The Different Types of Metallophores Produced by Salmonella enterica: A Review
Abstract
1. Introduction
2. Siderophores
2.1. Enterochelin (Enterobactin)
The Biosynthesis of Enterobactin
2.2. Interaction between Ent and the Immune System
2.2.1. Effect of Ent on Macrophages
2.2.2. The Interaction between Siderocalin and Enterobactin
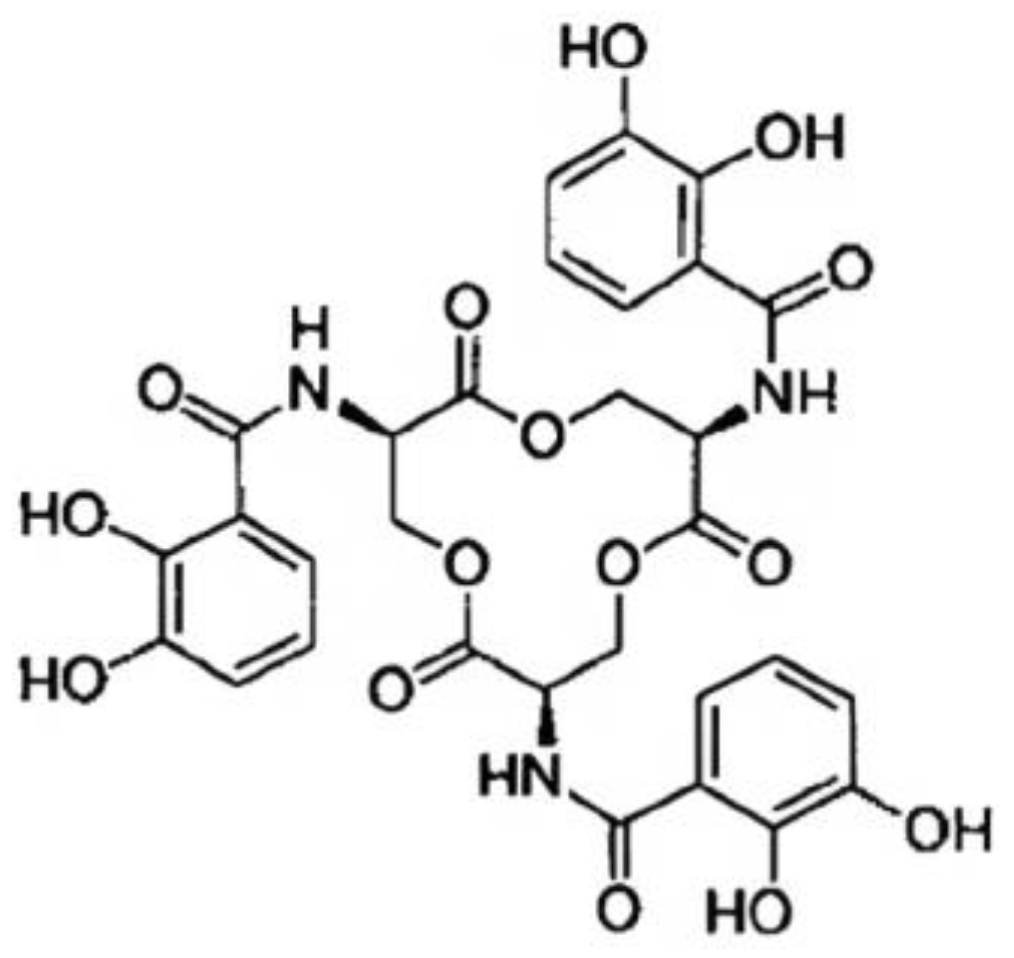
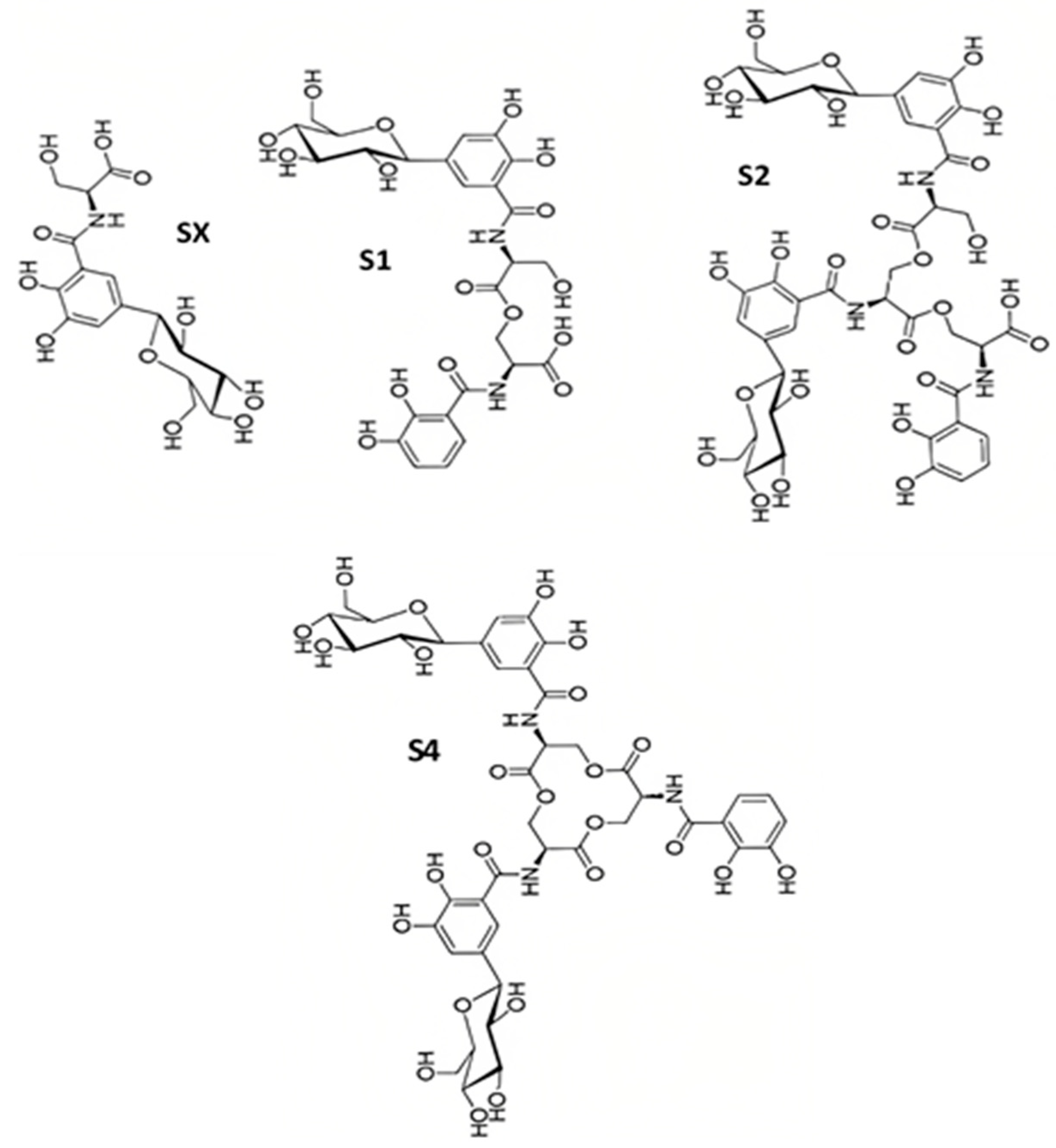
2.3. Salmochelins
Salmochelin Biosynthesis
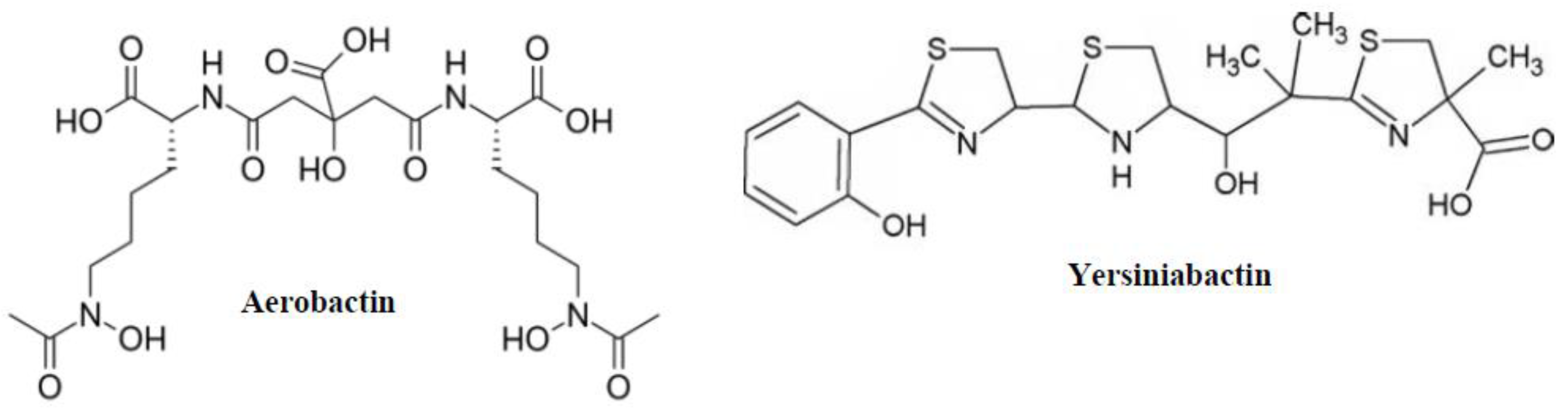
2.4. Aerobactin and Yersiniabactin
2.5. The Function of Siderophores
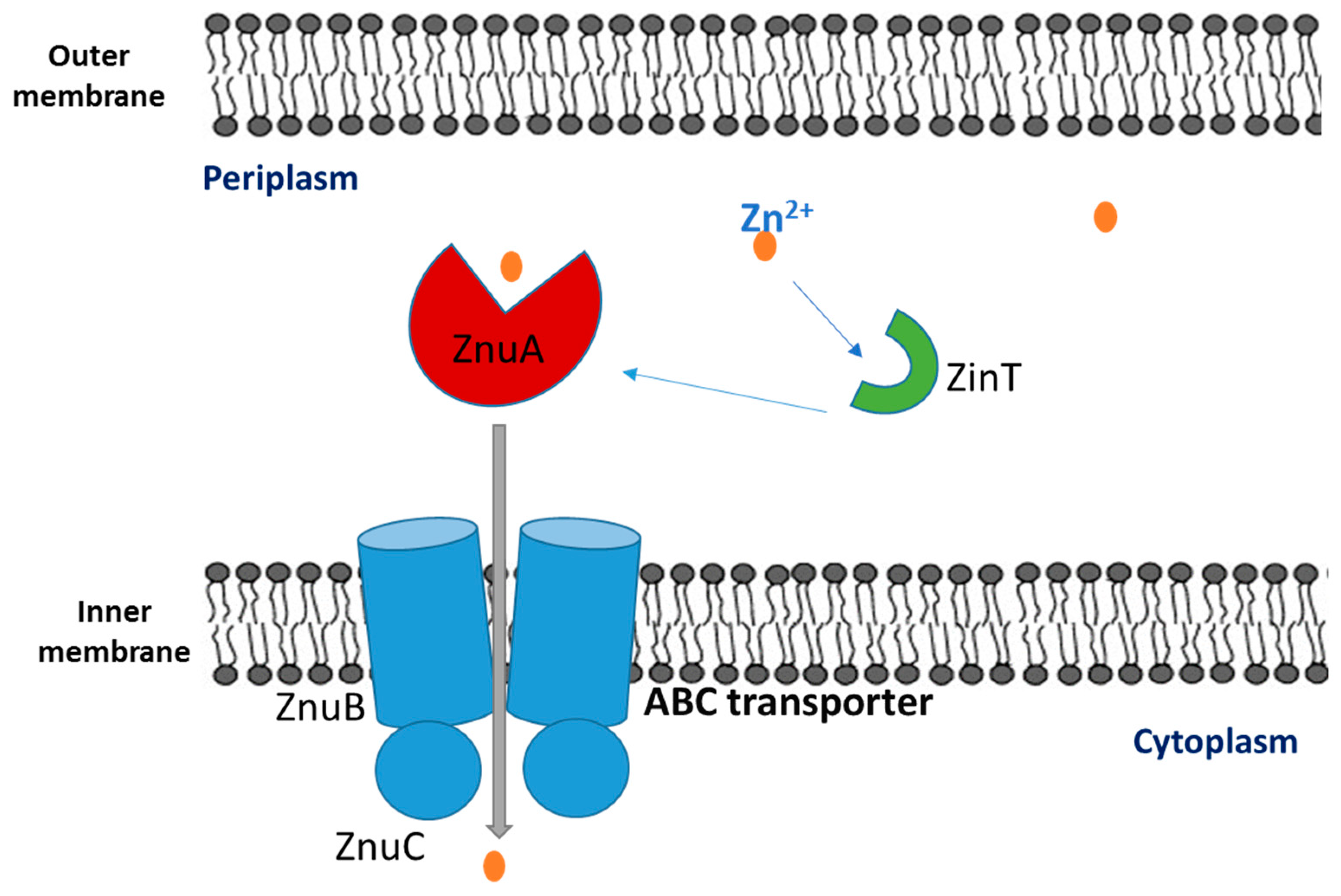
3. Zincophores
4. Up Taking Fungal Siderophores
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fardsanei, F.; Dallal, M.M.S.; Douraghi, M.; Memariani, H.; Bakhshi, B.; Salehi, T.Z.; Nikkhahi, F. Antimicrobial resistance, virulence genes and genetic relatedness of Salmonella enterica serotype Enteritidis isolates recovered from human gastroenteritis in Tehran, Iran. J. Glob. Antimicrob. Resist. 2018, 12, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Lou, L.; Zhang, P.; Piao, R.; Wang, Y. Salmonella Pathogenicity Island 1 (SPI-1) and Its Complex Regulatory Network. Front. Cell. Infect. Microbiol. 2019, 9, 270. [Google Scholar] [CrossRef]
- Takaya, A.; Yamamoto, T.; Tokoyoda, K. Humoral Immunity vs. Salmonella. Front. Immunol. 2020, 10, 3155. [Google Scholar] [CrossRef]
- He, Y.; Wang, J.; Zhang, R.; Chen, L.; Zhang, H.; Qi, X.; Chen, J. Epidemiology of foodborne diseases caused by Salmonella in Zhejiang Province, China, between 2010 and 2021. Front. Public Health 2023, 11, 1127925. [Google Scholar] [CrossRef]
- Foley, S.L.; Johnson, T.J.; Ricke, S.C.; Nayak, R.; Danzeisen, J. Salmonella pathogenicity and host adaptation in chicken-associated serovars. Microbiol. Mol. Biol. Rev. 2013, 77, 582–607. [Google Scholar] [CrossRef]
- Herekar, F.; Sarfaraz, S.; Imran, M.; Ghouri, N.; Shahid, S.; Mahesar, M. Clinical spectrum and outcomes of patients with different resistance patterns of Salmonella enterica. Pak. J. Med. Sci. 2022, 38, 356–361. [Google Scholar] [CrossRef]
