An Insight into Strain-Specificity of Streptomyces chrestomyceticus ADP4 and Identification of a Novel Peptide with Potential Antiviral Activities Against Significant Human Viruses, Including SARS-CoV2, HCV, and HIV
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganism, Culture Conditions, and Genomic DNA Preparation
2.2. Genome Sequencing, Assembly, and Annotation
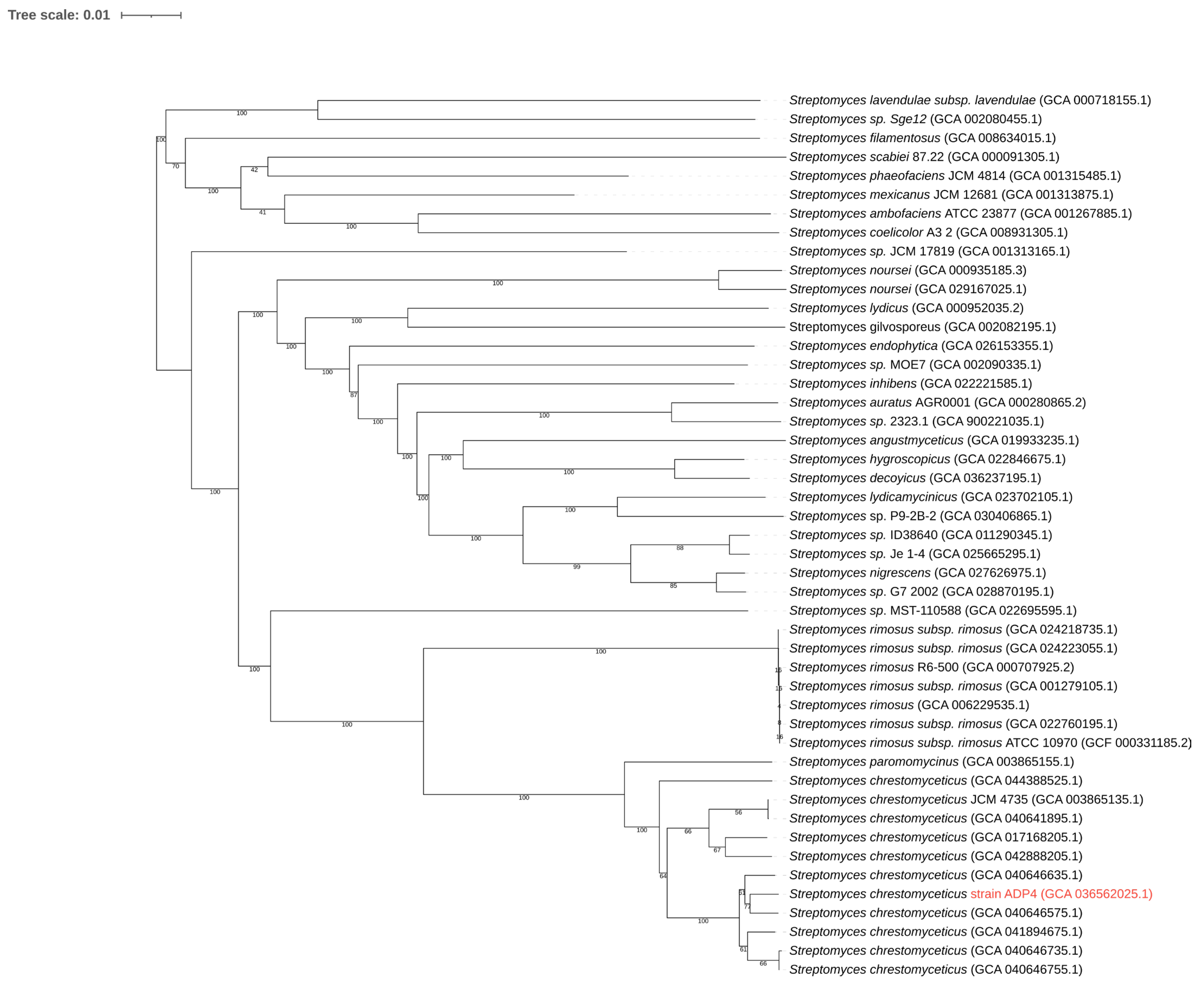
2.3. Phylogenetic Analysis
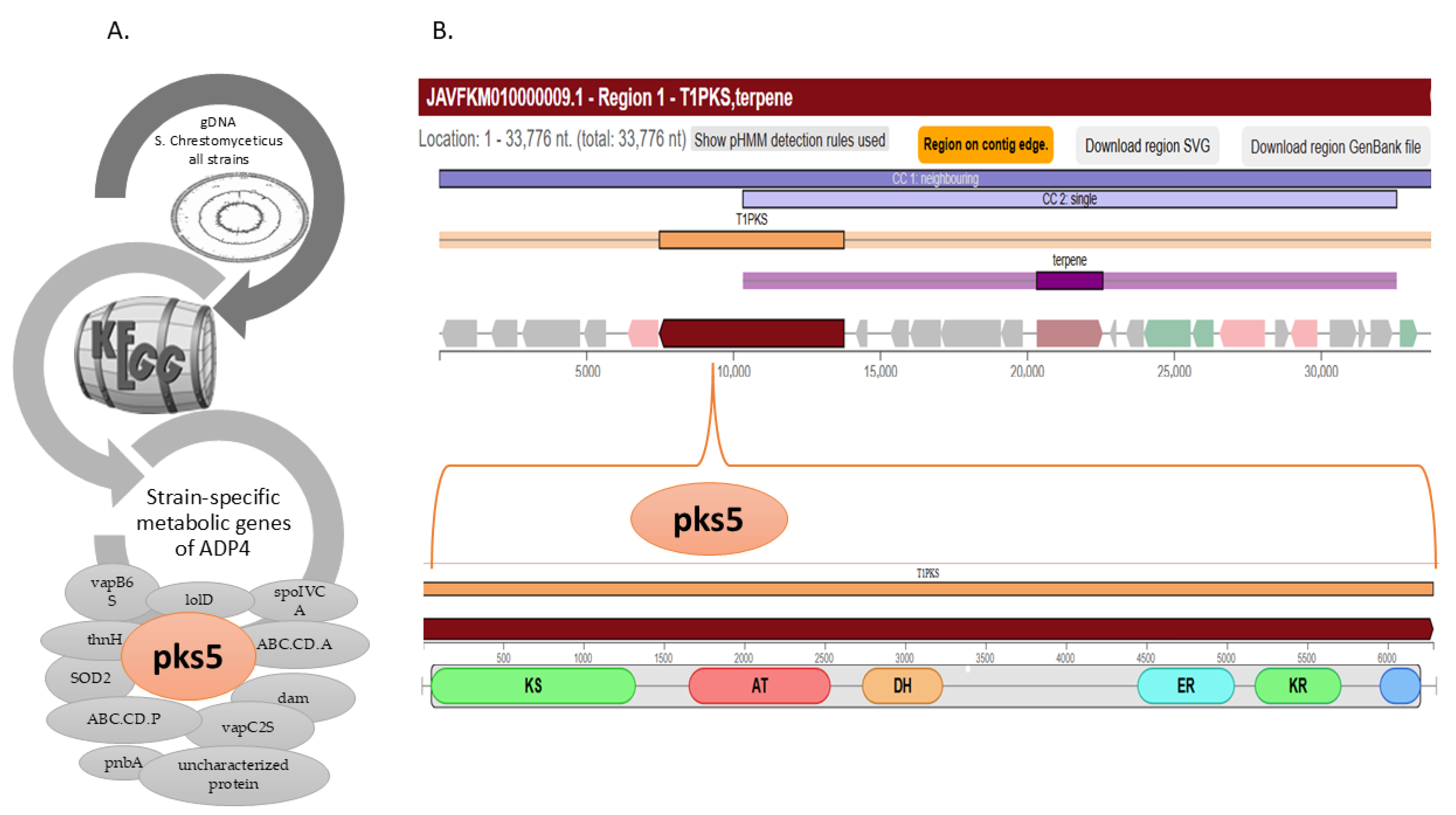
2.4. Analysis for the Specificity of ADP4
2.5. Antimicrobial Peptide Analysis
3. Results
3.1. Genome of S. chrestomyceticus Strain ADP4
3.2. Biosynthetic Gene Clusters of the Strain ADP4
3.3. Genome-Based Phylogeny
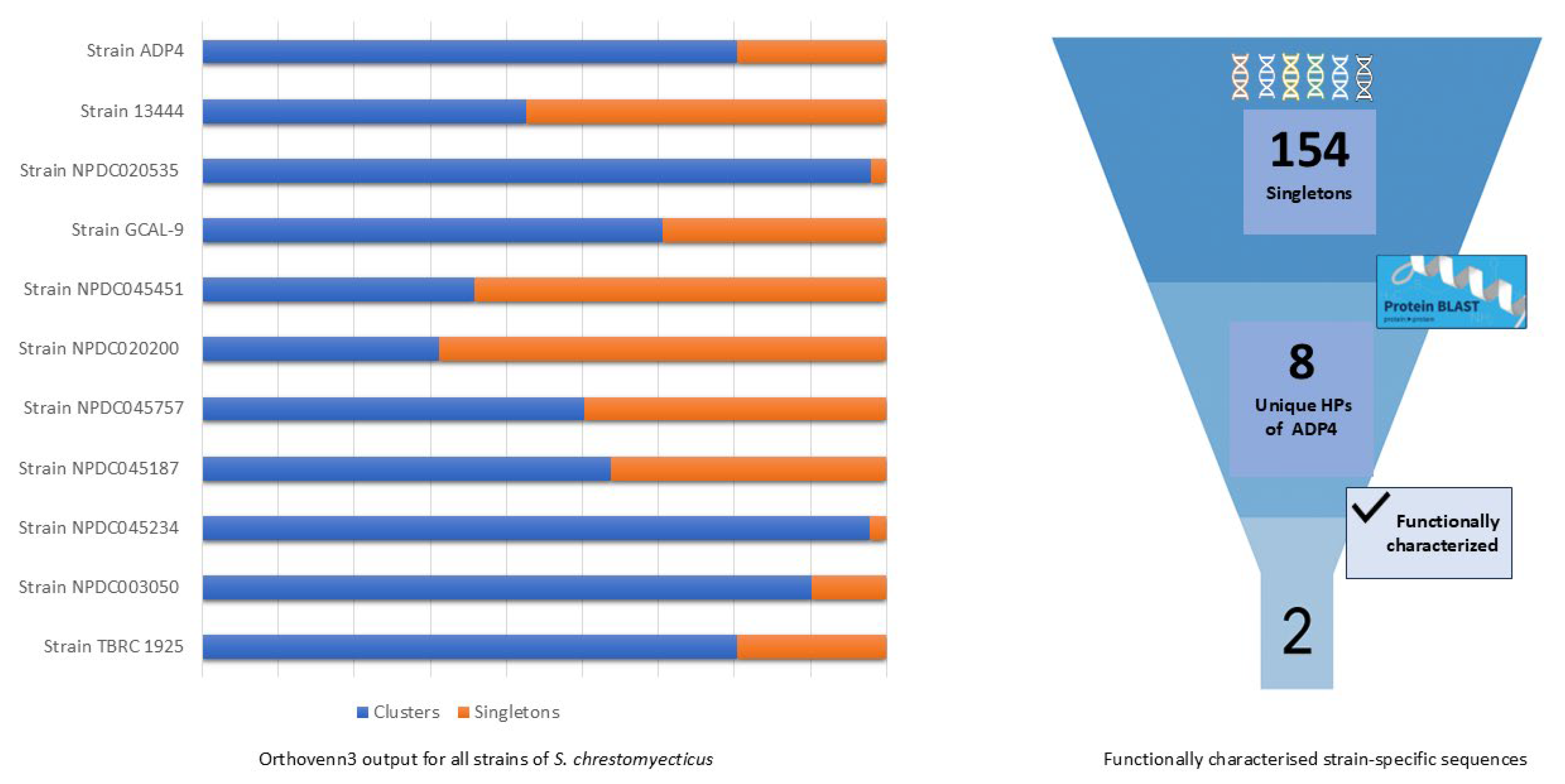
3.4. Comparative Genomics
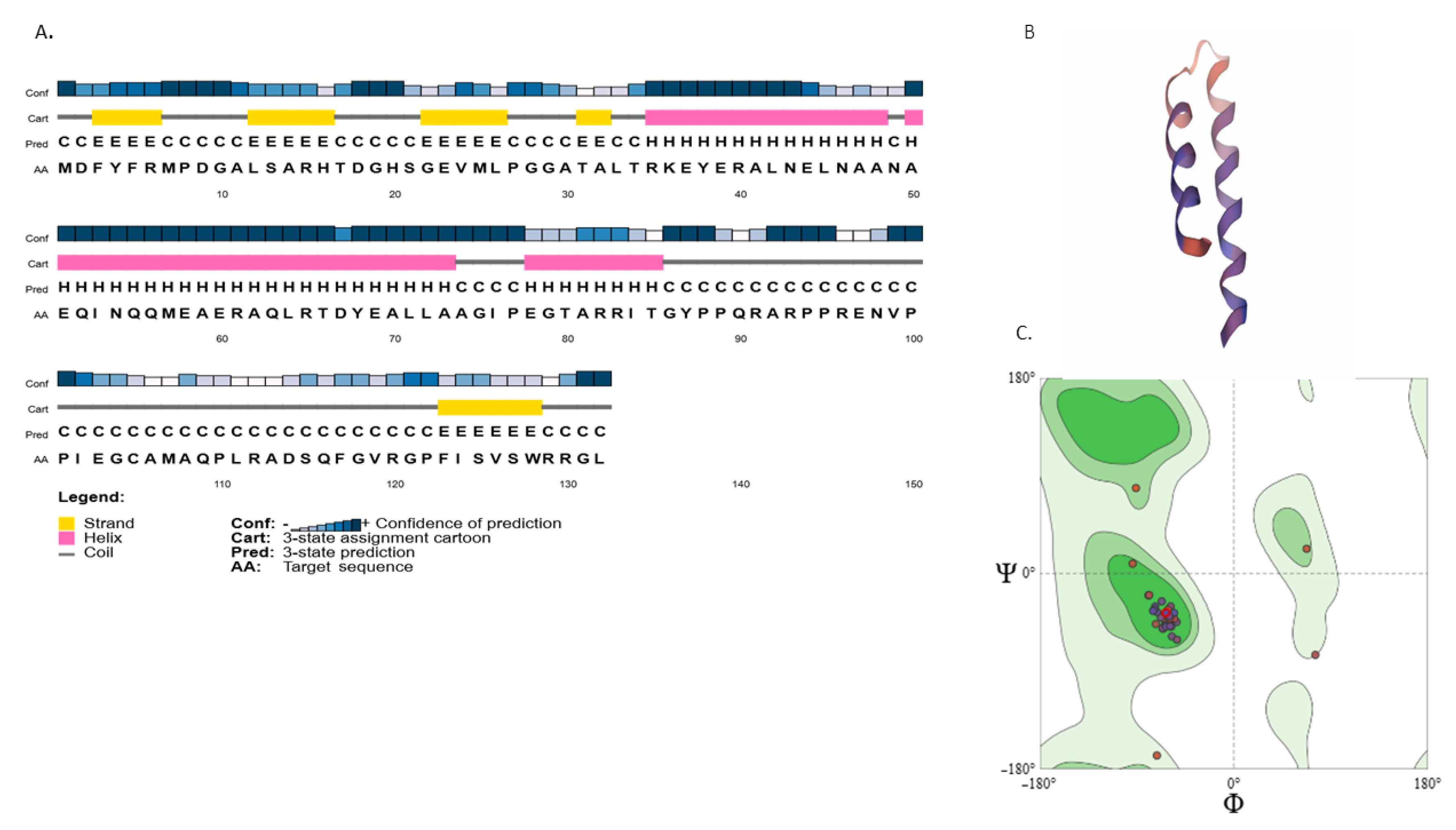
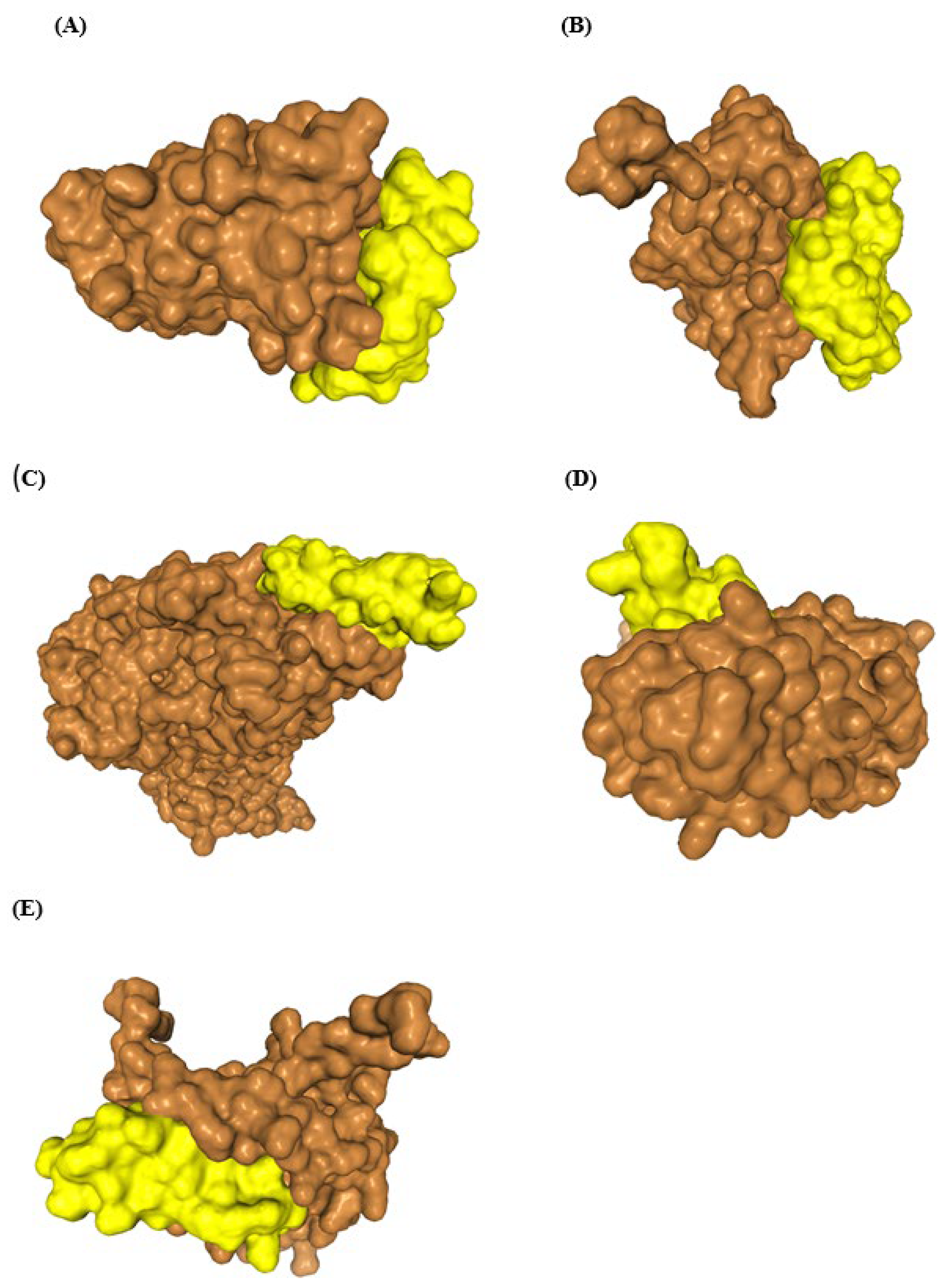
3.5. Characterization of ADP4-Specific Sequences and the Novel Antiviral Peptide
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Selim, M.S.M.; Abdelhamid, S.A.; Mohamed, S.S. Secondary metabolites and biodiversity of actinomycetes. J. Genet. Eng. Biotechnol. 2021, 19, 72. [Google Scholar] [CrossRef]
- Kim, S.; Lim, S.W.; Choi, J. Drug discovery inspired by bioactive small molecules from nature. Anim. Cells Syst. 2022, 26, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, W.; Wang, Z.; Yang, W.; Li, E.; Xia, X.; Yan, F.; Chiu, S. Emerging and reemerging infectious diseases: Global trends and new strategies for their prevention and control. Signal Transduct. Target Ther. 2024, 9, 223. [Google Scholar] [CrossRef]
- Helmi, N.R. Exploring the diversity and antimicrobial potential of actinomycetes isolated from different environments in Saudi Arabia: A systematic review. Front. Microbiol. 2025, 16, 1568899. [Google Scholar] [CrossRef]
- Srivastava, V.; Dubey, A.K. Anti-Biofilm Activity of the Metabolites of Streptomyces chrestomyceticus Strain ADP4 against Candida albicans. J. Biosci. Bioeng. 2016, 122, 434–440. [Google Scholar] [CrossRef]
