Isolation of Lactic Acid Bacteria from Raw Camel Milk in Saudi Arabia and Evaluation of Their Probiotic Potential
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Isolation of LAB
2.1.1. Raw Camel Milk Sampling
2.1.2. Isolation
2.2. Screening of Selected Bacterial Isolates for Probiotic Attributes
2.2.1. Antagonistic Activity
2.2.2. Resistance to Lysozyme
2.2.3. Acid pH and Bile Salt Tolerance
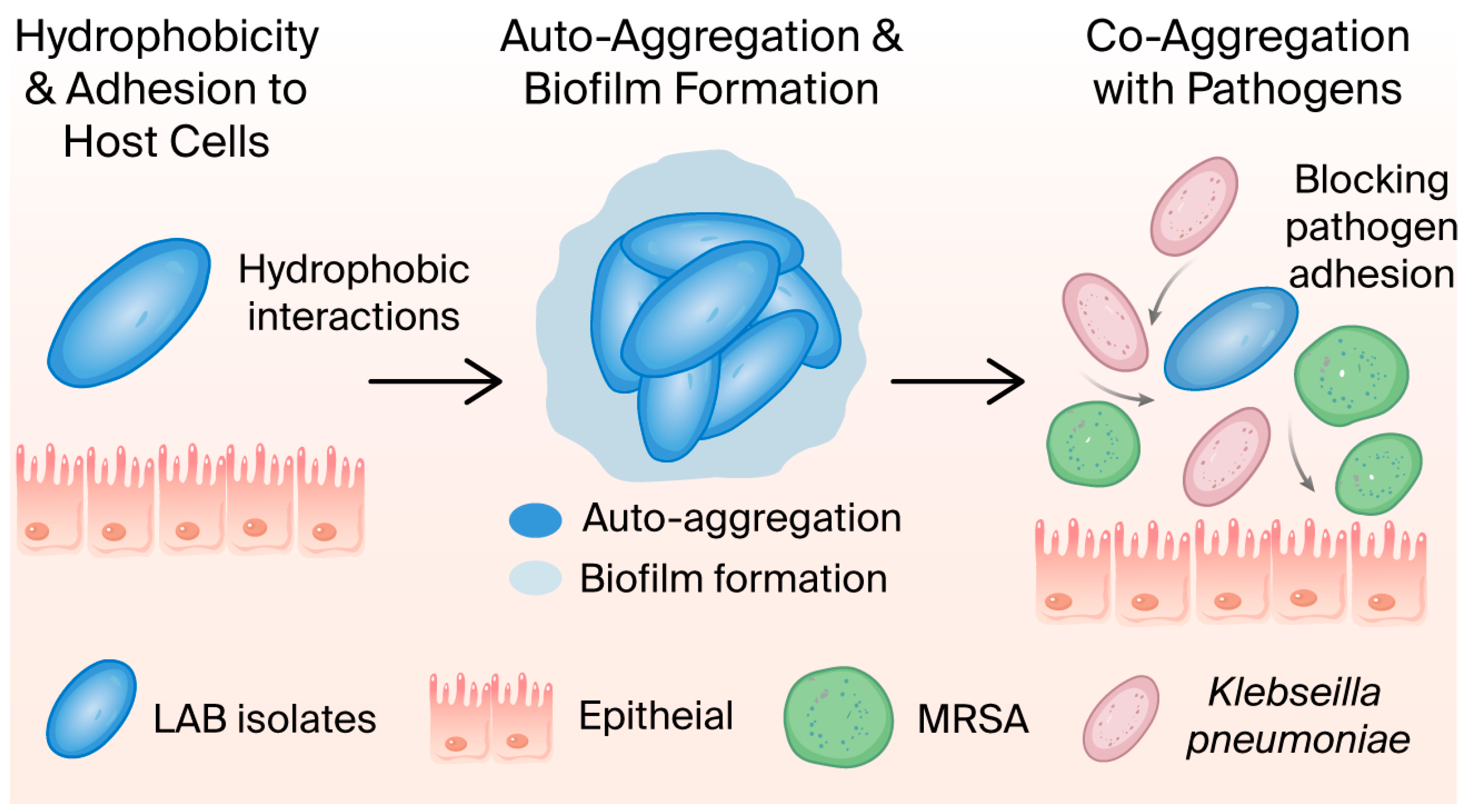
2.2.4. Cell Surface Hydrophobicity
2.2.5. LAB Isolates’ Aggregate Abilities
2.3. Characterization and Identification of Bacterial Isolates
2.3.1. Phenotypic Identification
2.3.2. Genotypic Identification
DNA Extraction and PCR
Phylogenetic Analysis
2.4. LAB Safety Assessment
2.4.1. Antibiotic Susceptibility
2.4.2. Hemolytic Activity Test
2.4.3. Gelatinase Activity
2.5. Statistical Analysis
3. Results
3.1. Isolation
3.2. Probiotic Qualities of LAB
3.2.1. Analyzing LAB Isolates for Antagonistic Activity and Lysozyme Tolerance
3.2.2. The Ability of Bacterial Isolates to Withstand Acidic Environments and Bile Salts
3.2.3. Cell Surface Hydrophobicity
3.2.4. LAB Isolates’ Aggregate Abilities
3.3. Phenotypic and Genotypic Identification of LAB Isolates
3.4. Safety Assessment
3.4.1. Hemolytic and Gelatinase Activity
3.4.2. Antibiotic Susceptibility
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LAB | Lactic acid bacteria |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| MRS | de Man–Rogosa–Sharpe |
| CFU | Colony-forming unit |
| OD | Optical density |
| ML | Maximum likelihood |
| AIC | Akaike Information Criterion |
| MP | Maximum parsimony |
| NCCLS | National Committee for Clinical Laboratory Standards |
| RBC | Red blood cell |
References
- Swelum, A.A.; El-Saadony, M.T.; Abdo, M.; Ombarak, R.A.; Hussein, E.O.S.; Suliman, G.; Alhimaidi, A.R.; Ammari, A.A.; Ba-Awadh, H.; Taha, A.E.; et al. Nutritional, antimicrobial and medicinal properties of Camel’s milk: A review. Saudi J. Biol. Sci. 2021, 28, 3126–3136. [Google Scholar] [CrossRef]
- BAHS. Basic Animal Husbandry Statistics. 2012. Available online: https://dahd.nic.in/related-links/publications-0 (accessed on 8 May 2025).