- Rabsch, W.; Tschäpe, H.; Bäumler, A.J. Non-typhoidal salmonellosis: Emerging problems. Microbes Infect. 2001, 3, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Uche, I.V.; MacLennan, C.A.; Saul, A. A systematic review of the incidence, risk factors and case fatality rates of invasive nontyphoidal Salmonella (iNTS) disease in Africa (1966 to 2014). PLoS Negl. Trop. Dis. 2017, 11, e0005118. [Google Scholar] [CrossRef] [PubMed]
- Ezzeddine, Z.; Ghssein, G. Towards new antibiotics classes targeting bacterial metallophores. Microb. Pathog. 2023, 29, 06221. [Google Scholar] [CrossRef]
- Awada, R.; Ghssein, G.; Roz, A.; Farhat, M.; Nehme, N.; Hassan, H.F. Prevalence of Campylobacter spp. in broilers in North Lebanon. Vet. World 2023, 16, 322–328. [Google Scholar] [CrossRef]
- McDermott, P.F.; Zhao, S.; Wagner, D.D.; Simjee, S.; Walker, R.D.; White, D.G. The food safety perspective of antibiotic resistance. Anim. Biotechnol. 2002, 13, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Rello, J.; Kalwaje Eshwara, V.; Conway-Morris, A.; Lagunes, L.; Alves, J.; Alp, E.; Zhang, Z.; Mer, M.; Investigators, T.S. Perceived differences between intensivists and infectious diseases consultants facing antimicrobial resistance: A global cross-sectional survey. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1235–1240. [Google Scholar] [CrossRef]
- Bloom, D.E.; Cadarette, D. Infectious disease threats in the twenty-first century: Strengthening the global response. Front. Immunol. 2019, 10, 549. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Sahl, H.-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. International Collaboration on Enteric Disease ‘Burden of Illness’ Studies. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Porcheron, G.; Garénaux, A.; Proulx, J.; Sabri, M.; Dozois, C.M. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: Correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front. Cell. Infect. Microbiol. 2013, 3, 90. [Google Scholar] [CrossRef]