- Singh, R.; Dubey, A.K. Differential Synthesis of Secondary Metabolites by Streptomyces chrestomyceticus Strain ADP4 in Response to Modulation in Nitrogen Source and Its Anti-Candida Activity. Proceedings 2020, 66, 5. [Google Scholar] [CrossRef]
- Singh, R.; Ali, M.; Dubey, A.K. Anti-Candida Attributes and In-Silico Drug-Likeness Properties of Phenyl 2′β, 6′β-Trimethyl Cyclohexyl Ketone and Phenyl Nonanyl Ether Produced by Streptomyces chrestomyceticus ADP4. J. Appl. Microbiol. 2023, 134, lxac024. [Google Scholar] [CrossRef]
- Singh, R.; Ali, M.; Dubey, A.K. Phenyl Pentyl Ketone and m-Isobutyl Methoxy Benzoate Produced by Streptomyces chrestomyceticus ADP4 Are Potent Antimicrobial Agents Displaying Broad Spectrum Activities. Indian J. Microbiol. 2023, 63, 181–189. [Google Scholar] [CrossRef]
- Singh, R.; Ali, M.; Dubey, A.K. Identification and Characterization of a Novel Decalin Derivative with Anti-Candida Activity from Streptomyces chrestomyceticus Strain ADP4. Arch. Microbiol. 2024, 206, 50. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Shukla, J.; Ali, M.; Dubey, A.K. A Novel Diterpenic Derivative Produced by Streptomyces chrestomyceticus ADP4 Is a Potent Inhibitor of Biofilm and Virulence Factors in Candida albicans and C. auris. J. Appl. Microbiol. 2024, 135, lxae139. [Google Scholar] [CrossRef]
- Singh, R.; Shukla, J.; Ali, M.; Dubey, A.K. A Novel Benzopyrone Derivative from Streptomyces chrestomyceticus ADP4 Inhibits Growth and Virulence Factors of Candida albicans. Curr. Microbiol. 2025, 82, 201. [Google Scholar] [CrossRef]
- Srivastava, V.; Singla, R.K.; Dubey, A.K. Inhibition of Biofilm and Virulence Factors of Candida albicans by Partially Purified Secondary Metabolites of Streptomyces chrestomyceticus Strain ADP4. Curr. Top. Med. Chem. 2018, 18, 925–945. [Google Scholar] [CrossRef]
