Exploring the Potential of Biocontrol Agent Against Root and Stem Rot Disease in Durian (Durio zibethinus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Pathogen Isolation
2.1.1. Plant Tissue Samples
2.1.2. Soil Sample
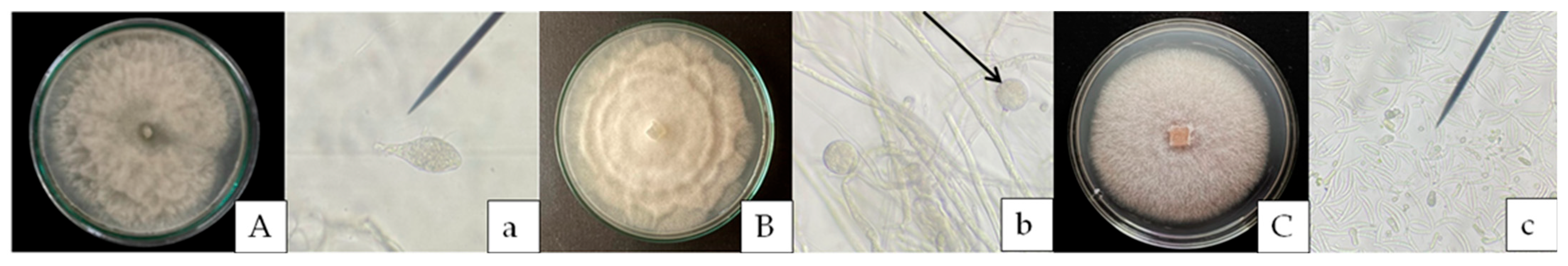
2.1.3. Morphological Investigation
2.2. Pathogenicity Test
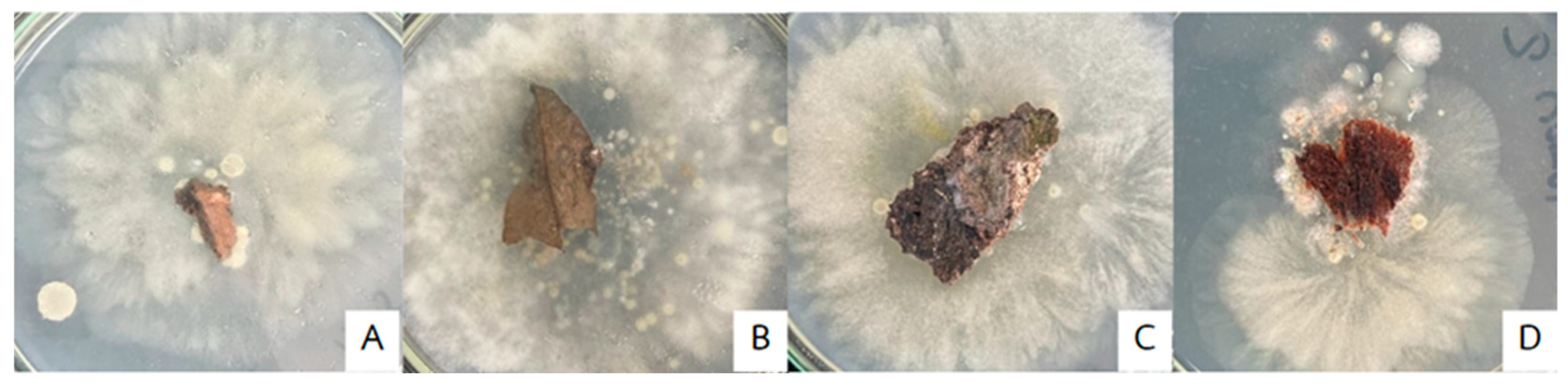
2.2.1. Detached Leaf Technique
2.2.2. Test on Durian Trees
2.3. Isolation of Bacteria
2.4. Testing the Efficacy of Isolated Bacteria in Inhibiting the Growth of the Mycelium of 3 Pathogens
- R1 = Radius of the fungal and oomycetes colony on the control plate
- R2 = Radius of the fungal and oomycetes colony on the test plate
2.5. Physiological, Biochemical, and Genetic Characterization of the Bacterial Isolates
2.5.1. Physiological
2.5.2. Biochemical
2.5.3. Molecular Analyses
2.6. Testing the Efficacy of 3-Selected Bacterial Strains Against P. palmivora in the Field
3. Results and Discussion
3.1. Isolation Pathogens Causing Root and Stem Rot in Durian
3.1.1. Plate Assay with Detached Leaf Technique
3.1.2. Plant Assay Test on Durian Trees
3.2. Isolation of Beneficial Bacteria
3.3. The Efficacy of Beneficial Bacteria Inhibited the Mycelium Pathogen Caused Durian Root and Stem Rot
3.4. Classification of Beneficial Bacteria
3.4.1. The Beneficial Bacteria Was Identified and Classified
3.4.2. Identification of Microorganisms Using Genetic Information
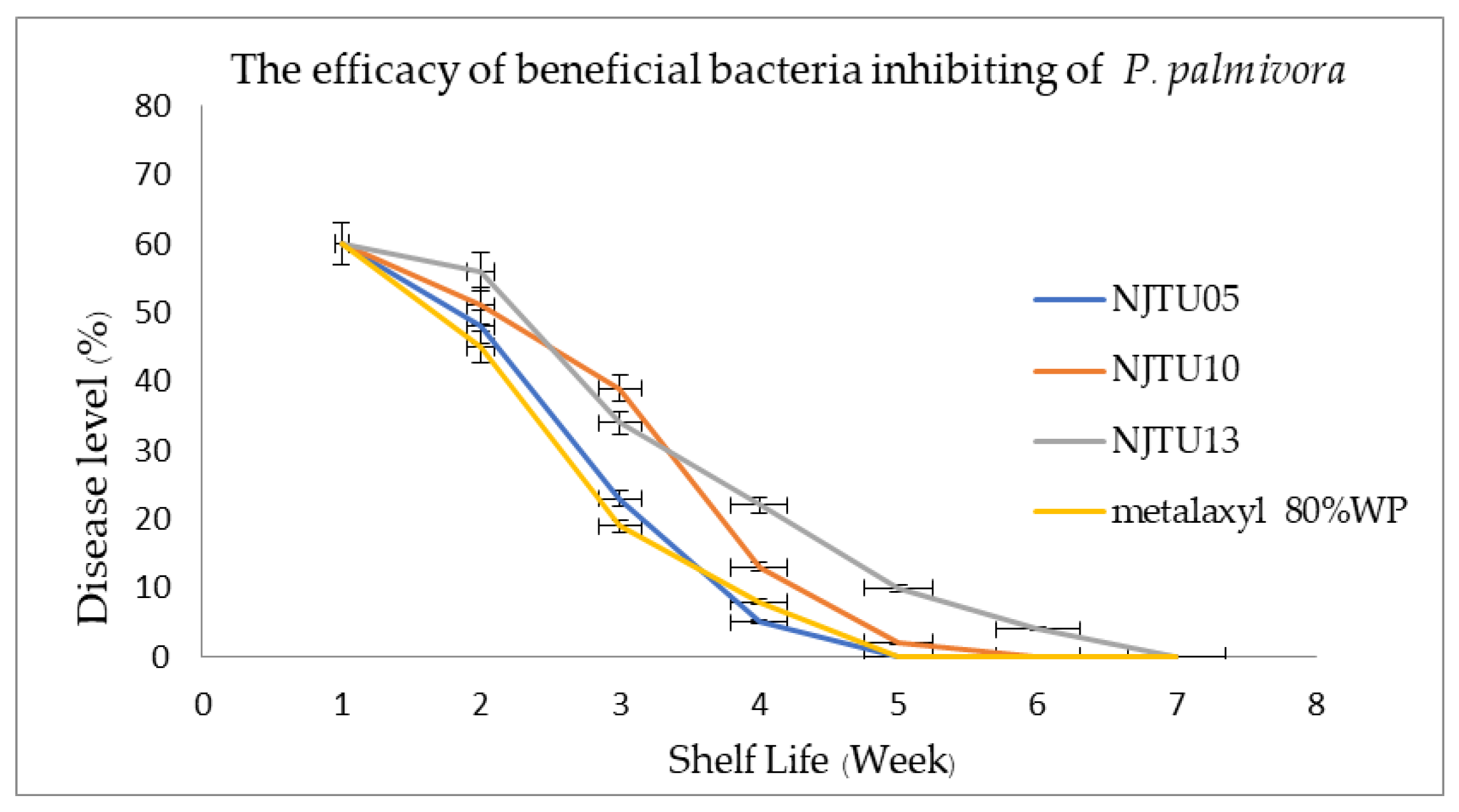
3.5. Efficacy of Three Selected Bacterial Strains Against P. palmivora in the Field
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Durian, the King of Thai Fruits, Is Loved by Foreigners. Available online: https://tpso.go.th/en (accessed on 13 February 2025).
- Angiosperm Phylogeny Group. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- Department of Agricultural Extension. Durian Production Statistics 2022. Ministry of Agriculture and Cooperatives. 2023. Available online: https://www.example.com (accessed on 16 June 2025).