- Ma, Z.; Jacobsen, F.E.; Giedroc, D.P. Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 2009, 109, 4644–4681. [Google Scholar] [CrossRef]
- Fukada, T.; Yamasaki, S.; Nishida, K.; Murakami, M.; Hirano, T. Zinc homeostasis and signaling in health and diseases: Zinc signaling. J. Biol. Inorg. Chem. 2011, 16, 1123–1134. [Google Scholar] [CrossRef]
- Hood, M.I.; Skaar, E.P. Nutritional immunity: Transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 2012, 10, 525–537. [Google Scholar] [CrossRef]
- Grüngreiff, K.; Reinhold, D.; Wedemeyer, H. The role of zinc in liver cirrhosis. Ann. Hepatol. 2016, 15, 7–16. [Google Scholar] [CrossRef]
- Abbaspour, N.; Hurrell, R.; Kelishadi, R. Review on iron and its importance for human health. J. Res. Med. Sci. 2014, 19, 164–174. [Google Scholar] [PubMed]
- Capdevila, D.A.; Edmonds, K.A.; Giedroc, D.P. Metallochaperones and metalloregulation in bacteria. Essays Biochem. 2017, 61, 177–200. [Google Scholar]
- Ghssein, G.; Ezzeddine, Z. The Key Element Role of Metallophores in the Pathogenicity and Virulence of Staphylococcus aureus: A Review. Biology 2022, 11, 1525. [Google Scholar] [CrossRef] [PubMed]
- Ghssein, G.; Matar, S.F. Chelating Mechanisms of Transition Metals by Bacterial Metallophores “Pseudopaline and Staphylopine”: A Quantum Chemical Assessment. Computation 2018, 6, 56. [Google Scholar] [CrossRef]
- Chaaban, T.; Mohsen, Y.; Ezzeddine, Z.; Ghssein, G. Overview of Yersinia pestis Metallophores: Yersiniabactin and Yersinopine. Biology 2023, 12, 598. [Google Scholar] [CrossRef] [PubMed]
- Saha, P.; Xiao, X.; Yeoh, B.S.; Chen, Q.; Katkere, B.; Kirimanjeswara, G.S.; Vijay-Kumar, M. The bacterial siderophore enterobactin confers survival advantage to Salmonella in macrophages. Gut Microbes 2019, 10, 412–423. [Google Scholar] [CrossRef]
- Fischbach, M.A.; Lin, H.; Liu, D.R.; Walsh, C.T. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat. Chem. Biol. 2006, 2, 132–138. [Google Scholar] [CrossRef]
- Loomis, L.D.; Raymond, K.N. Solution equilibria of enterobactin and metal enterobactin complexes. Inorg. Chem. 1991, 30, 906–911. [Google Scholar] [CrossRef]
