The Extracellular Vesicles Containing Inorganic Polyphosphate of Candida Yeast upon Growth on Hexadecane
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Fluorescent Microscopy
2.3. Scanning Electron Microscopy
2.4. Phosphate and Polyphosphate Extraction and Assay
2.5. The Treatment of Biomass with Polyphosphatase Ppx1
2.6. Amino Acid Sequences Alignments
2.7. Statistical Evaluation
3. Results and Discussion
3.1. Fluorescent and Scanning Electron Microscopy of Yeasts Cells
3.2. Polyphosphate Content in the Cells Cultivated on Glucose or Hexadecane
3.3. The Treatment with Polyphosphatase
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fukui, S.; Tanaka, A. Metabolism of Alkanes by Yeasts. In Proceedings of the Reactors and Reactions; Springer: Berlin/Heidelberg, Germany, 1981; pp. 217–237. [Google Scholar]
- Kogure, T.; Takagi, M.; Ohta, A. N-Alkane and Clofibrate, a Peroxisome Proliferator, Activate Transcription of ALK2 Gene Encoding Cytochrome P450alk2 through Distinct Cis-Acting Promoter Elements in Candida Maltosa. Biochem. Biophys. Res. Commun. 2005, 329, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, R.; Ohta, A. Genetic Features and Regulation of N-Alkane Metabolism in Yeasts. In Aerobic Utilization of Hydrocarbons, Oils and Lipids; Rojo, F., Ed.; Handbook of Hydrocarbon and Lipid Microbiology; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–13. ISBN 978-3-319-39782-5. [Google Scholar]
- Fukuda, R. Utilization of N-Alkane and Roles of Lipid Transfer Proteins in Yarrowia lipolytica. World J. Microbiol. Biotechnol. 2023, 39, 97. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, R. Metabolism of Hydrophobic Carbon Sources and Regulation of It in N-Alkane-Assimilating Yeast Yarrowia Lipolytica. Biosci. Biotechnol. Biochem. 2013, 77, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Finogenova, T.; Morgunov, I.; Kamzolova, S.; Chernyavskaya, O. Organic Acid Production by the Yeast Yarrowia Lipolytica: A Review of Prospects. Appl. Biochem. Microbiol. 2005, 41, 418–425. [Google Scholar] [CrossRef]
- Beier, A.; Hahn, V.; Bornscheuer, U.T.; Schauer, F. Metabolism of Alkenes and Ketones by Candida Maltosa and Related Yeasts. AMB Express 2014, 4, 75. [Google Scholar] [CrossRef]
- Mori, K.; Iwama, R.; Kobayashi, S.; Horiuchi, H.; Fukuda, R.; Ohta, A. Transcriptional Repression by Glycerol of Genes Involved in the Assimilation of N-Alkanes and Fatty Acids in Yeast Yarrowia Lipolytica. FEMS Yeast Res. 2013, 13, 233–240. [Google Scholar] [CrossRef]