- Faye, B. The Camel today: Assets and potentials. Anthropozoologica 2014, 49, 15–24. [Google Scholar] [CrossRef]
- Alhassani, W.E. Camel milk: Nutritional composition, therapeutic properties, and benefits for human health. Open Vet. J. 2024, 14, 3164–3180. [Google Scholar] [CrossRef]
- Alebie, G.; Yohannes, S.; Worku, A. Therapeutic applications of camel’s milk and urine against cancer: Current development efforts and future perspectives. J. Cancer Sci. Ther. 2017, 9, 468–478. [Google Scholar] [CrossRef]
- Food and Agricultural Organization of the United Nations and World Health Organization. Joint FAO/WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food; Food and Agricultural Organization of the United Nations, Rome, Italy, 2002. Available online: https://isappscience.org/wp-content/uploads/2019/04/probiotic_guidelines.pdf (accessed on 27 March 2025).
- Albarrak, S.M. Evaluation of somatic cells count, antioxidants and antimicrobial proteins in milk samples obtained from different breeds of the dromedary camel. Int. J. Vet. Sci. 2023, 12, 462–469. [Google Scholar] [CrossRef]
- Mahmoudi, I.; Moussa, O.B.; Khaldi, T.E.M.; Kebouchi, M.; Soligot, C.; Le Roux, Y.; Hassouna, M. Functional in vitro screening of Lactobacillus strains isolated strains from Tunisian camel raw milk toward their selection as probiotic. Small Rumin. Res. 2016, 137, 91–98. [Google Scholar] [CrossRef]
- Sharma, A.; Lavania, M.; Singh, R.; Lal, B. Identification and probiotic potential of lactic acid bacteria from camel milk. Saudi J. Biol. Sci. 2021, 28, 1622–1632. [Google Scholar] [CrossRef]
- Tola, G.B. Medicinal, Physiological, and Nutritional Benefits of Camel Milk. Preprints 2024, 2024111552. [Google Scholar] [CrossRef]
- Albarrak, S.M.; Al-Sobayil, F.A. Evaluation of protective proteins and cytokines in milk and serum of healthy camels during the first two months postpartum. Int. J. Vet. Sci. 2024, 13, 401–406. [Google Scholar] [CrossRef]
- Konuspayeva, G.; Faye, B. Recent Advances in Camel Milk Processing. Animals 2021, 11, 1045. [Google Scholar] [CrossRef]
- El-Hanafy, A.A.; Saad, Y.M.; Alkarim, S.A.; Almehdar, H.A.; Alzahrani, F.M.; Almatry, M.A.; Uversky, V.N.; Redwan, E.M. Yield and Composition Variations of the Milk from Different Camel Breeds in Saudi Arabia. Sci 2023, 5, 2. [Google Scholar] [CrossRef]
- Yang, Z.; Ni, L.; Tian, W.; Chi, H. Screening and Identification of Goat-Milk-Derived Lactic Acid Bacteria with Bacteriocin-like Activity and Probiotic Potentials. Microorganisms 2023, 11, 849. [Google Scholar] [CrossRef]
- Asha, M.N.; Chowdhury, S.R.; Hossain, H.; Rahman, A.; Al Emon, A.; Tanni, F.Y.; Islam, R.; Hossain, M.; Rahman, M. Antibacterial potential of lactic acid bacteria isolated from raw cow milk in Sylhet district, Bangladesh: A molecular approach. Vet. Med. Sci. 2024, 10, e1463. [Google Scholar] [CrossRef]
- Akhmetsadykova, S.; Baubekova, A.; Konuspayeva, G.; Akhmetsadykov, N.; Faye, B.; Loiseau, G. Lactic acid bacteria biodiversity in raw and fermented camel milk. Afr. J. Food Sci. Technol. 2015, 6, 84–88. [Google Scholar] [CrossRef]