- Raymond, K.N.; Dertz, E.A.; Kim, S.S. Enterobactin: An archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 2003, 100, 3584–3588. [Google Scholar] [CrossRef]
- Gehring, A.M.; Mori, I.; Walsh, C.T. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry 1998, 37, 2648–2659. [Google Scholar] [CrossRef]
- Walsh, C.; Liu, J.; Rusnak, F.; Sakaitani, M. Molecular studies on enzymes in chorismate metabolism and the enterobactin biosynthetic pathway. Chem. Rev. 1990, 90, 1105–1129. [Google Scholar] [CrossRef]
- Lambalot, R.H.; Gehring, A.M.; Flugel, R.S.; Zuber, P.; LaCelle, M.; Marahiel, M.A.; Walsh, C.T. A new enzyme superfamily—The phosphopantetheinyl transferases. Chem. Biol. 1996, 3, 923–936. [Google Scholar] [CrossRef]
- Furrer, J.L.; Sanders, D.N.; Hook-Barnard, I.G.; McIntosh, M.A. Export of the siderophore enterobactin in Escherichia coli: Involvement of a 43 kDa membrane exporter. Mol. Microbiol. 2002, 44, 1225–1234. [Google Scholar] [CrossRef]
- Bleuel, C.; Große, C.; Taudte, N.; Scherer, J.; Wesenberg, D.; Krauß, G.J.; Grass, G. TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J. Bacteriol. 2005, 187, 6701–6707. [Google Scholar] [CrossRef] [PubMed]
- Crosa, J.H.; Mey, A.R.; Payne, S.M. Iron Transport in Bacteria; ASM Press: Washington, DC, USA, 2004. [Google Scholar]
- Annamalai, R.; Jin, B.; Cao, Z.; Newton, S.M.; Klebba, P.E. Recognition of ferric catecholates by FepA. J. Bacteriol. 2004, 186, 3578–3589. [Google Scholar] [CrossRef]
- Buchanan, S.K.; Smith, B.S.; Venkatramani, L.; Xia, D.; Esser, L.; Palnitkar, M.; Deisenhofer, J. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 1999, 6, 56–63. [Google Scholar] [PubMed]
- Faraldo-Gomez, J.D.; Sansom, M.S. Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 2003, 4, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Fischbach, M.A.; Liu, D.R.; Walsh, C.T. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 2005, 127, 11075–11084. [Google Scholar] [CrossRef] [PubMed]
- Hantke, K. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 2001, 4, 172–177. [Google Scholar] [CrossRef]
