Unraveling Evolutionary Dynamics: Comparative Analysis of Chloroplast Genome of Cleomella serrulata from Leaf Extracts
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and DNA Extraction
2.2. DNA Sequencing, Mapping and Annotation
2.3. Phylogenetic Trees and ANI Calculations
3. Results and Discussion
3.1. Sampling and Identification
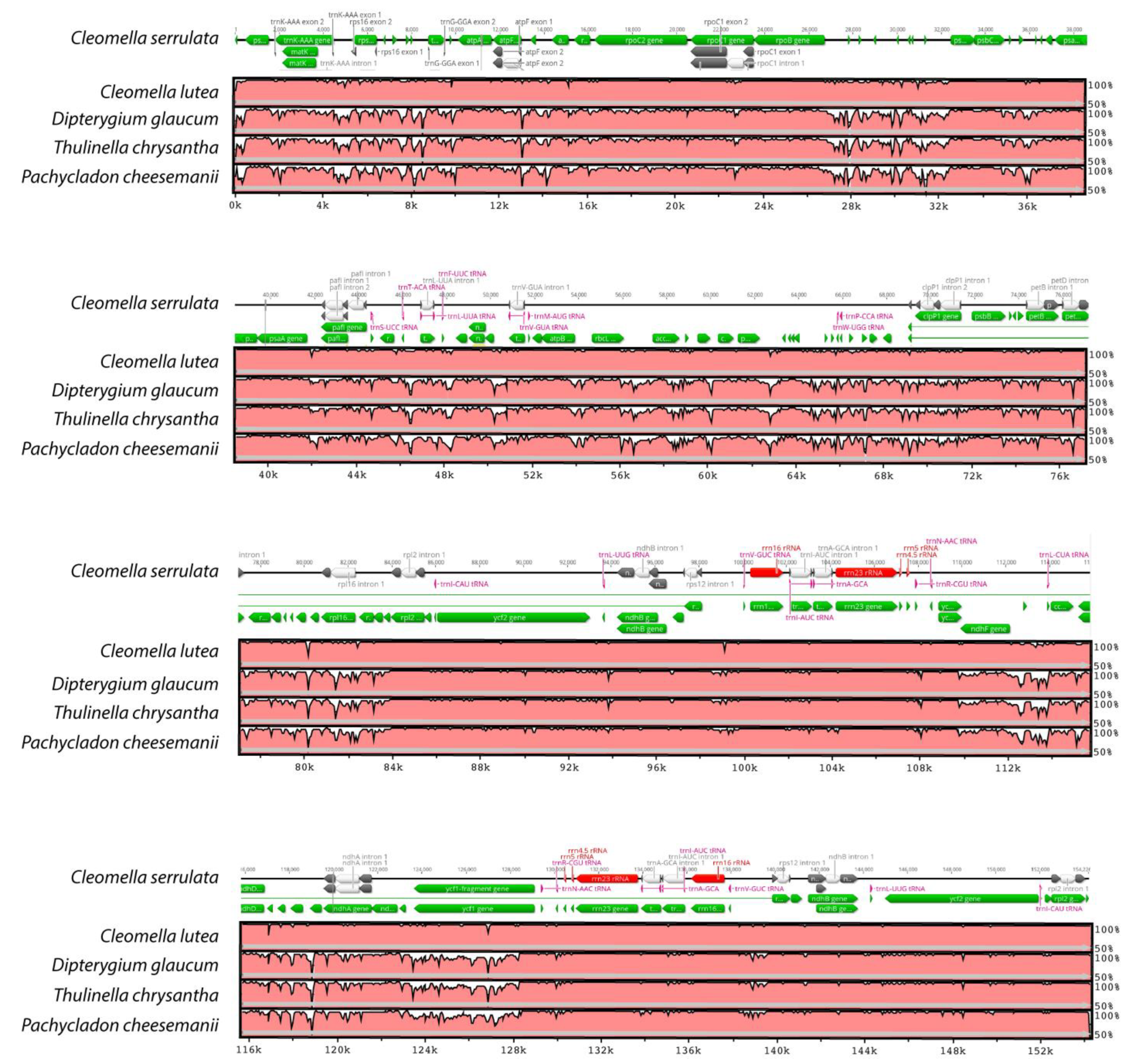
3.2. Genome Sequencing and Chloroplast Structural Analysis
3.3. Comparative Genomics
3.4. Chloroplast Versus Nuclear DNA Evolution
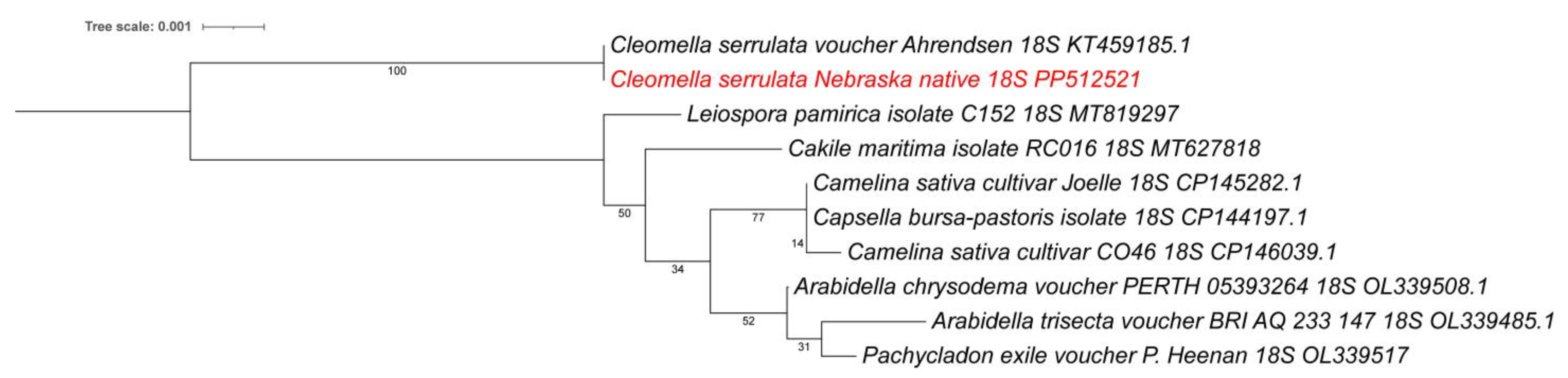
3.5. Phylogenetic Comparisons
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Riser, J.P.; Cardinal-McTeague, W.M.; Hall, J.C.; Hahn, W.J.; Sytsma, K.J.; Roalson, E.H. Phylogenetic relationships among the North American Cleomoids (Cleomaceae): A test of Iltis’s reduction series. Am. J. Bot. 2013, 100, 2102–2111. [Google Scholar] [CrossRef] [PubMed]
- Roalson, E.H. A revised synonymy, typification and key to species of Cleome sensu stricto (Cleomaceae). Phytotaxa 2021, 496, 54–68. [Google Scholar] [CrossRef]
- Barker, M.S.; Vogel, H.; Schranz, M.E. Paleopolyploidy in the Brassicales: Analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biol. Evol. 2009, 1, 391–399. [Google Scholar] [CrossRef]
- Cane, J.H. Breeding biologies, seed production and species-rich bee guilds of Cleome lutea and Cleome serrulata (Cleomaceae). Plant Species Biol. 2008, 23, 152–158. [Google Scholar] [CrossRef]
- Patchell, M.J.; Roalson, E.H.; Hall, J.C. Resolved phylogeny of Cleomaceae based on all three genomes. Taxon 2014, 63, 315–328. [Google Scholar] [CrossRef]
- Adams, K.R.; Stewart, J.D.; Baldwin, S.J. Pottery paint and other uses of Rocky Mountain Beeweed (Cleome serrulata pursh) in the Southwestern United States: Ethnographic Data, Archæological Record, and elemental composition. Kiva 2002, 67, 339–362. [Google Scholar] [CrossRef]
- Roalson, E.H.; Hall, J.C.; Riser, J.P., II; Cardinal-McTeague, W.M.; Cochrane, T.S.; Sytsma, K.J. A revision of generic boundaries and nomenclature in the North American Cleomoid clade (Cleomaceae). Phytotaxa 2015, 205, 129. [Google Scholar] [CrossRef][Green Version]
- Hall, J.C. Systematics of Capparaceae and Cleomaceae: An evaluation of the generic delimitations of capparis and cleome using plastid DNA sequence data this paper is one of a selection of papers published in the special issue on systematics research. Botany 2008, 86, 682–696. [Google Scholar] [CrossRef]