- Alhaj, O.A.; Jrad, Z.; Oussaief, O.; Jahrami, H.A.; Ahmad, L.; Alshuniaber, M.A.; Mehta, B.M. The characterization of Lactobacillus strains in camel and bovine milk during fermentation: A comparison study. Heliyon 2024, 10, e40162. [Google Scholar] [CrossRef]
- Benmechernene, Z.; Chentouf, H.F.; Bellil, Y.; Ghazi, F.; Quintela-Baluja, M.; Calo-Mata, P.; Barros-Velázquez, J. Technological Aptitude and Applications of Leuconostoc mesenteroides Bioactive Strains Isolated from Algerian Raw Camel Milk. BioMed Res. Int. 2013, 1, 418132. [Google Scholar] [CrossRef]
- Belkheir, K.; Laref, N. Autochthonous Lactic Acid Bacteria from Camelus dromedarius Milk. An Updated Systematic Review. Food Rev. Int. 2025, 41, 1576–1591. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Junior, W.J.L.; Sant’Ana, A.S.; Di Cagno, R.; Campanaro, S.; Gobbetti, M. Five years after the lactobacillus reclassification: Genomic and functional insights into Levilactobacillus, Lacticaseibacillus, Limosilactobacillus, Lactiplantibacillus, and Ligilactobacillus in fermented foods. Trends Food Sci. Technol. 2025, 163, 105118. [Google Scholar] [CrossRef]
- Alsulaimany, F.A.; Mohamed, M.M.; Al-Hejin, A.M.; Kabli, S. Molecular Identification of Goat’s Udder Microbes and Nutritional Value of Milk Using Dielectric Constants. Kafkas Univ. Vet. Fak. Derg. 2023, 29, 463–471. [Google Scholar] [CrossRef]
- Khelissa, S.; Chihib, N.-E.; Gharsallaoui, A. Conditions of nisin production by Lactococcus lactis subsp. lactis and its main uses as a food preservative. Arch. Microbiol. 2021, 203, 465–480. [Google Scholar] [CrossRef]
- Rezac, S.; Kok, C.R.; Heermann, M.; Hutkins, R. Fermented Foods as a Dietary Source of Live Organisms. Front. Microbiol. 2018, 9, 396129. [Google Scholar] [CrossRef]
- Younis, K.M.; Najem, A.H. Characterization and evaluation of probiotic strains isolated from different sources. Microbes Infect. Dis. 2025, (in press). [Google Scholar] [CrossRef]
- Ayivi, R.D.; Ibrahim, S.A. Lactic acid bacteria: An essential probiotic and starter culture for the production of yoghurt. Int. J. Food Sci. Technol. 2022, 57, 7008–7025. [Google Scholar] [CrossRef]
- Fijan, S.; Fijan, T. Current Trends in the Applications of Probiotics and Other Beneficial Microbes: Expanding Horizons. Appl. Microbiol. 2025, 5, 103. [Google Scholar] [CrossRef]
- Meral-Aktaş, H. Bacteriocin characterization of Enterococcus faecium isolates and evaluation of their in situ anti-Listerial activity in Beyaz cheese. Food Biosci. 2024, 61, 104741. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, X.; Niu, J.; Han, Z.; Bi, C.; Mehmood, K.; Farraj, D.A.A.; Alzaidi, I.; Iqbal, R.; Qin, J. Complete genome of multi-drug resistant Staphylococcus Aureus in bovine mastitic milk in Anhui, China. Pak. Vet. J. 2023, 43, 456–462. [Google Scholar] [CrossRef]
