Genome-Wide Association Study Reveals Marker–Trait Associations with Resistance to Pythium irregulare from Soybean Germplasm
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Inoculum Preparation
2.3. Resistance Evaluation and Assessment
2.4. Statistical Analysis
2.5. Genome-Wide Association Study
3. Results
3.1. Phenotypic Evaluation of Soybean Germplasm for P. irregulare Resistance
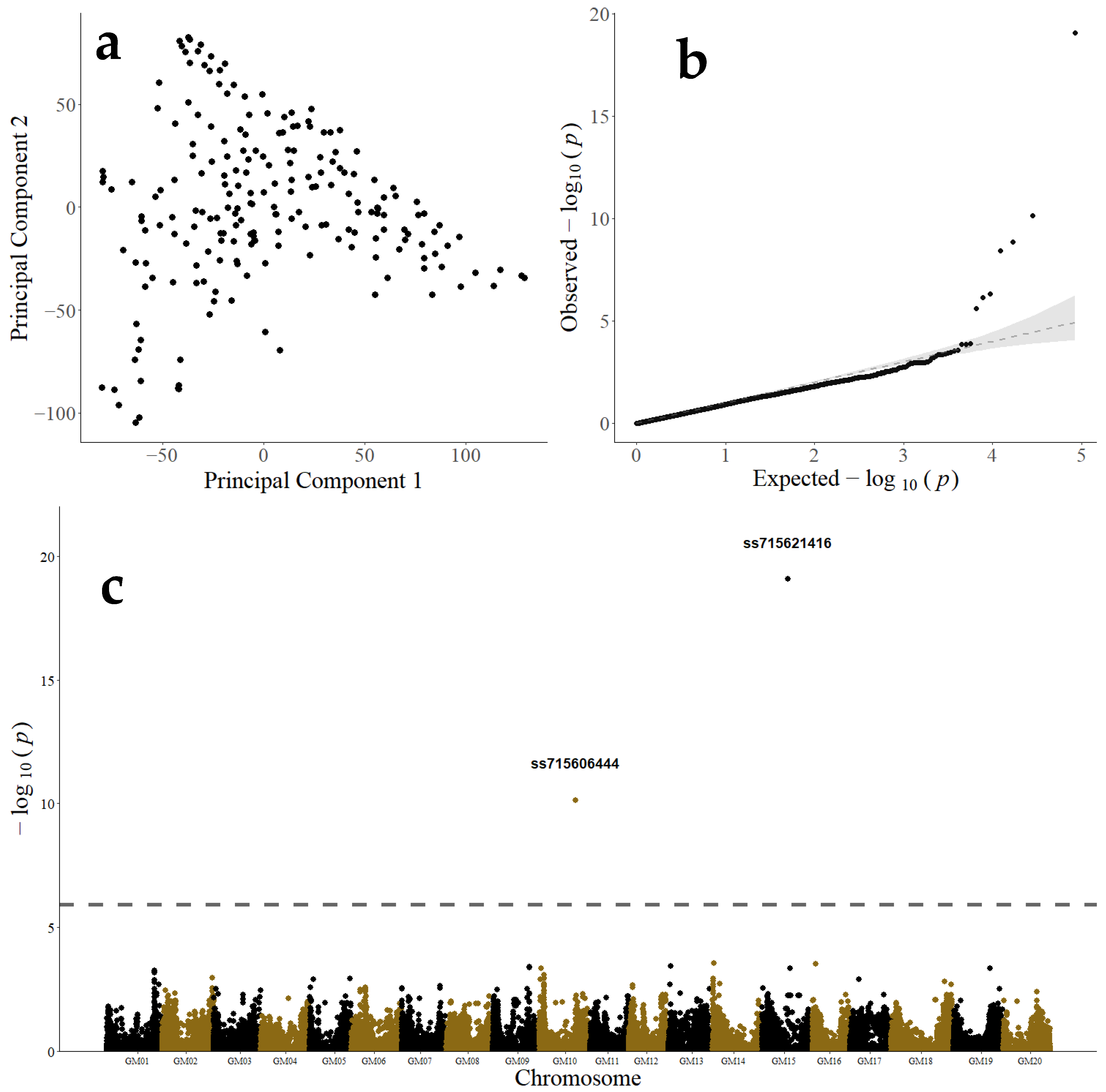
3.2. Genome-Wide Association Study and Candidate Gene Analysis of Pythium irregulare Resistance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allen, T.W.; Bradley, C.A.; Sisson, A.J.; Byamukama, E.; Chilvers, M.I.; Coker, C.M.; Collins, A.A.; Damicone, J.P.; Dorrance, A.E.; Dufault, N.S.; et al. Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2010 to 2014. Plant Health Prog. 2017, 18, 19–27. [Google Scholar] [CrossRef]
- Bradley, C.A.; Allen, T.W.; Sisson, A.J.; Bergstrom, G.C.; Bissonnette, K.M.; Bond, J.; Byamukama, E.; Chilvers, M.I.; Collins, A.A.; Damicone, J.P.; et al. Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2015 to 2019. Plant Health Prog. 2021, 22, 483–495. [Google Scholar] [CrossRef]
- Wrather, J.A.; Koenning, S.R. Estimates of disease effects on soybean yields in the United States 2003 to 2005. J. Nematol. 2006, 38, 173–180. [Google Scholar] [PubMed]
- Wrather, J.A.; Stienstra, W.C.; Koenning, S.R. Soybean disease loss estimates for the United States from 1996 to 1998. Can. J. Plant Pathol. 2001, 23, 122–131. [Google Scholar] [CrossRef]
- Winsor, S. Keep your eyes open for these wet-season soybean diseases. Crops Soils Mag. 2020, 53, 16–23. [Google Scholar] [CrossRef]
- Schaub, S.; Huber, R.; Finger, R. Tracking societal concerns on pesticides—A Google Trends analysis. Environ. Res. Lett. 2020, 15, 84049. [Google Scholar] [CrossRef]
- Broders, K.D.; Lipps, P.E.; Paul, P.A.; Dorrance, A.E. Evaluation of Fusarium graminearum associated with corn and soybean seed and seedling disease in Ohio. Plant Dis. 2007, 91, 1155–1160. [Google Scholar] [CrossRef] [PubMed]
- Matic, S.; Gilardi, G.; Gisi, U.; Gullino, M.L.; Garibaldi, A. Differentiation of Pythium spp. from vegetable crops with molecular markers and sensitivity to azoxystrobin and mefenoxam. Pest. Manag. Sci. 2019, 75, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Liu, X.; Du, X.; Li, G.; Li, C.; Zhao, D.; Liu, X. Sensitivity of Pythium spp. and Phytopythium spp. and tolerance mechanism of Pythium spp. to oxathiapiprolin. Pest Manag. Sci. 2020, 76, 3975–3981. [Google Scholar] [CrossRef]