- Ministry of Agriculture and Cooperatives. Durian Development Strategic Plan (2023–2027). 2023. Available online: https://www.moac.go.th (accessed on 16 June 2025).
- Saradee Story. Export Statistics of Thai Durian in 2021 and 2022. 2022. Available online: https://saradeestory.com/story/durian-exports64vs65/ (accessed on 29 June 2025).
- Suksiri, S.; Laipasu, P.; Soytong, K.; Poeaim, S. Isolation and identification of Phytophthora sp. and Pythium sp. from durian orchard in Chumphon province, Thailand. Int. J. Agric. Technol. 2018, 14, 389–402. [Google Scholar]
- Root Rot and Foot Rot. Available online: https://esc.doae.go.th (accessed on 18 January 2025).
- Jitfour, N.; Athinuwat, D.; Chuaboon, W. Brevibacillus formosusstrain NJTU05 powder formulation bioproduct to control root and stem rotdisease of durian. KKU Res. J. 2025, 53, 479–490. [Google Scholar]
- Pengnoo, A.; Lohlaeh, U.; Maduerehand, F.; Kaewmano, C.; Krualee, S.; Wongpisal, P.; Homhaul, W.; Boonyapipat, P.; Saeng-ngam, S.; Äkbärjan, A.; et al. Bacillus velezensis SM1: A promising biocontrol solution for Phytophthora durian root rot. Appl. Microbiol. 2025, 5, 21. [Google Scholar] [CrossRef]
- Danial, N.D.; Matthews, S.; Amaran, N.F.; Abd Kharim, N.A. Assessment of Streptomyces sp. and Bacillus sp. against Phytophthora palmivora, the causal agent of durian canker disease through different application methods at the nursery stage. J. Agrobiotechnol. 2024, 15, 18–24. [Google Scholar] [CrossRef]
- Detmanee, P.; Athinuwat, D.; Chueaboon, W. Characteristics and Efficacy of Antagonistic Microorganisms in Controlling Phytophthora palmivora, the Causal Agent of Root and Stem Rot in Durian. In Proceedings of the 15th National Plant Protection Conference, Rama Gardens Hotel, Bangkok, Thailand, 22–24 November 2022. [Google Scholar]