- Nozzolillo, C.; Amiguet, V.T.; Bily, A.C.; Harris, C.S.; Saleem, A.; Andersen, O.M.; Jordheim, M. Novel aspects of the flowers and floral pigmentation of two Cleome species (Cleomaceae), C. hassleriana and C. serrulata. Biochem. Syst. Ecol. 2010, 38, 361–369. [Google Scholar] [CrossRef]
- Patchell, M.J.; Bolton, M.C.; Mankowski, P.; Hall, J.C. Comparative floral development in Cleomaceae reveals two distinct pathways leading to monosymmetry. Int. J. Plant Sci. 2011, 172, 352–365. [Google Scholar] [CrossRef]
- Brown, N.J.; Parsley, K.; Hibberd, J.M. The future of C4 research—Maize, Flaveria or Cleome? Trends Plant Sci. 2005, 10, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Marshall, D.M.; Muhaidat, R.; Brown, N.J.; Liu, Z.; Stanley, S.; Griffiths, H.; Sage, R.F.; Hibberd, J.M. Cleome, a genus closely related to Arabidopsis, contains species spanning a developmental progression from C3 to C4 photosynthesism. Plant J. 2007, 51, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Voznesenskaya, E.V.; Koteyeva, N.K.; Chuong, S.D.X.; Ivanova, A.N.; Barroca, J.; Craven, L.A.; Edwards, G.E. Physiological, anatomical and biochemical characterisation of photosynthetic types in genus Cleome (Cleomaceae). Funct. Plant Biol. 2007, 34, 247–267. [Google Scholar] [CrossRef] [PubMed]
- Feodorova, T.A.; Voznesenskaya, E.V.; Edwards, G.E.; Roalson, E.H. Biogeographic patterns of diversification and the origins of C4 in Cleome (Cleomaceae). Syst. Bot. 2010, 35, 811–826. [Google Scholar] [CrossRef]
- Koteyeva, N.K.; Voznesenskaya, E.V.; Roalson, E.H.; Edwards, G.E. Diversity in forms of C-4 in the genus Cleome (Cleomaceae). Ann. Bot. 2011, 107, 269–283. [Google Scholar] [CrossRef]
- Schranz, M.E.; Mitchell-Olds, T. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell 2006, 18, 1152–1165. [Google Scholar] [CrossRef]
- Cheng, S.; Van den Bergh, E.; Zeng, P.; Zhong, X.; Xu, J.; Liu, X.; Hofberger, J.; Bruijn, S.; Bhide, A.S.; Bian, C.; et al. The Tarenaya hassleriana Genome Provides Insight into Reproductive Trait and Genome Evolution of Crucifers. Plant Cell 2013, 25, 2813–2830. [Google Scholar] [CrossRef]
- Brautigam, A.; Kajala, K.; Wullenweber, J.; Sommer, M.; Gagneul, D.; Weber, K.L.; Carr, K.M.; Gowik, U.; Mass, J.; Lercher, M.J.; et al. An mRNA Blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 2011, 155, 142–156. [Google Scholar] [CrossRef]
- Brautigam, A.; Mullick, T.; Schliesky, S.; Weber, A.P.M. Critical assessment of assembly strategies for non-model species mRNA-Seq data and application of next-generation sequencing to the comparison of C3 and C4 species. J. Exp. Bot. 2011, 62, 3093–3102. [Google Scholar] [CrossRef]