- Kogure, T.; Horiuchi, H.; Matsuda, H.; Arie, M.; Takagi, M.; Ohta, A. Enhanced Induction of Cytochromes P450alk That Oxidize Methyl-Ends of n-Alkanes and Fatty Acids in the Long-Chain Dicarboxylic Acid-Hyperproducing Mutant of Candida Maltosa. FEMS Microbiol. Lett. 2007, 271, 106–111. [Google Scholar] [CrossRef]
- Takagi, M.; Ohkuma, M.; Kobayashi, N.; Watanabe, M.; Yano, K. Purification of Cytochrome P-450«/A from n-Alkane-Grown Cells of Candida Maltosa, and Cloning and Nucleotide Sequencing of the Encoding Gene. Agric. Biol. Chem. 1989, 53, 2217–2226. [Google Scholar] [CrossRef]
- Schunck, W.H.; Kärgel, E.; Gross, B.; Wiedmann, B.; Mauersberger, S.; Köpke, K.; Kiessling, U.; Strauss, M.; Gaestel, M.; Müller, H.G. Molecular Cloning and Characterization of the Primary Structure of the Alkane Hydroxylating Cytochrome P-450 from the Yeast Candida Maltosa. Biochem. Biophys Res. Commun. 1989, 161, 843–850. [Google Scholar] [CrossRef]
- Mauersberger, S.; Ohkuma, M.; Schunck, W.-H.; Takagi, M. Candida Maltosa. In Nonconventional Yeasts in Biotechnology: A Handbook; Wolf, K., Ed.; Springer: Berlin/Heidelberg, Germany, 1996; pp. 411–580. ISBN 978-3-642-79856-6. [Google Scholar]
- Ohkuma, M.; Tanimoto, T.; Yano, K.; Takagi, M. CYP52 (Cytochrome P450alk) Multigene Family in Candida Maltosa: Molecular Cloning and Nucleotide Sequence of the Two Tandemly Arranged Genes. DNA Cell Biol. 1991, 10, 271–282. [Google Scholar] [CrossRef]
- Ohkuma, M.; Muraoka, S.; Tanimoto, T.; Fujii, M.; Ohta, A.; Takagi, M. CYP52 (Cytochrome P450alk) Multigene Family in Candida Maltosa: Identification and Characterization of Eight Members. DNA Cell Biol. 1995, 14, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Iwama, R.; Kobayashi, S.; Ishimaru, C.; Ohta, A.; Horiuchi, H.; Fukuda, R. Functional Roles and Substrate Specificities of Twelve Cytochromes P450 Belonging to CYP52 Family in N-Alkane Assimilating Yeast Yarrowia Lipolytica. Fungal. Genet. Biol. 2016, 91, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, S.; Tanaka, A.; Yamamura, M.; Teranishi, Y.; Fukui, S. Microbody of N-Alkane-Grown Yeast. Arch. Microbiol. 1977, 112, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mauersberger, S.; Kärgel, E.; Matyashova, R.N.; Müller, H.-G. Subcellular Organization of Alkane Oxidation in the Yeast Candida Maltosa. J. Basic Microbiol. 1987, 27, 565–582. [Google Scholar] [CrossRef]