- Tsakali, E.; Tsantes, A.G.; Houhoula, D.; Laliotis, G.P.; Batrinou, A.; Halvatsiotis, P.; Tsantes, A.E. The Detection of Bacterial Pathogens, including Emerging Klebsiella pneumoniae, Associated with Mastitis in the Milk of Ruminant Species. Appl. Sci. 2022, 13, 11484. [Google Scholar] [CrossRef]
- Alorainy, M.S.; Alhajuj, A.M.; Fathi, S.M. Biochemical identification, direct molecular detection, and antimicrobial agent resistance of Staphylococcus aureus isolated from raw camel milk in Al Madinah Region, Saudi Arabia. Int. J. Vet. Sci. 2023, 12, 740–745. [Google Scholar] [CrossRef]
- Jans, C.; Bugnard, J.; Njage, P.M.K.; Lacroix, C.; Meile, L. Lactic acid bacteria diversity of African raw and fermented camel milk products reveals a highly competitive, potentially health-threatening predominant microflora. LWT 2012, 47, 371–379. [Google Scholar] [CrossRef]
- Aljeldah, M.; Al Shammari, B.; Farrag, E.S.; Taha, E.M.; Mahmoud, S.Y. Prevalence of Multidrug-Resistant Methicillin-Resistant Staphylococcus aureus in Northeastern Saudi Hospitals. J. Pure Appl. Microbiol. 2022, 16, 1192–1199. [Google Scholar] [CrossRef]
- Al Yousef, S.A.; Mahmoud, S.Y.; Farrag, E.; Husseiny, S.; Bayoumi, F.; Ali, A. Clinical and Laboratory Profile of Urinary Tract Infections Associated with Extended Spectrum β-Lactamase Producing Escherichia coli and Klebsiella pneumoniae. Ann. Clin. Lab. Sci. 2016, 46, 393–400. [Google Scholar] [PubMed]
- Mahmoud, S.Y.M.; Atallah, A.A.; Badr, O.A.; Moustafa, M.M.A.; Esmael, A.; Ebrahim, N.; Aljeldah, M.; Al Shammari, B.; Alsafari, I.A.; Mohamed, S.A. Bioprospecting for Novel Probiotic Strains from Human Milk and Infants: Molecular, Biochemical, and Ultrastructural Evidence. Biology 2022, 11, 1405. [Google Scholar] [CrossRef]
- Schillinger, U.; Lücke, F.K. Antibacterial activity of Lactobacillus sake isolated from meat. Appl. Environ. Microbiol. 1989, 55, 1901–1906. [Google Scholar] [CrossRef]
- Kobierecka, P.A.; Wyszy’nska, A.K.; Aleksandrzak-Piekarczyk, T.; Kuczkowski, M.; Tuzimek, A.; Piotrowska, W.; Górecki, A.; Adamska, I.; Wieliczko, A.; Bardowski, J.; et al. In vitro characteristics of Lactobacillus spp. strains isolated from the chicken digestive tract and their role in the inhibition of Campylobacter colonization. Microbiol. Open 2017, 6, e00512. [Google Scholar] [CrossRef]
- Vinderola, C.; Reinheimer, J. Lactic acid starter and probiotic bacteria: A comparative ‘‘in vitro” study of probiotic characteristics and biological barrier resistance. Food Res. Int. 2003, 36, 895–904. [Google Scholar] [CrossRef]
- Reuben, R.; Roy, P.; Sarkar, S.; Alam, A.R.U.; Jahid, I. Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties. J. Dairy. Sci. 2020, 103, 1223–1237. [Google Scholar] [CrossRef]
- Armas, F.; Camperio, C.; Marianelli, C. In vitro assessment of the probiotic potential of Lactococcus lactis LMG 7930 against ruminant mastitis-causing pathogens. PLoS ONE 2017, 12, e0169543. [Google Scholar] [CrossRef] [PubMed]
- Axelsson, L. Lactic acid bacteria: Classification and physiology. In Lactic Acid Bacteria: Microbiological and Functional Aspects; CRC Press: Marcel Dekker, NY, USA, 2004; 66p. [Google Scholar]