- Yeoh, B.S.; Saha, P.; Xiao, X.; Singh, V.; Vijay-Kumar, M. Enterobactin, a metallophore, mitigates the immune responses of macrophages. J. Immunol. 2017, 198 (Suppl. S1), 113–121. [Google Scholar] [CrossRef]
- Ong, S.T.; Ho, J.Z.S.; Ho, B.; Ding, J.L. Iron-withholding strategy in innate immunity. Immunobiology 2006, 211, 295–314. [Google Scholar] [CrossRef] [PubMed]
- Devireddy, L.R.; Teodoro, L.R.; Richard, F.A.; Green, M.R. Induction of apoptosis by a secreted lipocalin that is transcriptionally regulated by IL-3 deprivation. Science 2001, 293, 829–834. [Google Scholar] [CrossRef]
- Goetz, D.H.; Holmes, M.A.; Borregaard, N.; Bluhm, M.E.; Raymond, K.N.; Strong, R.K. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 2002, 10, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Mori, K.; Li, J.Y.; Barasch, J. Iron, lipocalin, and kidney epithelia. Am. J. Phys. Renal Phys. 2003, 285, F9–F18. [Google Scholar]
- Yang, J.; Goetz, D.H.; Li, J.Y.; Wang, W.; Mori, K.; Setlik, D.; Du, T.; Erdjument-Bromage, H.; Tempst, P.; Strong, R.K.; et al. An iron delivery pathway mediated by a lipocalin. Mol. Cell 2002, 10, 1045–1056. [Google Scholar] [CrossRef]
- Kaplan, J. Mechanisms of cellular iron acquisition: Another iron in the fire. Cell 2002, 111, 603–606. [Google Scholar] [CrossRef]
- Flo, T.H.; Smith, K.D.; Sato, S.; Rodriguez, D.J.; Holmes, M.A.; Strong, R.K.; Akira, S.; Aderem, A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 2004, 432, 917–921. [Google Scholar] [CrossRef]
- Nairz, M.; Fritsche, G.; Brunner, P.; Talasz, H.; Hantke, K.; Weiss, G. Interferon-gamma limits the availability of iron for intramacrophage Salmonella typhimurium. Eur. J. Immunol. 2008, 38, 1923–1936. [Google Scholar] [CrossRef]
- Raffatellu, M.; George, M.D.; Akiyama, Y.; Hornsby, M.J.; Nuccio, S.P.; Paixao, T.A.; Butler, B.P.; Chu, H.; Santos, R.L.; Berger, T.; et al. Lipocalin-2resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 2009, 5, 476–486. [Google Scholar] [CrossRef]
- Hantke, K.; Nicholson, G.; Rabsch, W.; Winkelmann, G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. USA 2003, 100, 3677–3682. [Google Scholar] [CrossRef]