- Chueboon, W. Characteristics and Effectiveness Testing of Beneficial Bacteria for Controlling Erwinia carotovora subsp. Carotovora, the Cause of Soft Rot. Master’s Thesis, Kasetsart University, Bangkok, Thailand, 2008. [Google Scholar]
- Raza, W.; Ghazanfar, M.U.; Ullah, Z. Isolation and morphological characterization of Phytophthora palmivora on PDA medium. Plant Pathol. J. 2022, 38, 215–223. [Google Scholar]
- Salvamani, S.; Nawawi, N.M. Macroscopic and Microscopic Approaches for Identification of Fungi from Plant Soil of Cameron Highlands. Bioremediat. Sci. Technol. Res. 2014, 2, 14–18. [Google Scholar] [CrossRef]
- Vawdrey, L.; Martin, T.M.; De Faveri, J. A detached leaf bioassay to screen durian cultivars for susceptibility to Phytophthora palmivora. Australas. Plant Pathol. 2005, 34, 251–253. [Google Scholar] [CrossRef]
- Chatchaisiri, L.; Pongpisutta, R.; Rattanakreetakul, C.; Bincader, S. Pythium sp. Causing root of durian and efficacy of chemical fungicides to control. Agric. Sci. J. 2018, 49, 171–174. [Google Scholar]
- Datmanee, P.; Suknant, T.; Srisuwan, K.; Chuaboon, W. Isolation beneficial bacteria degrade heavy metal (Arsenic). In Proceedings of the 8th Academic Science and Technology Conference, Patumthani, Thailand, 26 March 2021. [Google Scholar]
- Somjinda, S.; Athinuwat, D.; Chueboon, W. Effects of Beneficial Bacteria in Controlling Colletotrichum capsici the Cause of Anthracnose in Chili Peppers. In Proceedings of the 10th RMUT Surin National Conference on Research and Innovation for Sustainable Development, Surin, Thailand, 19–20 September 2019. [Google Scholar]
- Jaiyen, K.; Laitalb, A.; Athinuwat, D.; Chueaboon, W. Selection and Testing of Beneficial Microorganisms for Controlling Major Diseases in Cannabis. In Proceedings of the 1st National Conference on Cannabis, Hemp, and Kratom, Nakhon Pathom, Thailand, 20 April 2023. [Google Scholar]