- Abd-Alla, M.H.; El-Enany, A.W.E. Production of acetone-butanol-ethanol from spoilage date palm (Phoenix dactylifera L.) fruits by mixed culture of Clostridium acetobutylicum and Bacillus subtilis. Biomass Bioenergy 2012, 42, 172–178. [Google Scholar] [CrossRef]
- Sanger, F.; Air, G.M.; Barrell, B.G.; Brown, N.L.; Coulson, A.R.; Fiddes, J.C.; Hutchison, C.; Slocombe, P.M.; Smith, M. Nucleotide sequence of bacteriophage φx174 DNA. Nature 1977, 265, 687–695. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Criscuolo, A.; Gribaldo, S. BMGE (block mapping and gathering with entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010, 10, 210. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Crandall, K.A. Modeltest: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef]
- Liasi, S.A.; Azmi, T.I.; Hassan, M.D.; Shuhaimi, M.; Rosforizan, M.; Ariff, A.B. Antimicrobial activity and anti-biotic sensitivity of three isolates of Lactic acid bacteria from fermented product. Malays. J. Microbiol. 2009, 5, 33–37. [Google Scholar] [CrossRef]
- National Committee for Clinical Laboratory Standards [NCCLS]. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 6th ed.; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2003. [Google Scholar]
- Wang, Y.; Zhou, J.; Xia, X.; Zhao, Y.; Shao, W. Probiotic potential of Lactobacillus paracasei FM-LP-4 isolated from Xinjiang camel milk yoghurt. Int. Dairy J. 2016, 62, 28–34. [Google Scholar] [CrossRef]
- Ribeiro, S.C.; Coelho, M.C.; Todorov, S.D.; Franco, B.D.G.M.; Dapkevicius, M.L.E.; Silva, C.C.G. Technological properties of bacteriocin-producing lactic acid bacteria isolated from Pico cheese an artisanal cow’s milk cheese. J. Appl. Microbiol. 2014, 116, 573–585. [Google Scholar] [CrossRef]
- Bouguerra, A.; Harzallah, D.; Yasin, I.S.M.; Silvaraj, S.; Bakli, S.; Rabhi, N.E.H. Characterization of probiotic lactic acid bacteria isolated from Algerian camel milk in vitro. Int. J. Sci. Res. 2020, 76, 244–256. [Google Scholar] [CrossRef]
- Yateem, A.; Balba, M.T.; Al-Surrayai, T.; Al-Mutairi, B.; Al-Daher, R. Isolation of lactic acid bacteria with probiotic potential from camel milk. Int. J. Dairy Sci. 2008, 3, 194–199. [Google Scholar] [CrossRef]
- Abbas, M.M.; Mahasneh, A.M. Isolation of Lactobacillus strains with probiotic potential from camel’s milk. Afr. J. Microbiol. Res. 2014, 8, 1645–1655. [Google Scholar] [CrossRef]
- Azat, R.; Liu, Y.; Li, W.; Kayir, A.; Lin, D.; Zhou, W.; Zheng, X. Probiotic properties of lactic acid bacteria isolated from traditionally fermented Xinjiang cheese. J. Zhejiang Univ.-Sci. B 2016, 17, 597–609. [Google Scholar] [CrossRef]
- World Health Organization (WHO). WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Champomier-Vergès, M.C.; Zagorec, M.; Fadda, S. Proteomics: A tool for understanding lactic acid bacteria adaptation to stressful environments. In Biotechnology of Lactic Acid Bacteria: Novel Applications; Mozzi, F., Raya, R.R., Vignolo, G.M., Eds.; Blackwell Publishing: Malden, MA, USA, 2010; pp. 57–72. [Google Scholar]