- Gandham, P.; Vadla, N.; Saji, A.; Srinivas, V.; Ruperao, P.; Selvanayagam, S.; Saxena, R.K.; Rathore, A.; Gopalakrishnan, S. Genome assembly, comparative genomics, and identification of genes/pathways underlying plant growth-promoting traits of an actinobacterial strain, Amycolatopsis sp. (BCA-696). Sci. Rep. 2024, 14, 15934. [Google Scholar] [CrossRef] [PubMed]
- Abraha, H.B.; Ramesha, R.M.; Ferdiansyah, M.K.; Son, H.; Kim, G.; Park, B.; Jeong, D.Y.; Kim, K.P. Genome Analysis of a Newly Sequenced B. subtilis SRCM117797 and Multiple Public B. subtilis Genomes Unveils Insights into Strain Diversification and Biased Core Gene Distribution. Curr. Microbiol. 2024, 81, 305. [Google Scholar] [CrossRef]
- Otani, H.; Udwary, D.W.; Mouncey, N.J. Comparative and pangenomic analysis of the genus Streptomyces. Sci. Rep. 2022, 12, 18909. [Google Scholar] [CrossRef]
- Minas, K.; McEwan, N.R.; Newbold, C.J.; Scott, K.P. Optimization of a high-throughput CTAB-based protocol for the extraction of qPCR-grade DNA from rumen fluid, plant and bacterial pure cultures. FEMS Microbiol. Lett. 2011, 325, 162–169. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 31 March 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Tanizawa, Y.; Fujisawa, T.; Nakamura, Y. DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 2018, 34, 1037–1039. [Google Scholar] [CrossRef] [PubMed]
- Haft, D.H.; Badretdin, A.; Coulouris, G.; DiCuccio, M.; Durkin, A.S.; Jovenitti, E.; Li, W.; Mersha, M.; O’Neill, K.R.; Virothaisakun, J.; et al. RefSeq and the prokaryotic genome annotation pipeline in the age of metagenomes. Nucleic Acids Res. 2024, 52, D762–D769. [Google Scholar] [CrossRef]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA—An ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Sun, J.; Lu, F.; Luo, Y.; Bie, L.; Xu, L.; Wang, Y. OrthoVenn3: An integrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res. 2023, 51, W397–W403. [Google Scholar] [CrossRef]