- Roth, M.G.; Webster, R.W.; Mueller, D.S.; Chilvers, M.I.; Faske, T.R.; Mathew, F.M.; Bradley, C.A.; Damicone, J.P.; Kabbage, M.; Smith, D.L. Integrated management of important soybean pathogens of the United States in changing climate. J. Integr. Pest Manag. 2020, 11, 17. [Google Scholar] [CrossRef]
- Molin, C.; Ribeiro, N.R.; Matsumoto, M.N.; Fernando Giasson, N.; Brollo, J.; Zanardo, B.; Pelissoni, M.; Capitanio, S.; Comín, T.; Deuner, C.C.; et al. Damping-off of soybean in southern Brazil can be associated with different species of Globisporangium spp. and Pythium spp. Plant Pathol. 2021, 70, 1686–1694. [Google Scholar] [CrossRef]
- Rojas, J.A.; Jacobs, J.L.; Napieralski, S.; Karaj, B.; Bradley, C.A.; Chase, T.; Esker, P.D.; Giesler, L.J.; Jardine, D.J.; Malvick, D.K.; et al. Oomycete species associated with soybean seedlings in North America—Part I: Identification and pathogenicity characterization. Phytopathology 2017, 107, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Broders, K.D.; Lipps, P.E.; Paul, P.A.; Dorrance, A.E. Characterization of Pythium spp. associated with corn and soybean seed and seedling disease in Ohio. Plant Dis. 2007, 91, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Reeves, E.R.; Kerns, J.P.; Cowger, C.; Shew, B.B. Pythium spp. associated with root rot and stunting of winter wheat in North Carolina. Plant Dis. 2021, 105, 986–996. [Google Scholar] [CrossRef] [PubMed]
- Rayside, P.A.; Mitchell, D.J.; Gallaher, R.N. Populations of Pythium spp. and associated microorganisms in a minimum tillage-multicropping system. Phytopathology 1982, 72, 996. [Google Scholar]
- Grijalba, P.E.; Ridao, A.d.C.; Steciow, M.M. Oomycetes species associated with soybean in Buenos Aires Province (Argentina). Phytoparasitica 2021, 49, 1027–1042. [Google Scholar] [CrossRef]
- Rizvi, S.S.A.; Yang, X.B. Fungi associated with soybean seedling disease in Iowa. Plant Dis. 1996, 80, 57–60. [Google Scholar] [CrossRef]
- Rojas, J.A.; Jacobs, J.L.; Napieralski, S.; Karaj, B.; Bradley, C.A.; Chase, T.; Esker, P.D.; Giesler, L.J.; Jardine, D.J.; Malvick, D.K.; et al. Oomycete species associated with soybean seedlings in North America-part II: Diversity and ecology in relation to environmental and edaphic factors. Phytopathology 2017, 107, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Bandara, A.Y.; Weerasooriya, D.K.; Bradley, C.A.; Allen, T.W.; Esker, P.D. Dissecting the economic impact of soybean diseases in the United States over two decades. PLoS ONE 2020, 15, e0231141. [Google Scholar] [CrossRef]