- Rahman, M.A.; Begum, M.F.; Alam, M.F. Screening of Trichoderma isolates as a biological control agent against Ceratocystis paradoxa causing pineapple disease of sugarcane. Mycol. Res. 2009, 37, 277–285. [Google Scholar]
- Farmer, K.; James, A.; Naraine, R.; Dolphin, G.; Sylvester, W.; Amadi, V.; Kotelnikova, S.V. Urinary tract infection Escherichia coli is related to the environmental Escherichia coli in their DNA barcoding and antibiotic resistance patterns in Grenada. Adv. Microbiol. 2016, 6, 33–46. [Google Scholar] [CrossRef]
- Valenza, G.; Ruoff, C.; Vogel, U.; Frosch, M.; Horn, M.A. Microbiological evaluation of the new VITEK 2 Neisseria-Hemophilus identification card. J. Clin. Microbiol. 2007, 45, 3493–3497. [Google Scholar] [CrossRef] [PubMed]
- Kongcharoensunthorn, W.; Treyasinha, R. PCR for screening of multidrug resistant Acinetobacter baumannii by MexA, MexB like gene, and 16–23S rRNA intergenic spacer region (ISR). BUU Sci. J. 2021, 21, 10–16. [Google Scholar]
- Guidelinrs for Surveillance for Plant Pests in Asia and the Pacific. Available online: https://www.aciar.gov.au/sites/default/files/legacy/node/2311/mn119_guidelines_for_surveillance_for_plant_pests__16233.pdf (accessed on 13 February 2025).
- Phongsuthat, N.; Phankamolsil, N.; Moonjuntha, P.; Amawan, S.; Intanoo, W.; Unartgam, J. Pathogenicity of cassava rot associated fungi and genetic variation of fungi. Thai Agric. Res. J. 2022, 40, 238–250. [Google Scholar]
- Agricultural Research Development Agency (ARDA). “Remove Eastern Durian Lessons 2022 for the Future Opportunities,” and New Challenges” and Training “Management and Control of Dry Twig and Root Rot Disease to Quality Durian Product”. Available online: https://en.arda.or.th/newlist-detail.php?id=990 (accessed on 13 February 2025).