- Vogel, F.; Kärgel, E.; Schunck, W.H. In Situ Localization of Cytochrome P-450, the First Enzyme Involved in Aliphatic Hydrocarbon Degradation in the Yeast Candida Maltosa. Prog. Histochem. Cytochem. 1991, 23, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, Y.; Song, Q.; Zhang, S.; Xie, L.; Yang, J. Transmembrane Transport Mechanism of N-Hexadecane by Candida Tropicalis: Kinetic Study and Proteomic Analysis. Ecotoxicol. Environ. Saf. 2021, 209, 111789. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, V.V.; Crowley, D.; Rogachevsky, V.V.; Negri, C.M.; Rusakova, T.G.; Kolesnikova, S.A.; Akhmetov, L.I. Microorganisms Form Exocellular Structures, Trophosomes, to Facilitate Biodegradation of Oil in Aqueous Media. FEMS Microbiol. Lett. 2011, 315, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, V.V.; Crowley, D.E.; Zvonarev, A.N.; Rusakova, T.G.; Negri, M.C.; Kolesnikova, S.A. Modifications of the Cell Wall of Yeasts Grown on Hexadecane and under Starvation Conditions. Yeast 2016, 33, 55–62. [Google Scholar] [CrossRef]
- Zvonarev, A.N.; Crowley, D.E.; Ryazanova, L.P.; Lichko, L.P.; Rusakova, T.G.; Kulakovskaya, T.V.; Dmitriev, V.V. Cell Wall Canals Formed upon Growth of Candida Maltosa in the Presence of Hexadecane Are Associated with Polyphosphates. FEMS Yeast Res. 2017, 17, fox026. [Google Scholar] [CrossRef]
- Zvonarev, A.; Farofonova, V.; Kulakovskaya, E.; Kulakovskaya, T.; Machulin, A.; Sokolov, S.; Dmitriev, V. Changes in Cell Wall Structure and Protein Set in Candida Maltosa Grown on Hexadecane. Folia Microbiol. 2021, 66, 247–253. [Google Scholar] [CrossRef]
- Anderson, J.; Mihalik, R.; Soll, D.R. Ultrastructure and Antigenicity of the Unique Cell Wall Pimple of the Candida Opaque Phenotype. J. Bacteriol. 1990, 172, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Staniszewska, M.; Bondaryk, M.; Swoboda-Kopec, E.; Siennicka, K.; Sygitowicz, G.; Kurzatkowski, W. Candida Albicans Morphologies Revealed by Scanning Electron Microscopy Analysis. Braz. J. Microbiol. 2013, 44, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Nett, J.E.; Sanchez, H.; Cain, M.T.; Ross, K.M.; Andes, D.R. Interface of Candida Albicans Biofilm Matrix-Associated Drug Resistance and Cell Wall Integrity Regulation. Eukaryot. Cell 2011, 10, 1660–1669. [Google Scholar] [CrossRef] [PubMed]
- Silva-Dias, A.; Miranda, I.M.; Branco, J.; Monteiro-Soares, M.; Pina-Vaz, C.; Rodrigues, A.G. Adhesion, Biofilm Formation, Cell Surface Hydrophobicity, and Antifungal Planktonic Susceptibility: Relationship among Candida spp. Front. Microbiol. 2015, 6, 205. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Negri, M.; Henriques, M.; Oliveira, R.; Williams, D.W.; Azeredo, J. Candida Glabrata, Candida Parapsilosis and Candida Tropicalis: Biology, Epidemiology, Pathogenicity and Antifungal Resistance. FEMS Microbiol. Rev. 2012, 36, 288–305. [Google Scholar] [CrossRef] [PubMed]