- Wrather, J.A.; Anderson, T.R.; Arsyad, D.M.; Gai, J.; Ploper, L.D.; Porta-Puglia, A.; Ram, H.H.; Yorinori, J.T. Soybean disease loss estimates for the top 10 soybean producing countries in 1994. Plant Dis. 1997, 81, 107–110. [Google Scholar] [CrossRef]
- Wrather, J.A.; Anderson, T.R.; Arsyad, D.M.; Tan, Y.; Ploper, L.D.; Porta-Puglia, A.; Ram, H.H.; Yorinori, J.T. Soybean disease loss estimates for the top ten soybean-producing countries in 1998. Can. J. Plant Pathol. 2001, 23, 115–121. [Google Scholar] [CrossRef]
- Broders, K.D.; Wallhead, M.W.; Austin, G.D.; Lipps, P.E.; Paul, P.A.; Mullen, R.W.; Dorrance, A.E. Association of soil chemical and physical properties with Pythium species diversity, community composition, and disease incidence. Phytopathology 2009, 2009, 957–967. [Google Scholar] [CrossRef]
- Kirkpatrick, M.T.; Rupe, J.C.; Rothrock, C.S. Soybean response to flooded soil conditions and the association with soilborne plant pathogenic genera. Plant Dis. 2006, 90, 592–596. [Google Scholar] [CrossRef]
- Lévesque, C.A.; De Cock, A.W.A.M. Molecular phylogeny and taxonomy of the genus Pythium. Mycol. Res. 2004, 108, 1363–1383. [Google Scholar] [CrossRef] [PubMed]
- Huzar-Novakowiski, J.; Dorrance, A.E. Genetic diversity and population structure of Pythium irregulare from soybean and corn production fields in Ohio. Plant Dis. 2018, 102, 1989–2000. [Google Scholar] [CrossRef]
- Bates, G.D.; Rothrock, C.S.; Rupe, J.C. Resistance of the soybean cultivar Archer to Pythium damping-off and root rot caused by several Pythium spp. Plant Dis. 2008, 92, 763–766. [Google Scholar] [CrossRef] [PubMed]
- Rod, K.S.; Walker, D.R.; Bradley, C.A. Evaluation of major ancestors of North American soybean cultivars for resistance to three Pythium species that cause seedling blight. Plant Dis. 2018, 102, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Paul, C.; Walker, D.R. Aggressiveness of isolates of five Pythium species on seeds and seedlings of six North American soybean cultivars. Can. J. Plant Pathol. 2022, 44, 596–614. [Google Scholar] [CrossRef]
- Molin, C.; Ribeiro, N.R.; Matsumoto, M.N.; Brollo, J.; Bordignon, K.; dos Santos de Britto, J.; Balbinotti, M.; Barbieri, M.; Deuner, C.C.; Huzar-Novakowiski, J. Soybean cultivars adapted to southern Brazil differ in their reaction to Globisporangium irregulare, G. ultimum var. sporangiiferum and Pythium conidiophorum. Plant Pathol. 2022, 71, 1932–1943. [Google Scholar] [CrossRef]