- Swiatczak, J.; Kalwasinska, A.; Wojciechowska, A.; Brzezinska, M.S. Physiological properties and genomic insights into the plant growth promoting rhizobacterium Brevibacillus laterosporus K75 isolated from maize rhizosphere. J. Sci. Food Agric. 2023, 103, 1432–1444. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Guo, L.; Zeng, H.; Yang, X.; Yuan, J.; Shi, H.; Xiong, Y.; Chen, M.; Han, L.; Qiu, D. Purification and characterization of a novel antimicrobial peptide from Brevibacillus laterosporus A60. J. Pept. Sci. 2012, 33, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Sivakorn, N.; Trakul-Sukrat, P.; Songsaeng, P.; Worakuldamrongchai, S. Control of Durian Root and Crown Rot Diseases Using Bioagents Derived from Bacillus Subtilis; Plant Protection Research and Development Office: Bangkok, Thailand, 2011. [Google Scholar]




| Isolates | Sample | Location of Isolation | Characteristics |
|---|---|---|---|
| TNP01 | soil | Tha Mai, Chanthaburi | Rosaceous |
| TNP02 | leaf | Tha Mai, Chanthaburi | Rosaceous |
| TNP03 | soil | Tha Mai, Chanthaburi | Rosaceous |
| TNP04 | root | Tha Mai, Chanthaburi | Chrysanthemum |
| TNP17 | soil | Tha Mai, Chanthaburi | Chrysanthemum |
| TNP18 | soil | Tha Mai, Chanthaburi | Radial |
| MNP11 | root | Makham, Chanthaburi | Chrysanthemum |
| MNP13 | soil | Makham, Chanthaburi | Radial |
| MNP19 | leaf | Makham, Chanthaburi | Rosaceous |
| MNP20 | root | Makham, Chanthaburi | Rosaceous |
| KNP05 | leaf | Khao Khitchakut, Chanthaburi | Chrysanthemum |
| KNP06 | soil | Khao Khitchakut, Chanthaburi | Radial |
| KNP07 | soil | Khao Khitchakut, Chanthaburi | Rosaceous |
| KNP09 | root | Khao Khitchakut, Chanthaburi | Rosaceous |
| KNP10 | root | Khao Khitchakut, Chanthaburi | Chrysanthemum |
| KNP16 | leaf | Khao Khitchakut, Chanthaburi | Rosaceous |
| KNP21 | soil | Khao Khitchakut, Chanthaburi | Radial |
| Group | Name | Colony | Gram Reaction |
|---|---|---|---|
| A | NJTU01 | Round, convex, wavy edges, and milky white | + |
| B | NJTU02 | Round, convex, smooth edges, and dull yellow | − |
| C | NJTU03 | Round, smooth surface, smooth edges, and opaque color | + |
| D | NJTU04 | Round, shiny, small smooth edges, and clear green | + |
| E | NJTU05 | Round, shiny, small smooth edges, and clear tan | + |
| F | NJTU10 | Round, smooth, and large wavy edges | + |
| G | NJTU13 | Round, with serrated edges, rough surface, and dark opaque color | + |
| H | NJTU14 | Cylinder, slightly wavy edge, and cloudy white | + |
| Treatment | Inhibition of Pathogen (%) | ||
|---|---|---|---|
| P. palmivora | Fusarium sp. | Pythium sp. | |
| Control (ddH2O) | 0.00 ± 0.00 d | 0.00 ± 0.00 d | 0.00 ± 0.00 d |
| B. subtilis strain TU-089 | 68.52 ± 2.14 b | 57.22 ± 1.98 c | 22.59 ± 5.64 c |
| B. megaterium strain KP14 | 71.30 ± 3.02 ab | 69.26 ± 5.26 b | 26.67 ± 5.11 c |