- Dižová, S.; Černáková, L.; Bujdáková, H. The Impact of Farnesol in Combination with Fluconazole on Candida Albicans Biofilm: Regulation of ERG20, ERG9, and ERG11 Genes. Folia Microbiol. 2018, 63, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Suchodolski, J.; Derkacz, D.; Muraszko, J.; Panek, J.J.; Jezierska, A.; Łukaszewicz, M.; Krasowska, A. Fluconazole and Lipopeptide Surfactin Interplay During Candida Albicans Plasma Membrane and Cell Wall Remodeling Increases Fungal Immune System Exposure. Pharmaceutics 2020, 12, 314. [Google Scholar] [CrossRef]
- Yeh, Y.-C.; Wang, H.-Y.; Lan, C.-Y. Candida Albicans Aro1 Affects Cell Wall Integrity, Biofilm Formation and Virulence. J. Microbiol. Immunol. Infect. 2020, 53, 115–124. [Google Scholar] [CrossRef]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The Fungal Cell Wall: Candida, Cryptococcus, and Aspergillus Species. Front. Microbiol. 2019, 10, 2993. [Google Scholar] [CrossRef]
- Blasi, B.; Poyntner, C.; Rudavsky, T.; Prenafeta-Boldú, F.X.; Hoog, S.D.; Tafer, H.; Sterflinger, K. Pathogenic Yet Environmentally Friendly? Black Fungal Candidates for Bioremediation of Pollutants. Geomicrobiol. J. 2016, 33, 308–317. [Google Scholar] [CrossRef]
- Lavrin, T.; Konte, T.; Kostanjšek, R.; Sitar, S.; Sepčič, K.; Prpar Mihevc, S.; Žagar, E.; Župunski, V.; Lenassi, M.; Rogelj, B.; et al. The Neurotropic Black Yeast Exophiala Dermatitidis Induces Neurocytotoxicity in Neuroblastoma Cells and Progressive Cell Death. Cells 2020, 9, 963. [Google Scholar] [CrossRef] [PubMed]
- Rao, N.N.; Gómez-García, M.R.; Kornberg, A. Inorganic Polyphosphate: Essential for Growth and Survival. Annu. Rev. Biochem. 2009, 78, 605–647. [Google Scholar] [CrossRef] [PubMed]
- Ikeh, M.; Ahmed, Y.; Quinn, J. Phosphate Acquisition and Virulence in Human Fungal Pathogens. Microorganisms 2017, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.L.; Gomes, F.M.; Girard-Dias, W.; Almeida, F.P.; Albuquerque, P.C.; Kretschmer, M.; Kronstad, J.W.; Frases, S.; de Souza, W.; Rodrigues, M.L.; et al. Phosphorus-Rich Structures and Capsular Architecture in Cryptococcus Neoformans. Future Microbiol. 2017, 12, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Bowring, B.G.; Sethiya, P.; Desmarini, D.; Lev, S.; Tran Le, L.; Bahn, Y.-S.; Lee, S.-H.; Toh-e, A.; Proschogo, N.; Savage, T.; et al. Dysregulating PHO Signaling via the CDK Machinery Differentially Impacts Energy Metabolism, Calcineurin Signaling, and Virulence in Cryptococcus Neoformans. mBio 2023, 14, e0355122. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Pan, H. Functions of CaPhm7 in the Regulation of Ion Homeostasis, Drug Tolerance, Filamentation and Virulence in Candida Albicans. BMC Microbiol. 2018, 18, 49. [Google Scholar] [CrossRef] [PubMed]