- Ellis, M.L.; McHale, L.K.; Paul, P.A.; Martin, S.K.S.; Dorrance, A.E. Soybean germplasm resistant to Pythium irregulare and molecular mapping of resistance quantitative trait loci derived from the Soybean accession PI 424354. Crop Sci. 2013, 53, 1008–1021. [Google Scholar] [CrossRef]
- Clevinger, E.M.; Biyashev, R.; Lerch-Olson, E.; Yu, H.; Quigley, C.; Song, Q.; Dorrance, A.E.; Robertson, A.E.; Saghai Maroof, M.A. Identification of quantitative disease resistance loci toward four Pythium species in soybean. Front. Plant Sci. 2021, 12, 644746. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Li, W.; McCoy, A.G.; Wang, K.; Jacobs, J.; Zhang, N.; Huo, X.; Wani, S.H.; Gu, C.; Chilvers, M.I.; et al. Identification and characterization of pleiotropic and epistatic QDRL conferring partial resistance to Pythium irregulare and P. sylvaticum in soybean. Theor. Appl. Genet. 2022, 135, 3571–3582. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Wani, S.H.; Collins, P.J.; Wen, Z.; Gu, C.; Chilvers, M.I.; Wang, D. Mapping quantitative trait loci for tolerance to Pythium irregulare in Soybean (Glycine max L.). G3 Genes Genomes Genet. 2018, 8, 3155–3161. [Google Scholar] [CrossRef] [PubMed]
- Scott, K.; Balk, C.; Veney, D.; McHale, L.K.; Dorrance, A.E. Quantitative disease resistance loci towards Phytophthora sojae and three species of Pythium in six soybean nested association mapping populations. Crop Sci. 2019, 59, 605–623. [Google Scholar] [CrossRef]
- Stasko, A.K.; Wickramasinghe, D.; Nauth, B.J.; Acharya, B.; Ellis, M.L.; Taylor, C.G.; McHale, L.K.; Dorrance, A.E. High-density mapping of resistance QTL toward Phytophthora sojae, Pythium irregulare, and Fusarium graminearum in the same soybean population. Crop Sci. 2016, 56, 2476–2492. [Google Scholar] [CrossRef]
- Robideau, G.P.; De Cock, A.W.A.M.; Coffey, M.D.; Voglmayr, H.; Brouwer, H.; Bala, K.; Chitty, D.W.; DÉSaulniers, N.; Eggertson, Q.A.; Gachon, C.M.M.; et al. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol. Ecol. Resour. 2011, 11, 1002–1011. [Google Scholar] [CrossRef]
- Detranaltes, C.; Ma, J.; Cai, G. Identification of soybean germplasm and associated molecular markers with resistance to Fusarium graminearum. Agronomy 2023, 13, 2376. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Kuznetsova, A.; Brockhoff, P.B.; Christensen, R.H.B. lmerTest package: Tests in linear mixed effects models. J. Stat. Softw. 2017, 82, 26. [Google Scholar] [CrossRef]