| NJTU05 | 77.96 ± 1.39 a | 76.67 ± 0.45 a | 53.70 ± 1.89 b |
| NJTU10 | 63.70 ± 2.50 c | 67.59 ± 0.94 b | 47.22 ± 3.93 b |
| NJTU13 | 76.48 ± 1.83 a | 69.07 ± 4.72 b | 24.44 ± 7.63 c |
| metalaxyl | 75.19 ± 5.37 a | 73.89 ± 2.77 a | 77.22 ± 1.20 a |
| F-test | * | * | * |
| C.V. (%) | 22.27 | 23.46 | 19.69 |
| Characteristics | Reactions 1 | Characteristics | Reactions 1 |
|---|---|---|---|
| Gram reaction | +ve | D-mannitol | − |
| β-xylosidase | − | D-mannose | − |
| L-lysine arylamidase | − | D-melezitose | − |
| L-aspartate arylamidase | − | N-acetyl-D-glucosamine | − |
| Leucine arylamidase | + | Palatinose | − |
| Phenylalanine arylamidase | + | L-rhamnose | − |
| L-proline arylamidase | + | β-glucosidase | − |
| β-galactosidase | + | β-mannosidase | − |
| L-pyrrolidonyl arylamidase | + | Phosphoryl choline | − |
| α-galactosidase | − | Pyruvate | − |
| Alanine arylamidase | − | α-glucosidase | − |
| Tyrosine arylamidase | + | D-tagatose | − |
| β-N-acetly-glucosaminidase | + | D-trehalose | − |
| Ala-Phe-Pro arylamidase | + | Inulin | − |
| Cyclodextrine | − | D-glucose | − |
| D-galactose | − | D-ribose | − |
| Glycogene | − | Putrescine assimilation | − |
| Myo-inositol | − | Growth in 6.5% NaCl | − |
| Methyl-α-D-glucopyranoside acidification | − | Kanamycin resistance | − |
| Ellman | + | Oleandomycin resistance | − |
| Methyl-D-xyloside | − | Esculin hydrolyse | − |
| α-monnosidase | − | Tetrazolium red | − |
| Maltotriose | − | Polymixin_B resistance | − |
| Glycine arylamidase | + | Glycine arylamidase | + |
| 1+ve | =Gram positive bacteria | ID Message Confidence level | %Probability |
| + | =Positive reaction | Excellent | 96 to 99 |
| − | =Negative reaction | Very Good | 93 to 95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Datmanee, P.; Jitfour, N.; Athinuwat, D.; Chuaboon, W. Exploring the Potential of Biocontrol Agent Against Root and Stem Rot Disease in Durian (Durio zibethinus). Int. J. Plant Biol. 2025, 16, 75. https://doi.org/10.3390/ijpb16030075
Datmanee P, Jitfour N, Athinuwat D, Chuaboon W. Exploring the Potential of Biocontrol Agent Against Root and Stem Rot Disease in Durian (Durio zibethinus). International Journal of Plant Biology. 2025; 16(3):75. https://doi.org/10.3390/ijpb16030075
Chicago/Turabian StyleDatmanee, Ponchanok, Nattarika Jitfour, Dusit Athinuwat, and Wilawan Chuaboon. 2025. "Exploring the Potential of Biocontrol Agent Against Root and Stem Rot Disease in Durian (Durio zibethinus)" International Journal of Plant Biology 16, no. 3: 75. https://doi.org/10.3390/ijpb16030075
APA StyleDatmanee, P., Jitfour, N., Athinuwat, D., & Chuaboon, W. (2025). Exploring the Potential of Biocontrol Agent Against Root and Stem Rot Disease in Durian (Durio zibethinus). International Journal of Plant Biology, 16(3), 75. https://doi.org/10.3390/ijpb16030075