- Konopka, K.; Neilands, J.B. Effect of serum albumin on siderophore-mediated utilization of transferrin iron. Biochemistry 1984, 23, 2122–2127. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Lin, H.; Fischbach, M.A.; Liu, D.R.; Walsh, C.T.; Groves, J.T. Enzymatic tailoring of the bacterial siderophore enterobactin alters membrane partitioning and iron acquisition. ACS Chem. Biol. 2006, 1, 29–32. [Google Scholar] [CrossRef]
- Bister, B.; Bischoff, D.; Nicholson, G.J.; Valdebenito, M.; Schneider, K.; Winkelmann, G.; Hantke, K.; Süssmuth, R.D. The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica. Biometals 2004, 17, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Bäumler, A.J.; Tsolis, R.M.; van der Velden, A.W.M.; Stojiljkovic, I.; Anic, S.; Heffron, F. Identification of a new iron regulated locus of Salmonella typhi. Gene 1996, 183, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.I.; Valdebenito, M.; Hantke, K. Salmochelin, the long-overlooked catecholate siderophore of Salmonella. Biometals 2009, 22, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, M.A.; Lin, H.; Liu, D.R.; Walsh, C.T. In vitro characterization of IroB, a pathogen-associated C-glycosyltransferase. Proc. Natl. Acad. Sci. USA 2005, 102, 571–576. [Google Scholar] [CrossRef]
- Crouch, M.L.V.; Castor, M.; Karlinsey, J.E.; Kalhorn, T.; Fang, F.C. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2008, 67, 971–983. [Google Scholar] [CrossRef]
- Bäumler, A.J.; Norris, T.L.; Lasco, T.; Voight, W.; Reissbrodt, R.; Rabsch, W.; Heffron, F. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J. Bacteriol. 1998, 180, 1446–1453. [Google Scholar] [CrossRef]
- Zhu, M.; Valdebenito, M.; Winkelmann, G.; Hantke, K. Functions of the siderophore esterases IroD and IroE in iron-salmochelin utilization. Microbiology 2005, 151, 2363–2372. [Google Scholar] [CrossRef]
- Lim, D.; Kim, K.; Song, M.; Jeong, J.H.; Chang, J.H.; Kim, S.R.; Hong, C.; Im, S.S.; Park, S.H.; Lee, J.C.; et al. Transcriptional regulation of Salmochelin glucosyltransferase by Fur in Salmonella. Biochem. Biophys. Res. Commun. 2020, 529, 70–76. [Google Scholar] [CrossRef]