- Song, Q.; Hyten, D.L.; Jia, G.; Quigley, C.V.; Fickus, E.W.; Nelson, R.L.; Cregan, P.B. Fingerprinting soybean germplasm and its utility in genomic research. G3 Genes Genomes Genet. 2015, 5, 1999–2006. [Google Scholar] [CrossRef]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 2019, 8, giy154. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Molina, A.; Miedes, E.; Bacete, L.; Rodríguez, T.; Mélida, H.; Denancé, N.; Sánchez-Vallet, A.; Rivière, M.-P.; López, G.; Freydier, A.; et al. Arabidopsis cell wall composition determines disease resistance specificity and fitness. Proc. Natl. Acad. Sci. USA 2021, 118, e2010243118. [Google Scholar] [CrossRef] [PubMed]
- Chiniquy, D.; Underwood, W.; Corwin, J.; Ryan, A.; Szemenyei, H.; Lim, C.C.; Stonebloom, S.H.; Birdseye, D.S.; Vogel, J.; Kliebenstein, D.; et al. PMR5, an acetylation protein at the intersection of pectin biosynthesis and defense against fungal pathogens. Plant J. 2019, 100, 1022–1035. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB transcription factors as regulators of secondary metabolism in plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wong, S.-M. A conserved carboxylesterase inhibits tobacco mosaic virus (TMV) accumulation in Nicotiana benthamiana plants. Viruses 2020, 12, 195. [Google Scholar] [CrossRef] [PubMed]
- Ko, M.; Cho, J.H.; Seo, H.-H.; Lee, H.-H.; Kang, H.-Y.; Nguyen, T.S.; Soh, H.C.; Kim, Y.S.; Kim, J.-I. Constitutive expression of a fungus-inducible carboxylesterase improves disease resistance in transgenic pepper plants. Planta 2016, 244, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T. Function and application of a non-ester-hydrolyzing carboxylesterase discovered in tulip. Biosci. Biotechnol. Biochem. 2017, 81, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.-H.; Park, A.R.; Lee, H.-H.; Park, S.; Han, Y.-J.; Hoang, Q.T.N.; Choi, G.J.; Kim, J.-C.; Kim, Y.S.; Kim, J.-I. A fungus-inducible pepper carboxylesterase exhibits antifungal activity by decomposing the outer layer of fungal cell walls. Mol. Plant-Microbe Interact. 2018, 31, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Duan, W.; Wei, C.; Chen, K.; Grierson, D.; Zhang, B. Genome-wide identification and functional analysis of carboxylesterase and methylesterase gene families in peach (Prunus persica L. Batsch). Front. Plant Sci. 2019, 10, 1511. [Google Scholar] [CrossRef]