- Mohite, O.S.; Jørgensen, T.S.; Booth, T.J.; Charusanti, P.; Phaneuf, P.V.; Weber, T.; Palsson, B.O. Pangenome mining of the Streptomyces genus redefines species’ biosynthetic potential. Genome Biol. 2025, 26, 9. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, K.T.; Tiedje, J.M. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. USA 2004, 101, 3160–3165. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Zhang, Z.; Yang, T.; Chen, M.; Li, J.; Chen, F.; Yang, J.; Li, W.; Zhang, B.; Zhang, Z.; et al. Comparative genomics analysis of Streptomyces species reveals their adaptation to the marine environment and their diversity at the genomic level. Front. Microbiol. 2016, 7, 998. [Google Scholar] [CrossRef]
- Ishino, Y.; Krupovic, M.; Forterre, P. History of CRISPR-Cas from Encounter with a Mysterious Repeated Sequence to Genome Editing Technology. J. Bacteriol. 2018, 200, e00580-17. [Google Scholar] [CrossRef]
- Loureiro, A.; da Silva, G.J. CRISPR-Cas: Converting a Bacterial Defence Mechanism into a State-of-the-Art Genetic Manipulation Tool. Antibiotics 2019, 8, 18. [Google Scholar] [CrossRef]
- Qiu, Y.; Wang, S.; Chen, Z.; Guo, Y.; Song, Y. An Active Type I-E CRISPR-Cas System Identified in Streptomyces avermitilis. PLoS ONE 2016, 11, e0149533. [Google Scholar] [CrossRef]
- Hua, H.M.; Xu, J.F.; Huang, X.S.; Zimin, A.A.; Wang, W.F.; Lu, Y.H. Low-Toxicity and High-Efficiency Streptomyces Genome Editing Tool Based on the Miniature Type V-F CRISPR/Cas Nuclease AsCas12f1. J. Agric. Food Chem. 2024, 72, 5358–5367. [Google Scholar] [CrossRef]
- Lobb, B.; Tremblay, B.J.; Moreno-Hagelsieb, G.; Doxey, A.C. An assessment of genome annotation coverage across the bacterial tree of life. Microb. Genom. 2020, 6, e000341. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Pan, H.-X.; Tang, G.-L. Newly Discovered Mechanisms of Antibiotic Self-Resistance with Multiple Enzymes Acting at Different Locations and Stages. Antibiotics 2023, 12, 35. [Google Scholar] [CrossRef]