- Ouwehand, A.C.; Vesterlund, S. Antimicrobial components from lactic acid bacteria. In Lactic Acid Bacteria Microbiology and Functional Aspects, 3rd ed.; Salminen, S., von Wright, A., Ouwehand, A., Eds.; CRC Press: Marcel Dekker, NY, USA, 2004; pp. 375–395. [Google Scholar]
- Kokwe, N.H.; Tshabuse, F.; Swalaha, F.M. In Vitro Characterization and Safety Assessment of Streptococcus salivarius, Levilactobacillus brevis and Pediococcus pentosaceus Isolated from the Small Intestine of Broiler Breeders. Microorganisms 2025, 13, 1231. [Google Scholar] [CrossRef]
- Meena, K.K.; Joshi, M.; Rao, P.; Panwar, N.L.; Gupta, L.; Meena, S. Evaluation of probiotic properties of lactic acid bacteria isolated from sheep milk and its suitability as a starter in a milk-cereal-based beverage. Food Biomacromol. 2025, 2, 69–83. [Google Scholar] [CrossRef]
- Sahoo, T.K.; Jena, P.K.; Nagar, N.; Patel, A.K.; Seshadri, S. In vitro evaluation of probiotic properties of lactic acid bacteria from the gut of Labeo rohita and Catla catla. Probiotics Antimicrob. Proteins 2015, 7, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Del Re, B.; Sgorbati, B.; Miglioli, M.; Palenzona, D. Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett. Appl. Microbiol. 2000, 31, 438–442. [Google Scholar] [CrossRef]
- Yao, P.; Mohd Esah, E.; Zhao, C. Regulatory mechanisms and applications of Lactobacillus biofilms in the food industry. Front. Microbiol. 2025, 15, 1465373. [Google Scholar] [CrossRef]
- Espeche, M.C.; Otero, M.C.; Sesma, F.; Nader-Macias, M.E. Screening of surface properties and antagonistic substances production by lactic acid bacteria isolated from the mammary gland of healthy and mastitic cows. Vet. Microbiol. 2009, 135, 346–357. [Google Scholar] [CrossRef]
- Espeche, M.C.; Pellegrino, M.; Frola, I.; Larriestra, A.; Bogni, C.; Nader-Macías, M.E.F. Lactic acid bacteria from raw milk as potentially beneficial strains to prevent bovine mastitis. Anaerobe 2012, 18, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Puniya, M.; Ravinder, K.M.; Panwar, H.; Kumar, N. Screening of lactic acid bacteria of different origin for their probiotic potential. J. Food Process. Technol. 2016, 7, 545. [Google Scholar] [CrossRef]
- Han, S.; Lu, Y.; Xie, J.; Fei, Y.; Zheng, G.; Wang, Z.; Liu, J.; Lv, L.; Ling, Z.; Berglund, B.; et al. Probiotic Gastrointestinal Transit and Colonization After Oral Administration: A Long Journey. Front. Cell. Infect. Microbiol. 2021, 11, 609722. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Iliev, I.; Goyal, A. Leuconostoc mesenteroides NRRL B-1149 as probiotic and its dextran with anticancer properties. J. Biosci. Biotechnol. 2014, 3, 79–87. [Google Scholar]
- Ansari, F.; Pourjafar, H.; Samakkhah, S.A.; Mirzakhani, E. An overview of probiotic camel milk as a nutritional beverage: Challenges and perspectives. Food Sci. Nutr. 2024, 12, 6123–6141. [Google Scholar] [CrossRef]