- Anderson, J.; Akond, M.; Kassem, M.A.; Meksem, K.; Kantartzi, S.K. Quantitative trait loci underlying resistance to sudden death syndrome (SDS) in MD96-5722 by ‘Spencer’ recombinant inbred line population of soybean. 3 Biotech 2015, 5, 203–210. [Google Scholar] [CrossRef][Green Version]
- Lin, F.; Chhapekar, S.S.; Vieira, C.C.; Da Silva, M.P.; Rojas, A.; Lee, D.; Liu, N.; Pardo, E.M.; Lee, Y.-C.; Dong, Z.; et al. Breeding for disease resistance in soybean: A global perspective. Theor. Appl. Genet. 2022, 135, 3773–3872. [Google Scholar] [CrossRef] [PubMed]
- Luckew, A.S.; Swaminathan, S.; Leandro, L.F.; Orf, J.H.; Cianzio, S.R. ‘MN1606SP’ by ‘Spencer’ filial soybean population reveals novel quantitative trait loci and interactions among loci conditioning SDS resistance. Theor. Appl. Genet. 2017, 130, 2139–2149. [Google Scholar] [CrossRef] [PubMed]
- Rolling, W.; Lake, R.; Dorrance, A.E.; McHale, L.K. Genome-wide association analyses of quantitative disease resistance in diverse sets of soybean [Glycine max (L.) Merr.] plant introductions. PLoS ONE 2020, 15, e0227710. [Google Scholar] [CrossRef]
- Van, K.; Rolling, W.; Biyashev, R.M.; Matthiesen, R.L.; Abeysekara, N.S.; Robertson, A.E.; Veney, D.J.; Dorrance, A.E.; McHale, L.K.; Saghai Maroof, M.A. Mining germplasm panels and phenotypic datasets to identify loci for resistance to Phytophthora sojae in soybean. Plant Genome 2021, 14, e20063. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhou, B.; Zhao, J.; Guo, N.; Zhang, B.; Yang, F.; Chen, S.; Gai, J.; Xing, H. Identification of quantitative trait loci for partial resistance to Phytophthora sojae in soybean. Plant Breed. 2011, 130, 144–149. [Google Scholar] [CrossRef]
- Wu, X.-l.; Zhou, B.; Sun, S.; Zhao, J.-M.; Chen, S.-y.; Gai, J.-y.; Xing, H. Genetic analysis and mapping of resistance to Phytophthora sojae of Pm14 in soybean. Sci. Agric. Sin. 2011, 44, 456–460. [Google Scholar]
- Lee, S.; Rouf Mian, M.A.; McHale, L.K.; Sneller, C.H.; Dorrance, A.E. Identification of quantitative trait loci conditioning partial resistance to Phytophthora sojae in soybean PI 407861A. Crop Sci. 2013, 53, 1022–1031. [Google Scholar] [CrossRef]
- Ludke, W.H.; Schuster, I.; Silva, F.L.d.; Montecelli, T.D.N.; Soares, B.d.A.; Oliveira, A.B.d.; Volpato, L. SNP markers associated with soybean partial resistance to Phytophthora sojae. Crop Breed. Appl. Biotechnol. 2019, 19, 31–39. [Google Scholar] [CrossRef]