- Kulakovskaya, E.; Zvonarev, A.; Kulakovskaya, T. PHM6 and PHM7 Genes Are Essential for Phosphate Surplus in the Cells of Saccharomyces Cerevisiae. Arch. Microbiol. 2023, 205, 47. [Google Scholar] [CrossRef]
- Liu, N.-N.; Acosta-Zaldívar, M.; Qi, W.; Diray-Arce, J.; Walker, L.A.; Kottom, T.J.; Kelly, R.; Yuan, M.; Asara, J.M.; Lasky-Su, J.A.; et al. Phosphoric Metabolites Link Phosphate Import and Polysaccharide Biosynthesis for Candida Albicans Cell Wall Maintenance. mBio 2020, 11, e03225-19. [Google Scholar] [CrossRef]
- Ohkuma, M.; Hwang, C.W.; Masuda, Y.; Nishida, H.; Sugiyama, J.; Ohta, A.; Takagi, M. Evolutionary Position of N-Alkane-Assimilating Yeast Candida Maltosa Shown by Nucleotide Sequence of Small-Subunit Ribosomal RNA Gene. Biosci. Biotechnol. Biochem. 1993, 57, 1793–1794. [Google Scholar] [CrossRef]
- Olanbiwoninu, A.A.; Popoola, B.M. Biofilms and Their Impact on the Food Industry. Saudi. J. Biol. Sci. 2023, 30, 103523. [Google Scholar] [CrossRef]
- Li, X.; Zhang, K.; Xue, Y. Continuous Immobilization of Lead by Biomineralization in a Fluidized-Bed Biofilm Reactor. J. Clean. Prod. 2022, 379, 134765. [Google Scholar] [CrossRef]
- An, H.; Tian, T.; Wang, Z.; Jin, R.; Zhou, J. Role of Extracellular Polymeric Substances in the Immobilization of Hexavalent Chromium by Shewanella Putrefaciens CN32 Unsaturated Biofilms. Sci. Total Environ. 2022, 810, 151184. [Google Scholar] [CrossRef] [PubMed]
- Yunda, E.; Gutensohn, M.; Ramstedt, M.; Björn, E. Methylmercury Formation in Biofilms of Geobacter Sulfurreducens. Front. Microbiol. 2023, 14, 1079000. [Google Scholar] [CrossRef] [PubMed]
- Hermida, L.; Agustian, J. The Application of Conventional or Magnetic Materials to Support Immobilization of Amylolytic Enzymes for Batch and Continuous Operation of Starch Hydrolysis Processes. Rev. Chem. Eng. 2022. [Google Scholar] [CrossRef]
- Kim, J.-H.; Lee, E.-S.; Song, K.-J.; Kim, B.-M.; Ham, J.-S.; Oh, M.-H. Development of Desiccation-Tolerant Probiotic Biofilms Inhibitory for Growth of Foodborne Pathogens on Stainless Steel Surfaces. Foods 2022, 11, 831. [Google Scholar] [CrossRef] [PubMed]
- Bouaziz, A.; Houfani, A.A.; Baoune, H. Enzymology of Microbial Biofilms. In Ecological Interplays in Microbial Enzymology; Maddela, N.R., Abiodun, A.S., Prasad, R., Eds.; Environmental and Microbial Biotechnology; Springer Nature: Singapore, 2022; pp. 117–140. ISBN 978-981-19015-5-3. [Google Scholar]