- Belknap, K.C.; Park, C.J.; Barth, B.M.; Andam, C.P. Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci. Rep. 2020, 10, 2003. [Google Scholar] [CrossRef]
- Shi, Y.M.; Hirschmann, M.; Shi, Y.N.; Ahmed, S.; Abebew, D.; Tobias, N.J.; Grün, P.; Crames, J.J.; Pöschel, L.; Kuttenlochner, W.; et al. Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria. Nat. Chem. 2022, 14, 701–712. [Google Scholar] [CrossRef]
- Alam, K.; Mazumder, A.; Sikdar, S.; Zhao, Y.-M.; Hao, J.; Song, C.; Wang, Y.; Sarkar, R.; Islam, S.; Zhang, Y.; et al. Streptomyces: The biofactory of secondary metabolites. Front. Microbiol. 2022, 13, 968053. [Google Scholar] [CrossRef]
- Vicente, C.M.; Thibessard, A.; Lorenzi, J.N.; Benhadj, M.; Hôtel, L.; Gacemi-Kirane, D.; Lespinet, O.; Leblond, P.; Aigle, B. Comparative genomics among closely related Streptomyces strains revealed specialized metabolite biosynthetic gene cluster diversity. Antibiotics 2018, 7, 86. [Google Scholar] [CrossRef]
- Cimermancic, P.; Medema, M.H.; Claesen, J.; Kurita, K.; Wieland Brown, L.C.; Mavrommatis, K.; Pati, A.; Godfrey, P.A.; Koehrsen, M.; Clardy, J.; et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 2014, 158, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.N.; Guo, G.Q.; Krishnan, A.; Ramesh, M.; Katari, N.K.; Shahbaaz, M.; Abdellattif, M.H.; Singh, S.K.; Dua, K. Peptides-Based Therapeutics: Emerging Potential Therapeutic Agents for COVID-19. Therapie 2022, 77, 319–328. [Google Scholar] [CrossRef]
- Neghabi Hajigha, M.; Hajikhani, B.; Vaezjalali, M.; Samadi Kafil, H.; Kazemzadeh Anari, R.; Goudarzi, M. Antiviral and antibacterial peptides: Mechanisms of action. Heliyon 2024, 10, e40121. [Google Scholar] [CrossRef] [PubMed]
