processes on Soil Microorganisms: Insights from Functional Metagenomics Analysis" Journal of Xenobiotics 14, no. 3: 970-988. https://doi.org/10.3390/jox14030054
APA StyleHejazirad, S. P., de Abreu, C. M., Carneiro, G. H. F., Gomes, C. R., Spinola Filho, P. R. d. C., da Costa, M. R., & Santos, J. B. d. (2024). The Impact of Metolachlor Applications and Phytoremediation Processes on Soil Microorganisms: Insights from Functional Metagenomics Analysis. Journal of Xenobiotics, 14(3), 970-988. https://doi.org/10.3390/jox14030054