- De Paula, A.; Jeronymo-Ceneviva, A.; Silva, L.; Todorov, S.; Franco, B.M.; Choiset, Y.; Haertlé, T.; Chobert, J.-M.; Dousset, X.; Penna, A. Leuconostoc mesenteroides SJRP55: A potential probiotic strain isolated from Brazilian water buffalo mozzarella cheese. Probiotics Antimicrob. Proteins 2014, 6, 186–197. [Google Scholar] [CrossRef]
- Colombo, M.; Nero, L.A.; Todorov, S.D. Safety profiles of beneficial lactic acid bacteria isolated from dairy systems. Braz. J. Microbiol. 2020, 51, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Morandi, S.; Cremonesi, P.; Silvetti, T.; Brasca, M. Technological characterisation, antibiotic susceptibility and antimicrobial activity of wild-type Leuconostoc strains isolated from north Italian traditional cheeses. J. Dairy Res. 2013, 80, 457–466. [Google Scholar] [CrossRef]
- Hemme, D.; Foucaud-Scheunemann, C. Leuconostoc, characteristics, use in dairy technology and prospects in functional foods. Int. Dairy J. 2004, 14, 467–494. [Google Scholar] [CrossRef]
- Moreno, M.R.F.; Sarantinopoulos, P.; Tsakalidou, E.; De Vuyst, L. The role and application of enterococci in food and health. Int. J. Food Microbiol. 2006, 106, 1–24. [Google Scholar] [CrossRef]
- Gueimonde, M.; Sánchez, B.; de los Reyes-Gavilán, C.G.; Margolles, A. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013, 4, 51661. [Google Scholar] [CrossRef]
- Fraqueza, M.J. Antibiotic resistance of lactic acid bacteria isolated from dry-fermented sausages. Int. J. Food Microbiol. 2015, 212, 76–88. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar] [CrossRef]
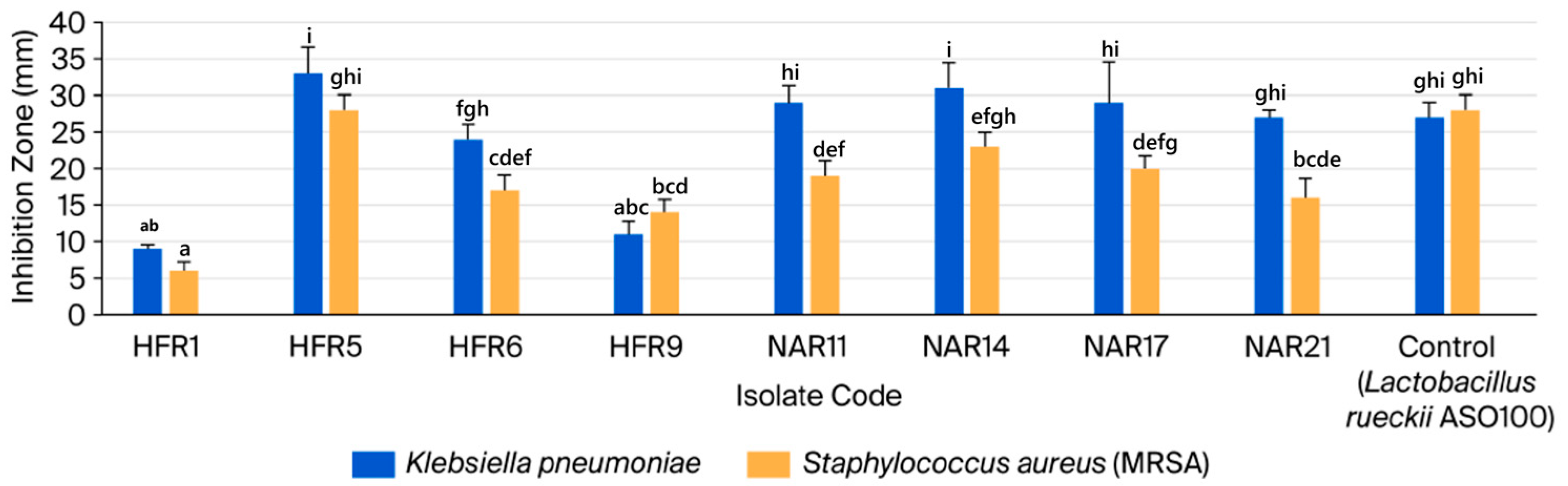
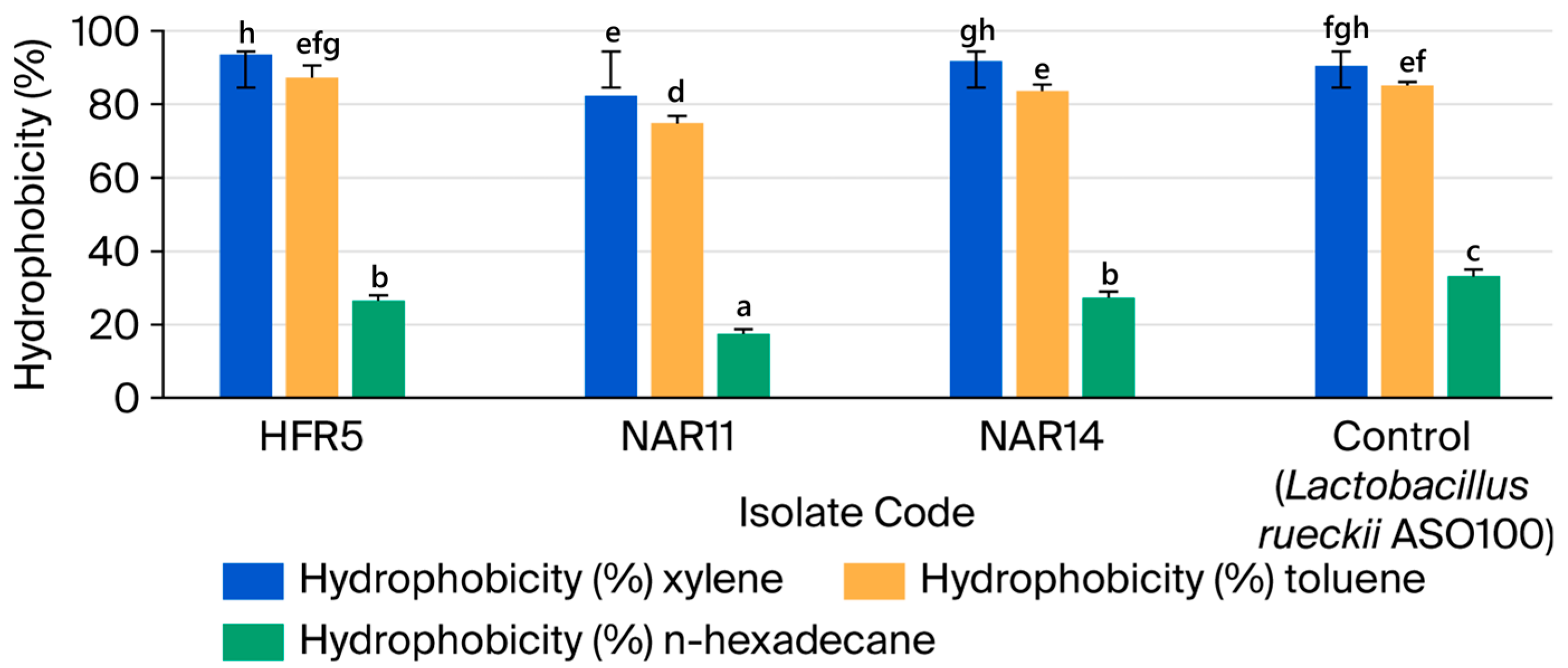
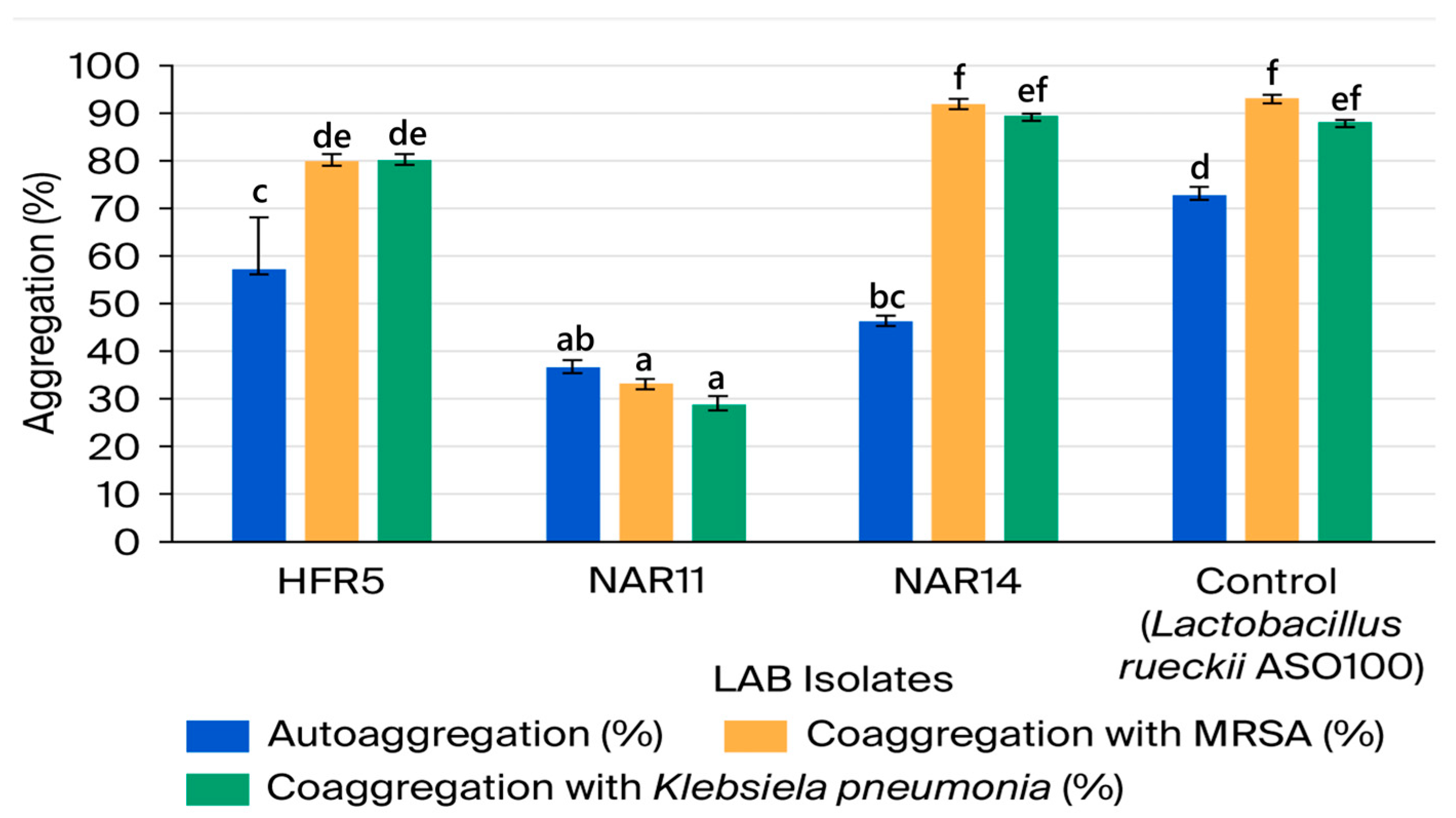
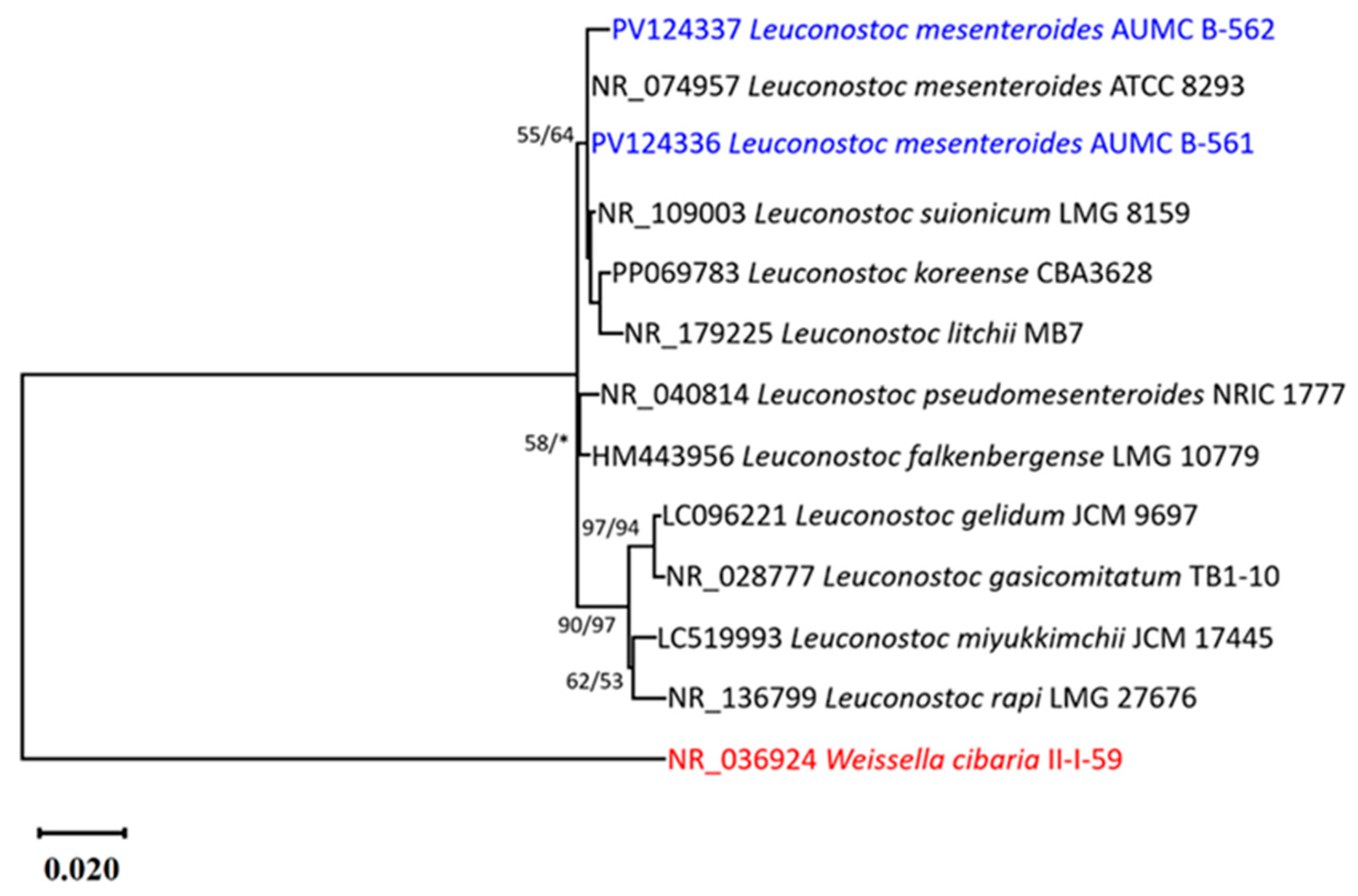
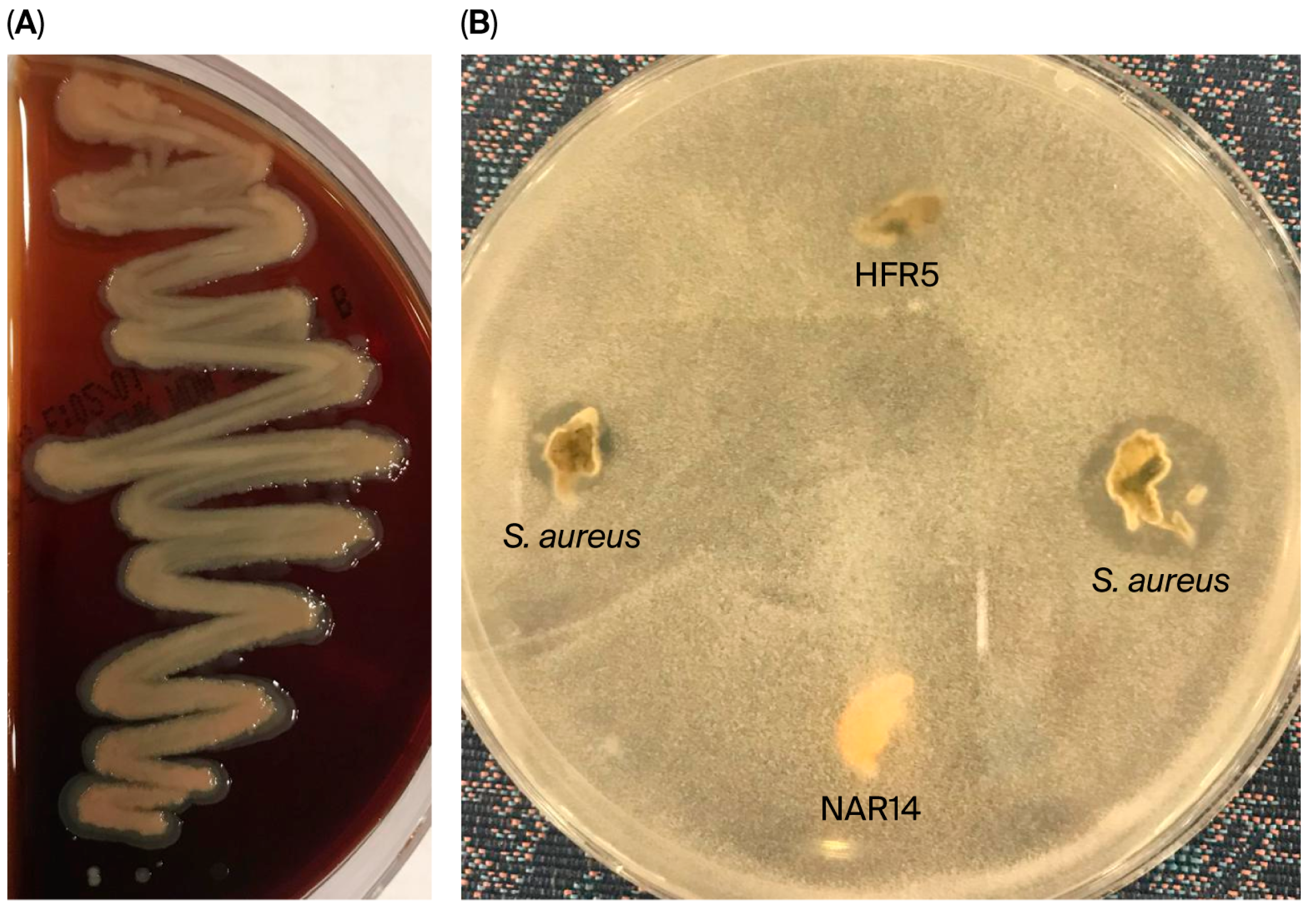
| No. | Isolate Code | Camel Color | Cell Morphology | Gram Stain | Endospore Stain | Catalase Production | Facultatively Anaerobic or Microaerophilic |
|---|---|---|---|---|---|---|---|
| 1 | HFR1 | Brown | Cocci | + | Non | − | + |
| 2 | HFR5 | Brown | Cocci | + | Non | − | + |
| 3 | HFR6 | Brown | Cocci | + | Non | − | + |
| 4 | HFR7 | Brown | Rods | + | Non | − | − |
| 5 | HFR9 | Brown | Cocci | + | Non | − | + |
| 6 | HFR10 | Brown | Rods | + | Non | − | − |
| 7 | NAR11 | Black | Cocci | + | Non | − | + |
| 8 | NAR14 | Black | Cocci | + | Non | − | + |
| 9 | NAR15 | Black | Rods | + | Non | − | − |
| 10 | NAR17 | Black | Cocci | + | Non | − | + |
| 11 | NAR18 | Black | Rods | + | Non | − | − |
| 12 | NAR21 | Black | Cocci | + | Non | − | + |
| Isolate Code | Inhibition Zone Caused by Various Concentrations of Lysozyme (mg/mL) | ||||||
|---|---|---|---|---|---|---|---|
| 0.1 | 0.2 | 0.4 | 0.6 | 0.8 | 1 | 2 | |
| HFR5 | − | − | − | − | − | − | − |
| HFR6 | − | − | − | + | ++ | ++ | +++ |
| NAR11 | − | − | − | − | − | − | − |
| NAR14 | − | − | − | − | − | − | − |
| NAR17 | − | − | − | + | ++ | ++ | +++ |
| NAR21 | − | − | − | + | + | ++ | +++ |