- Alzahrani, D.; Albokhari, E.; Yaradua, S.; Abba, A. Complete chloroplast genome sequences of Dipterygium glaucum and Cleome chrysantha and other Cleomaceae Species, comparative analysis and phylogenetic relationships. Saudi J. Biol. Sci. 2021, 28, 2476–2490. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2010; Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 31 May 2024).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comp. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Kim, J.I.; Jeong, Y.-S.; Yi, G. AGORA: Organellar genome annotation from the amino acid and nucleotide references. Bioinformatics 2018, 34, 2661–2663. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res 2019, 47, 256–259. [Google Scholar] [CrossRef]
- Mayor, C.; Brudno, M.; Schwartz, J.R.; Poliakov, A.; Rubin, E.M.; Frazer, K.A.; Pachter, L.S.; Dubchak, I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 2000, 16, 1046–1047. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Pax, F.; Hoffmann, K. Capparidaceae. In Die Natürlichen Pflanzenfamilien, 2nd ed.; Engler, A., Prantl, K., Eds.; Engelmann: Leipzig, Germany, 1936; Volume 17, pp. 146–233. [Google Scholar]
- Airy Shaw, H.K. Diagnoses of new families, new names, etc., for the seventh edition of Willis’s ‘Dictionary’. Kew Bull. 1965, 18, 249–273. [Google Scholar] [CrossRef]
- Hutchinson, J. The Genera of Flowering Plants; Clarendon Press: Oxford, UK, 1967. [Google Scholar]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasquez, M.K.; Stock, E.K.; Terrell, K.J.; Ramirez, J.; Kyndt, J.A. Unraveling Evolutionary Dynamics: Comparative Analysis of Chloroplast Genome of Cleomella serrulata from Leaf Extracts. Int. J. Plant Biol. 2024, 15, 914-926. https://doi.org/10.3390/ijpb15030065
Vasquez MK, Stock EK, Terrell KJ, Ramirez J, Kyndt JA. Unraveling Evolutionary Dynamics: Comparative Analysis of Chloroplast Genome of Cleomella serrulata from Leaf Extracts. International Journal of Plant Biology. 2024; 15(3):914-926. https://doi.org/10.3390/ijpb15030065
Chicago/Turabian StyleVasquez, Madelynn K., Emma K. Stock, Kaziah J. Terrell, Julian Ramirez, and John A. Kyndt. 2024. "Unraveling Evolutionary Dynamics: Comparative Analysis of Chloroplast Genome of Cleomella serrulata from Leaf Extracts" International Journal of Plant Biology 15, no. 3: 914-926. https://doi.org/10.3390/ijpb15030065
APA StyleVasquez, M. K., Stock, E. K., Terrell, K. J., Ramirez, J., & Kyndt, J. A. (2024). Unraveling Evolutionary Dynamics: Comparative Analysis of Chloroplast Genome of Cleomella serrulata from Leaf Extracts. International Journal of Plant Biology, 15(3), 914-926. https://doi.org/10.3390/ijpb15030065