- Sheng, Y.; Chen, Z.; Wu, W.; Lu, Y. Engineered Organic Nanoparticles to Combat Biofilms. Drug Discov. Today 2023, 28, 103455. [Google Scholar] [CrossRef] [PubMed]
- Eldarov, M.A.; Baranov, M.V.; Dumina, M.V.; Shgun, A.A.; Andreeva, N.A.; Trilisenko, L.V.; Kulakovskaya, T.V.; Ryasanova, L.P.; Kulaev, I.S. Polyphosphates and Exopolyphosphatase Activities in the Yeast Saccharomyces Cerevisiae under Overexpression of Homologous and Heterologous PPN1 Genes. Biochem. Mosc. 2013, 78, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Vagabov, V.M.; Trilisenko, L.V.; Kulaev, I.S. Dependence of Inorganic Polyphosphate Chain Length on the Orthophosphate Content in the Culture Medium of the Yeast Saccharomyces Cerevisiae. Biochemistry 2000, 65, 349–354. [Google Scholar]
- Heinonen, J.K.; Lahti, R.J. A New and Convenient Colorimetric Determination of Inorganic Orthophosphate and Its Application to the Assay of Inorganic Pyrophosphatase. Anal. Biochem. 1981, 113, 313–317. [Google Scholar] [CrossRef]
- Andreeva, N.; Ledova, L.; Ryazanova, L.; Tomashevsky, A.; Kulakovskaya, T.; Eldarov, M. Ppn2 Endopolyphosphatase Overexpressed in Saccharomyces Cerevisiae: Comparison with Ppn1, Ppx1, and Ddp1 Polyphosphatases. Biochimie 2019, 163, 101–107. [Google Scholar] [CrossRef]
- Serafim, L.S.; Lemos, P.C.; Levantesi, C.; Tandoi, V.; Santos, H.; Reis, M.A.M. Methods for Detection and Visualization of Intracellular Polymers Stored by Polyphosphate-Accumulating Microorganisms. J. Microbiol. Methods 2002, 51, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, N.; Ryazanova, L.; Dmitriev, V.; Kulakovskaya, T.; Kulaev, I. Adaptation of Saccharomyces Cerevisiae to Toxic Manganese Concentration Triggers Changes in Inorganic Polyphosphates. FEMS Yeast Res. 2013, 13, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Lichko, L.; Kulakovskaya, T. Polyphosphatase PPX1 of Saccharomyces Cerevisiae as a Tool for Polyphosphate Assay. Adv. Enzym. Res. 2015, 3, 93. [Google Scholar] [CrossRef]
- Hothorn, M.; Neumann, H.; Lenherr, E.D.; Wehner, M.; Rybin, V.; Hassa, P.O.; Uttenweiler, A.; Reinhardt, M.; Schmidt, A.; Seiler, J.; et al. Catalytic Core of a Membrane-Associated Eukaryotic Polyphosphate Polymerase. Science 2009, 324, 513–516. [Google Scholar] [CrossRef] [PubMed]
- Guan, Z.; Chen, J.; Liu, R.; Chen, Y.; Xing, Q.; Du, Z.; Cheng, M.; Hu, J.; Zhang, W.; Mei, W.; et al. The Cytoplasmic Synthesis and Coupled Membrane Translocation of Eukaryotic Polyphosphate by Signal-Activated VTC Complex. Nat. Commun. 2023, 14, 718. [Google Scholar] [CrossRef] [PubMed]