- Zakaryan, H.; Chilingaryan, G.; Arabyan, E.; Serobian, A.; Wang, G. Natural antimicrobial peptides as a source of new antiviral agents. J. Gen. Virol. 2021, 102, 001661. [Google Scholar] [CrossRef]
| Strain | ADP4 | NPDC020535 | NPDC045187 | NPDC045234 | NPDC045757 | NBRC 13444 | TBRC 1925 | NPDC003050 | NPDC020200 | GCAL-9 | NPDC045451 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Attribute | |||||||||||
| Genome (Mb) | 9.64 | 9.31 | 9.51 | 9.67 | 9.68 | 9.38 | 9.34 | 9.86 | 9.74 | 9.52 | 9.76 |
| Number of CDSs | 8378 | 8145 | 8299 | 8513 | 8465 | 8346 | 8096 | 8623 | 8430 | 8182 | 8589 |
| GC content (%) | 72.0 | 72.1 | 72.0 | 72.1 | 72.1 | 72.0 | 72.1 | 72.0 | 72.1 | 72.1 | 72.1 |
| DNA scaffolds | 68 | 35 | 447 | 83 | 72 | 1 | 66 | 52 | 67 | 246 | 66 |
| Gap ratio (%) | 0.002 | 0.003 | 0.005 | 0.008 | 0.004 | 0.0 | 0.0 | 0.0 | 0.005 | 0.0 | 0.004 |
| Coding ratio (%) | 86.2 | 87.6 | 83.3 | 86.4 | 86.4 | 87.2 | 87.3 | 86.1 | 87.0 | 84.8 | 86.0 |
| N50 (bp) | 345,478 | 693,272 | 77,237 | 254,561 | 254,010 | 377,418 | 597,445 | 668,145 | 329,609 | 84,471 | 393,997 |
| Number of CRISPR loci | 13 | 6 | 3 | 2 | 3 | 6 | 4 | 13 | 10 | 2 | 3 |
| Number of rRNAs | 8 | 2 | 2 | 2 | 3 | 21 | 2 | 2 | 2 | 4 | 2 |
| Number of tRNAs | 90 | 92 | 85 | 88 | 94 | 92 | 94 | 96 | 111 | 87 | 89 |
| Sequence Name | NCBI Accession No. | Size (aa) | InterPro | DeepProLoc | TargetP | PSIPRED | CDD-BLAST | DeepTMHMM | Blastp Significant Hit (Cutoff 1 × 10−5) | DB with No Prediction for Any HP |
|---|---|---|---|---|---|---|---|---|---|---|
| HP1 | WP_331790072.1 | 143 | CC, SP, TM | Extracellular | SP | SP, extracellular | - | SP | - | Pfam, CATH |
| HP2 | WP_331790253.1 | 132 | IDR | Cytoplasmic | Other | Cytoplasmic | Partial match, 36% | - | HP hits with only 47% and less identity | |
| HP3 | WP_331789357.1 | 60 | IDR | Cytoplasmic Membrane | Other | Cytoplasmic | - | - | - | |