| Control (Lactobacillus rueckii ASO100) | − | − | − | − | − | − | − |
| Isolate | Control (Log10 CFU mL−1) | pH 2 | pH 3 | pH 4 | pH 5 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Log10 CFU mL−1 | Viability (%) | Log10 CFU mL−1 | Viability (%) | Log10 CFU mL−1 | Viability (%) | Log10 CFU mL−1 | Viability (%) | ||
| HFR5 | 8.3 | 6.6 | 79.15 c | 7.1 | 85.53 e | 7.6 | 91.16 fg | 8.0 | 96.38 h |
| NAR11 | 7.8 | 2.13 | 27.34 a | 5.7 | 73.11 b | 6.7 | 85.89 e | 7.1 | 91.02 fg |
| NAR14 | 8.5 | 6.1 | 71.76 b | 7.0 | 82.34 d | 7.6 | 89.40 f | 7.93 | 93.33 g |
| * Control | 8.8 | 6.5 | 73.86 b | 7.5 | 85.22 e | 8.6 | 97.72 h | 8.7 | 98.48 h |
| Isolate | Control (Log10 CFU mL−1) | Concentration of Bile Salt (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | |||||||
| Log10 CFU mL−1 | Viability (%) | Log10 CFU mL−1 | Viability (%) | Log10 CFU mL−1 | Viability (%) | Log10 CFU mL−1 | Viability (%) | Log10 CFU mL−1 | Viability (%) | ||
| HFR5 | 8.3 | 7.8 | 93.91 n | 7.13 | 85.94 m | 6.4 | 76.70 k | 3.8 | 45.84 g | 2.06 | 24.89 c |
| NAR11 | 7.8 | 6.8 | 87.17 m | 5.6 | 72.27 j | 4.0 | 51.27 h | 2.0 | 25.63 c | 1.33 | 17.08 a |
| NAR14 | 8.5 | 8.2 | 96.47 o | 6.9 | 81.17 l | 6.13 | 72.15 j | 3.33 | 39.21 f | 2.4 | 28.23 d |
| * Control | 8.8 | 8.1 | 95.29 no | 7.3 | 86.27 m | 5.5 | 64.07 i | 3.03 | 35.68 e | 1.63 | 19.21 b |
| Antibiotic (µg/disc) | Symbol | Inhibition Zone Diameter (mm) | |||
|---|---|---|---|---|---|
| L. mesenteroides (HFR5) | L. mesenteroides (NAR14) | ||||
| mm | Reaction | mm | Reaction | ||
| Kanamycin | K | 13 | R | 14 | R |
| Cefotaxime | CTX | 23 | S | 25 | S |
| Penicillin | P | 21 | S | 23 | S |
| Vancomycin | VA | 3 | R | 4 | R |
| Moxifloxacin | MFX | 25 | S | 25 | S |
| Chloramphenicol | C | 16 | I | 19 | I |
| Erythromycin | E | 25 | S | 23 | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhejaili, M.; Farrag, E.; Mahmoud, S.; Abd-Alla, A.-E.; Elsharouny, T. Isolation of Lactic Acid Bacteria from Raw Camel Milk in Saudi Arabia and Evaluation of Their Probiotic Potential. Microbiol. Res. 2025, 16, 248. https://doi.org/10.3390/microbiolres16120248
Alhejaili M, Farrag E, Mahmoud S, Abd-Alla A-E, Elsharouny T. Isolation of Lactic Acid Bacteria from Raw Camel Milk in Saudi Arabia and Evaluation of Their Probiotic Potential. Microbiology Research. 2025; 16(12):248. https://doi.org/10.3390/microbiolres16120248
Chicago/Turabian StyleAlhejaili, Mohammed, Eman Farrag, Sabry Mahmoud, Abd-Ellah Abd-Alla, and Tarek Elsharouny. 2025. "Isolation of Lactic Acid Bacteria from Raw Camel Milk in Saudi Arabia and Evaluation of Their Probiotic Potential" Microbiology Research 16, no. 12: 248. https://doi.org/10.3390/microbiolres16120248
APA StyleAlhejaili, M., Farrag, E., Mahmoud, S., Abd-Alla, A.-E., & Elsharouny, T. (2025). Isolation of Lactic Acid Bacteria from Raw Camel Milk in Saudi Arabia and Evaluation of Their Probiotic Potential. Microbiology Research, 16(12), 248. https://doi.org/10.3390/microbiolres16120248