- Gerasimaitė, R.; Mayer, A. Enzymes of Yeast Polyphosphate Metabolism: Structure, Enzymology and Biological Roles. Biochem. Soc. Trans. 2016, 44, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Liebana-Jordan, M.; Brotons, B.; Falcon-Perez, J.M.; Gonzalez, E. Extracellular Vesicles in the Fungi Kingdom. Int. J. Mol. Sci. 2021, 22, 7221. [Google Scholar] [CrossRef]
- Oliveira, B.T.M.; Dourado, T.M.H.; Santos, P.W.S.; Bitencourt, T.A.; Tirapelli, C.R.; Colombo, A.L.; Almeida, F. Extracellular Vesicles from Candida Haemulonii Var. Vulnera Modulate Macrophage Oxidative Burst. J. Fungi 2023, 9, 562. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, Z.; Liu, Y.; Liao, B.; Zong, Y.; Shi, Y.; Liao, M.; Wang, J.; Zhou, X.; Cheng, L.; et al. Extracellular Vesicles of Candida Albicans Regulate Its Own Growth through the L-Arginine/Nitric Oxide Pathway. Appl. Microbiol. Biotechnol. 2023, 107, 355–367. [Google Scholar] [CrossRef]
- Macias-Paz, I.U.; Pérez-Hernández, S.; Tavera-Tapia, A.; Luna-Arias, J.P.; Guerra-Cárdenas, J.E.; Reyna-Beltrán, E. Candida albicans the main opportunistic pathogenic fungus in humans. Rev. Argent Microbiol. 2023, 55, 189–198. [Google Scholar] [CrossRef]
- Branco, J.; Miranda, I.M.; Rodrigues, A.G. Candida parapsilosis Virulence and Antifungal Resistance Mechanisms: A Comprehensive Review of Key Determinants. J. Fungi 2023, 9, 80. [Google Scholar] [CrossRef]
- Dos Santos, M.M.; Ishida, K. We need to talk about Candida tropicalis: Virulence factors and survival mechanisms. Med. Mycol. 2023, 61, myad075. [Google Scholar] [CrossRef]
- Martín, J.F. Interaction of calcium responsive proteins and transcriptional factors with the PHO regulon in yeasts and fungi. Front. Cell Dev. Biol. 2023, 11, 1225774. [Google Scholar] [CrossRef]
- Lucas, C.; Silva, C. The Extracellular Matrix of Yeasts: A Key Player in the Microbial Biology Change of Paradigm. Front. Biosci.-Elite 2023, 15, 13. [Google Scholar] [CrossRef]
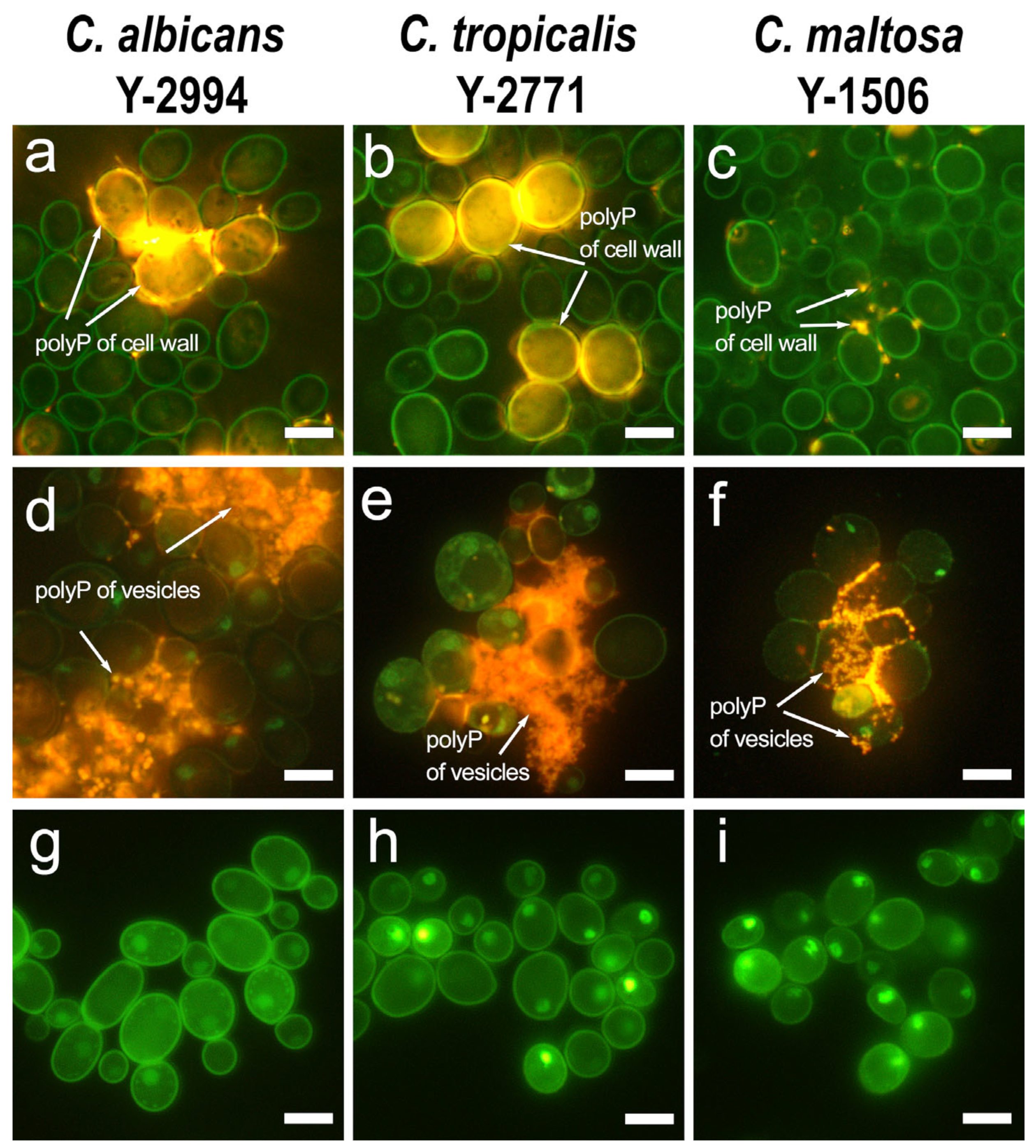
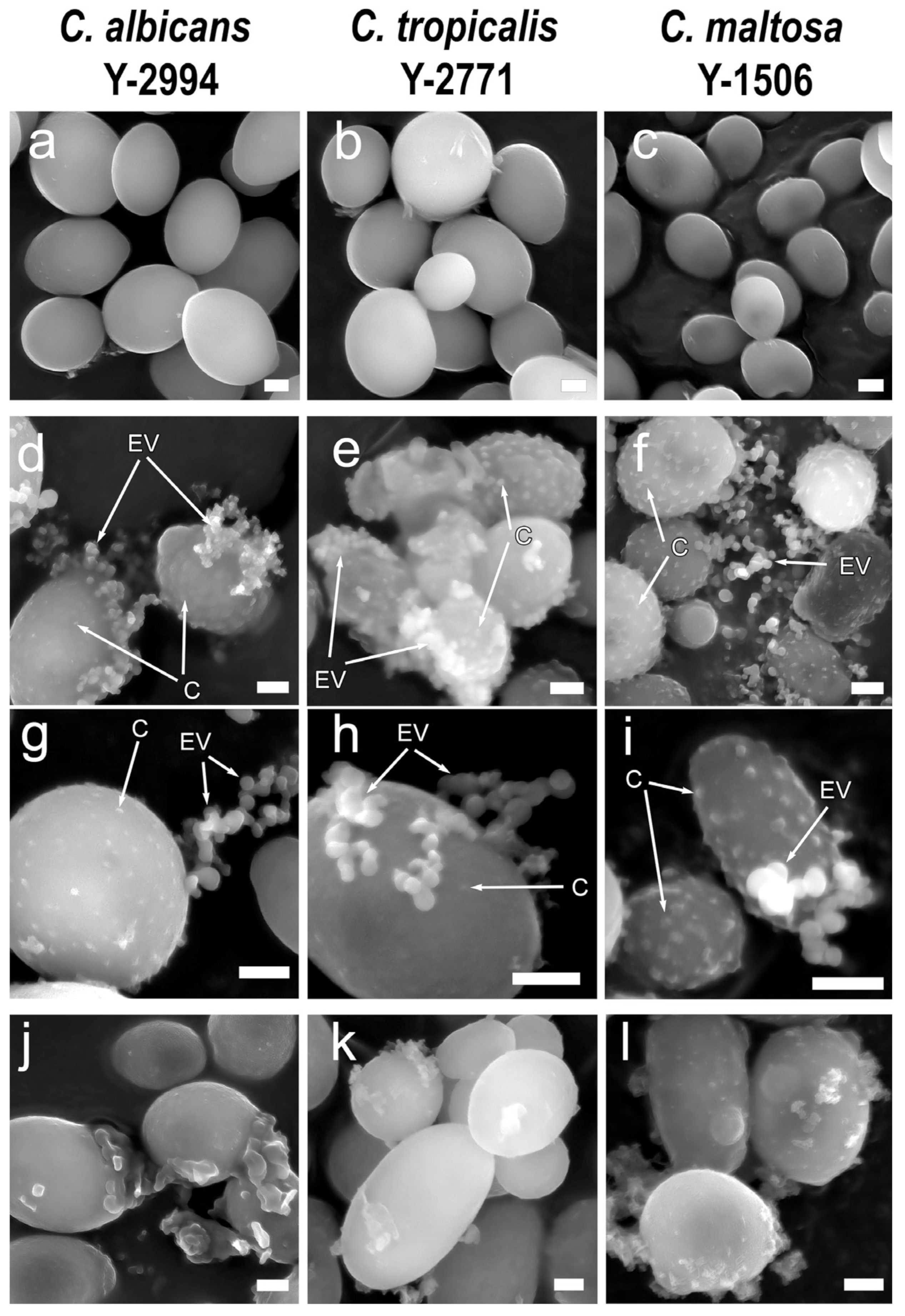

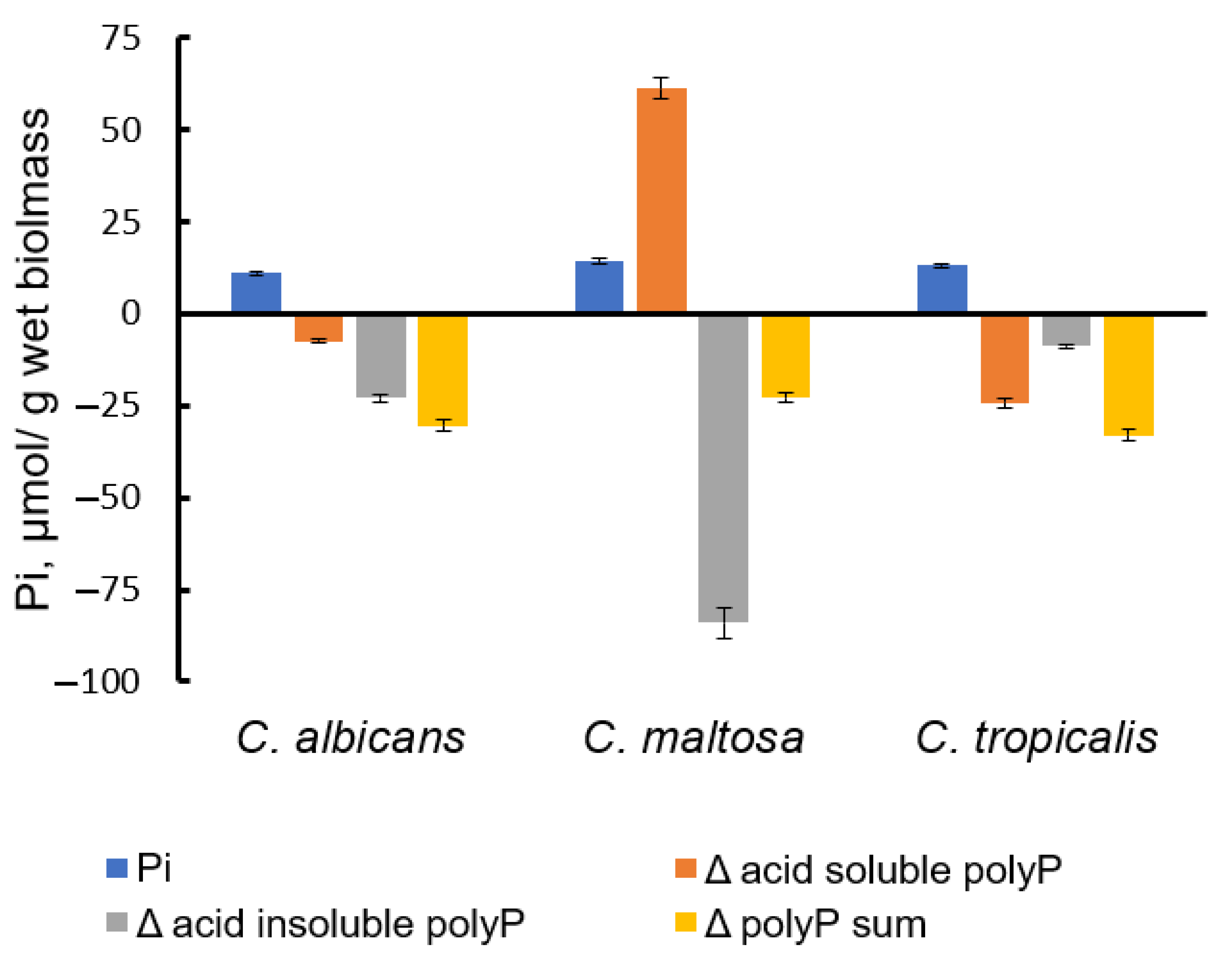
| Glucose | Hexadecane | |||||
|---|---|---|---|---|---|---|
| Yeast Species | Pi | Acid Soluble polyP | Acid Insoluble polyP | Pi | Acid Soluble polyP | Acid Insoluble polyP |
| C albicans | 10.6 ± 0.20 | 22.0 ± 1.93 | 115 ± 4.1 | 27.6 ± 1.625 | 34.0 ± 1.26 | 59.1 ± 13.4 |
| C. tropicalis | 10.3 ± 0.25 | 3.93 ± 0.66 | 30.5 ± 1.0 | 14.9 ± 0.23 | 10.9 ± 0.91 | 44.8 ± 4.97 |
| C. maltosa | 15.8 ± 2.4 | 8.02 ± 2.04 | 79.6 ± 1.1 | 16.3 ± 1.2 | 11.5 ± 4.24 | 124 ± 9.9 |
| Species | C. albicans | C. tropicalis | C. maltosa | |
|---|---|---|---|---|