| HP4 | WP_331786462.1 | 58 | CC, SP, TM | Cytoplasmic | mTP | Cytoplasmic | - | - | Transposase | |
| HP5 | WP_331786362.1 | 70 | No match | Cytoplasmic Membrane | Other | Cytoplasmic | - | - | - | |
| HP6 | WP_331788561.1 | 84 | No match | Extracellular | Other | Extracellular | - | - | - | |
| HP7 | WP_331789350.1 | 90 | No match | Cytoplasmic | Other | Cytoplasmic | - | - | - | |
| HP8 | WP_331786751.1 | 65 | No match | Extracellular | Other | Extracellular | - | - | - |
| Based on DBAASP AVP Activity Prediction | HP2 | ||
|---|---|---|---|
| Viral pathogen | Name | Class | Predictive value |
| Hepatitis C virus (HCV) | Active | 0.8 | |
| Zika virus (ZIKV) | Active | 0.53 | |
| West Nile virus (WNV) | Not Active | 0.54 | |
| SARS-CoV-2 | Active | 0.64 | |
| SARS-CoV | Active | 0.65 | |
| Japanese encephalitis virus (JEV) | Active | 0.74 | |
| HIV-1 | Active | 0.53 | |
| DENV-2 | Active | 0.55 | |
| DENV-1 | Active | 0.6 | |
| Fungal pathogen | Candida albicans Saccharomyces cerevisiae | Not active | - |
| Bacterial pathogen | Gram-positive Gram-negative | Not active | - |
| Hemolytic activity prediction of peptide: Human erythrocytes | Not active | - | |
| AVP | Enfuvirtide | C34 Peptide | hBD-1 | Tifuvirtide | HP2-AP | |
|---|---|---|---|---|---|---|
| Size of peptide | In amino acids | 36 | 34 | 36 | 39 | 31 |
| Type of peptide | Synthetic/natural | Natural | Synthetic | Natural | Synthetic | Natural |
| Physiochemical properties | Molecular weight (MW) | 4448.16 | 4182.11 | 3931.78 | 4992.48 | 3558.74 |
| Toxicity | hERG blocker | No | No | No | No | No |
| AMES toxicity | No | No | No | No | No | |
| Rat oral acute toxicity | No | No | No | No | No | |
| Carcinogenicity | No | No | No | No | No | |
| LC50DM | 5.392 | 5.238 | 6.096 | 4.987 | ||
| Lipophilicity | Log Po/w | −0.919 | −1.382 | −1.643 | −0.544 | −4.55 |
| Water solubility | Log S (ESOL) | −3.814 | −4.02 | −3.06 | −3.785 | −3.535 |