- Sun, S.; Kim, M.Y.; Van, K.; Lee, Y.-W.; Li, B.; Lee, S.-H. QTLs for resistance to Phomopsis seed decay are associated with days to maturity in soybean (Glycine max). Theor. Appl. Genet. 2013, 126, 2029–2038. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhao, X.; Qu, Y.; Teng, W.; Qiu, L.; Zheng, H.; Wang, Z.; Han, Y.; Li, W. Loci and candidate genes in soybean that confer resistance to Fusarium graminearum. Theor. Appl. Genet. 2019, 132, 431–441. [Google Scholar] [CrossRef]
- Ellis, M.L.; Wang, H.; Paul, P.A.; Martin, S.K.S.; McHale, L.K. Identification of soybean genotypes resistant to Fusarium graminearum and genetic mapping of resistance quantitative trait loci in the cultivar conrad. Crop Sci. 2012, 52, 2224–2233. [Google Scholar] [CrossRef]
- Silva, M.P.; Klepadlo, M.; Gbur, E.E.; Pereira, A.; Mason, R.E.; Rupe, J.C.; Bluhm, B.H.; Wood, L.; Mozzoni, L.A.; Chen, P. QTL mapping of charcoal rot resistance in PI 567562A soybean accession. Crop Sci. 2019, 59, 474–479. [Google Scholar] [CrossRef]
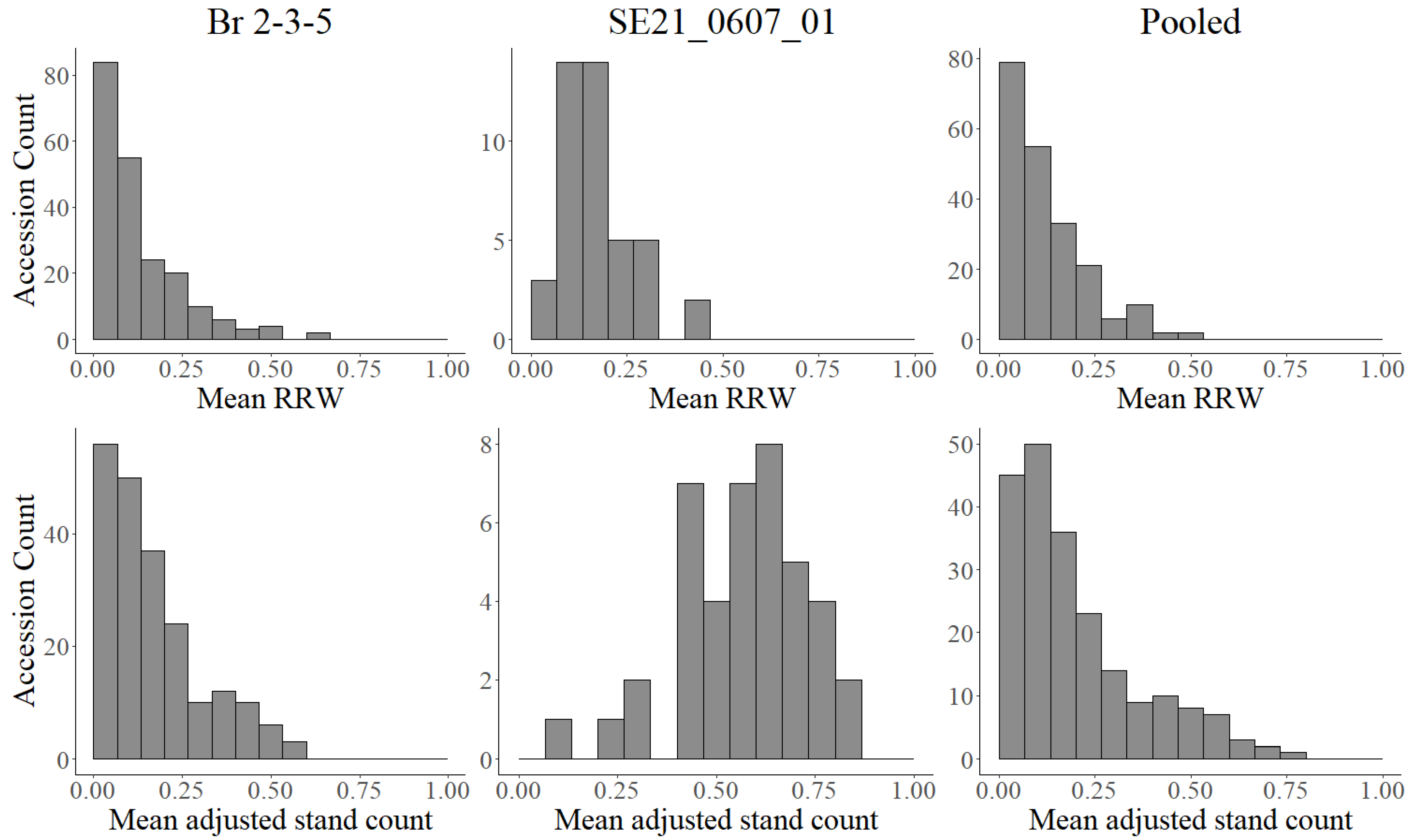
| Isolate Br 2-3-5 | Isolate SE21_0607_01 | Pooled | ||||||
|---|---|---|---|---|---|---|---|---|
| Estimator | Effect | p-Value a | Estimator | Effect | p-Value a | Estimator | Effect | p-Value a |
| Intercept | 0.126 | 3.16 × 10−1 | Intercept | 0.176 | 5.74 × 10−3 | Intercept | 0.114 | 2.89 × 10−1 |
| PI 490766 | 0.521 | 4.54 × 10−4 * | PI 548520 | 0.256 | 1.08 × 10−3 * | PI 548520 | 0.358 | 4.02 × 10−3 * |
| PI 547459 | 0.488 | 1.00 × 10−3 * | PI 578503 | 0.253 | 1.28 × 10−3 * | FC 29333 | 0.373 | 9.14 × 10−3 * |
| PI 591511 | 0.401 | 6.82 × 10−3 * | PI 84987 A | 0.139 | 6.96 × 10−2 | PI 84946-2 | 0.321 | 2.49 × 10−2 * |
| PI 547460 | 0.390 | 8.51 × 10−3 * | PI 632418 | 0.135 | 7.75 × 10−2 | PI 490766 | 0.273 | 2.81 × 10−2 * |
| PI 548520 | 0.385 | 9.33 × 10−3 * | PI 548521 | 0.131 | 8.80 × 10−2 | PI 547459 | 0.271 | 2.94 × 10−2 * |
| FC 29333 | 0.381 | 1.01 × 10−2 * | PI 603549 | 0.108 | 1.58 × 10−1 | PI 548362 | 0.310 | 3.00 × 10−2 * |
| PI 548360 | 0.331 | 2.52 × 10−2 * | PI 567258 | 0.104 | 1.74 × 10−1 | PI 548360 | 0.245 | 4.82 × 10−2 * |
| PI 84946-2 | 0.329 | 2.63 × 10−2 * | PI 438239 B | 0.088 | 2.46 × 10−1 | PI 591511 | 0.242 | 5.14 × 10−2 |
| PI 548362 | 0.318 | 3.15 × 10−2 * | PI 88468 | 0.085 | 2.63 × 10−1 | PI 547460 | 0.237 | 5.64 × 10−2 |