| VTC4 (SGD:S000003549) | Accession number | XP_711583.1 | XP_002545726.1 | EMG48228.1 |
| Query Cover, % | 99 | 99 | 99 | |
| Percent identity | 60.82 | 60.27 | 60.95 | |
| PPX1 (SGD:S000001244) | Accession number | KHC58625.1 | XP_002547193.1 | EMG50694.1 |
| Query Cover, % | 97 | 90 | 95 | |
| Percent identity | 32.78 | 34.36 | 34.2 | |
| PPN2 (SGD:S000005161) | Accession number | KHC62723.1 | XP_002550886.1 | EMG47562.1 |
| Query Cover, % | 95 | 95 | 96 | |
| Percent identity | 29.01 | 30 | 29.14 | |
| PPN1 (SGD:S000002860) | Accession number | KHC31142.1 | XP_002550589.1 | EMG47375.1 |
| Query Cover, % | 94 | 93 | 87 | |
| Percent identity | 41.01 | 40.59 | 42.95 |
| Species | C. tropicalis | C. maltosa | |
|---|---|---|---|
| VTC4 (XP_711583.1) | Accession number | XP_002545726.1 | EMG48228.1 |
| Query Cover, % | 99 | 99 | |
| Percent identity | 82.71 | 86.85 | |
| PPX1 (KHC58625.1) | Accession number | XP_002547193.1 | EMG50694.1 |
| Query Cover, % | 100 | 100 | |
| Percent identity | 61.94 | 63.03 | |
| PPN2 (KHC62723.1) | Accession number | XP_002550886.1 | EMG47562.1 |
| Query Cover, % | 100 | 100 | |
| Percent identity | 59.95 | 59.44 | |
| PPN1 (KHC31142.1) | Accession number | XP_002550589.1 | EMG47375.1 |
| Query Cover, % | 97 | 99 | |
| Percent identity | 65.1 | 64.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zvonarev, A.N.; Trilisenko, L.V.; Farofonova, V.V.; Kulakovskaya, E.V.; Abashina, T.N.; Dmitriev, V.V.; Kulakovskaya, T. The Extracellular Vesicles Containing Inorganic Polyphosphate of Candida Yeast upon Growth on Hexadecane. J. Xenobiot. 2023, 13, 529-543. https://doi.org/10.3390/jox13040034
Zvonarev AN, Trilisenko LV, Farofonova VV, Kulakovskaya EV, Abashina TN, Dmitriev VV, Kulakovskaya T. The Extracellular Vesicles Containing Inorganic Polyphosphate of Candida Yeast upon Growth on Hexadecane. Journal of Xenobiotics. 2023; 13(4):529-543. https://doi.org/10.3390/jox13040034
Chicago/Turabian StyleZvonarev, Anton N., Ludmila V. Trilisenko, Vasilina V. Farofonova, Ekaterina V. Kulakovskaya, Tatiana N. Abashina, Vladimir V. Dmitriev, and Tatiana Kulakovskaya. 2023. "The Extracellular Vesicles Containing Inorganic Polyphosphate of Candida Yeast upon Growth on Hexadecane" Journal of Xenobiotics 13, no. 4: 529-543. https://doi.org/10.3390/jox13040034
APA StyleZvonarev, A. N., Trilisenko, L. V., Farofonova, V. V., Kulakovskaya, E. V., Abashina, T. N., Dmitriev, V. V., & Kulakovskaya, T. (2023). The Extracellular Vesicles Containing Inorganic Polyphosphate of Candida Yeast upon Growth on Hexadecane. Journal of Xenobiotics, 13(4), 529-543. https://doi.org/10.3390/jox13040034