| Pharmacokinetics | GI absorption | Low | Low | Low | Low | Low |
| Drug likeliness | Lipinski | Rejected | Rejected | Rejected | Rejected | Rejected |
| Pfizer rule | Accepted | Accepted | Accepted | Accepted | Accepted | |
| Excretion | T1/2 | 2.841 | 2.597 | 4.176 | 2.994 | 2.367 |
| CLplasma | −0.25 | −0.061 | 0.152 | −0.475 | 0.179 | |
| Accession no. (Drug bank/Uniprot) | DB00109 (FDA-approved) | P19550 (628–661 residue) | P60022 | DB05413 (under trial) | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verma, V.; Mohan, M.K.; Dubey, A.K. An Insight into Strain-Specificity of Streptomyces chrestomyceticus ADP4 and Identification of a Novel Peptide with Potential Antiviral Activities Against Significant Human Viruses, Including SARS-CoV2, HCV, and HIV. Microbiol. Res. 2025, 16, 249. https://doi.org/10.3390/microbiolres16120249
Verma V, Mohan MK, Dubey AK. An Insight into Strain-Specificity of Streptomyces chrestomyceticus ADP4 and Identification of a Novel Peptide with Potential Antiviral Activities Against Significant Human Viruses, Including SARS-CoV2, HCV, and HIV. Microbiology Research. 2025; 16(12):249. https://doi.org/10.3390/microbiolres16120249
Chicago/Turabian StyleVerma, Varsha, Medicherla Krishna Mohan, and Ashok K. Dubey. 2025. "An Insight into Strain-Specificity of Streptomyces chrestomyceticus ADP4 and Identification of a Novel Peptide with Potential Antiviral Activities Against Significant Human Viruses, Including SARS-CoV2, HCV, and HIV" Microbiology Research 16, no. 12: 249. https://doi.org/10.3390/microbiolres16120249
APA StyleVerma, V., Mohan, M. K., & Dubey, A. K. (2025). An Insight into Strain-Specificity of Streptomyces chrestomyceticus ADP4 and Identification of a Novel Peptide with Potential Antiviral Activities Against Significant Human Viruses, Including SARS-CoV2, HCV, and HIV. Microbiology Research, 16(12), 249. https://doi.org/10.3390/microbiolres16120249