| PI 639559 B | 0.268 | 6.97 × 10−2 | PI 548360 | 0.085 | 2.66 × 10−1 | PI 639559 B | 0.260 | 6.86 × 10−2 |
| Isolate | Predictor | SS | MS | NDF | DDF | F-Value | p-Value a |
|---|---|---|---|---|---|---|---|
| Br 2-3-5 | Accession | 10.089 | 0.04874 | 207 | 414 | 1.4958 | 3.120 × 10−4 * |
| SE21_0607_01 | Accession | 1.083 | 0.0258 | 42 | 84 | 2.9961 | 9.530 × 10−6 * |
| Pooled | Accession | 9.057 | 0.0438 | 207 | 539 | 1.4333 | 6.728 × 10−4 * |
| Marker Name | Chromosome | Position (bp) a | Major/Minor Allele | MAF b | p-Value c | RRW Effect d |
|---|---|---|---|---|---|---|
| ss715621416 | 15 | 26,089,343 | C/A | 0.087 | 3.56 × 10−15 | 0.172 |
| ss715606444 | 10 | 36,778,506 | C/T | 0.082 | 1.53 × 10−6 | 0.093 |
| SNP Marker | Major/Minor Allele | Effect | PI 548520 | FC 29333 | PI 84946-2 | PI 490766 | PI 547459 | PI 548362 | PI 548360 |
|---|---|---|---|---|---|---|---|---|---|
| ss715621416 | C/A | 0.172 | C | A | C | A | A | C | C |
| ss715606444 | C/T | 0.093 | T | C | C | T | T | T | C |
| Sum of allelic effects: | 0.265 | 0.093 | 0.172 | 0 | 0.265 | 0.265 | 0.093 | 0 | |
| Pooled mean RRW: | 0.472 ± 0.12 | 0.507 ± 0.35 | 0.455 ± 0.37 | 0.386 ± 0.16 | 0.384 ± 0.16 | 0.444 ± 0.30 | 0.359 ± 0.06 | ||
| Pooled mean adjusted stand count: | 0.794 ± 0.15 | 0.467 ± 0.24 | 0.222 ± 0.18 | 0.597 ± 0.10 | 0.594 ± 0.17 | 0.444 ± 0.24 | 0.581 ± 0.09 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Detranaltes, C.; Ma, J.; Cai, G. Genome-Wide Association Study Reveals Marker–Trait Associations with Resistance to Pythium irregulare from Soybean Germplasm. Int. J. Plant Biol. 2024, 15, 769-782. https://doi.org/10.3390/ijpb15030056
Detranaltes C, Ma J, Cai G. Genome-Wide Association Study Reveals Marker–Trait Associations with Resistance to Pythium irregulare from Soybean Germplasm. International Journal of Plant Biology. 2024; 15(3):769-782. https://doi.org/10.3390/ijpb15030056
Chicago/Turabian StyleDetranaltes, Christopher, Jianxin Ma, and Guohong Cai. 2024. "Genome-Wide Association Study Reveals Marker–Trait Associations with Resistance to Pythium irregulare from Soybean Germplasm" International Journal of Plant Biology 15, no. 3: 769-782. https://doi.org/10.3390/ijpb15030056
APA StyleDetranaltes, C., Ma, J., & Cai, G. (2024). Genome-Wide Association Study Reveals Marker–Trait Associations with Resistance to Pythium irregulare from Soybean Germplasm. International Journal of Plant Biology, 15(3), 769-782. https://doi.org/10.3390/ijpb15030056