- Balbontín, R.; Villagra, N.; Pardos de la Gándara, M.; Mora, G.; Figueroa-Bossi, N.; Bossi, L. Expression of IroN, the salmochelin siderophore receptor, requires mRNA activation by RyhB small RNA homologues. Mol. Microbiol. 2016, 100, 139–155. [Google Scholar] [CrossRef]
- Zhang, Z.; Gosset, G.; Barabote, R.; Gonzalez, C.S.; Cuevas, W.A.; Saier, M.H. Functional interactions between the carbon and iron utilization regulators, Crp and Fur, in Escherichia coli. J. Bacteriol. 2005, 187, 980–990. [Google Scholar] [CrossRef]
- Oves-Costales, D.; Kadi, N.; Challis, G.L. The long-overlooked enzymology of a nonribosomal peptide synthetase-independent pathway for virulence-conferring siderophore biosynthesis. Chem. Commun. 2009, 43, 6530–6541. [Google Scholar] [CrossRef]
- De Lorenzo, V.; Martinez, J. Aerobactin production as a virulence factor: A reevaluation. Eur. J. Clin.Microbiol. Infect. Dis. 1988, 7, 621–629. [Google Scholar] [CrossRef]
- Köster, W. ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12. Res. Microbiol. 2001, 152, 291–301. [Google Scholar] [CrossRef]
- Oelschlaeger, T.; Zhang, D.; Schubert, S.; Carniel, E.; Rabsch, W.; Karch, H.; Hacker, J. The high-pathogenicity island is absent in human pathogens of Salmonella enterica subspecies I but present in isolates of subspecies III and VI. J. Bacteriol. 2003, 185, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Wellawa, D.H.; Allan, B.; White, A.P.; Köster, W. Iron-Uptake Systems of Chicken-Associated Salmonella Serovars and Their Role in Colonizing the Avian Host. Microorganisms 2020, 8, 1203. [Google Scholar] [CrossRef] [PubMed]
- Bogomolnaya, L.M.; Tilvawala, R.; Elfenbein, J.R.; Cirillo, J.D.; Andrews-Polymenis, H.L. Linearized Siderophore Products Secreted via MacAB Efflux Pump Protect Salmonella enterica Serovar Typhimurium from Oxidative Stress. mBio 2020, 11, e00528-20. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C.; Kettle, A.J.; Hampton, M.B. Reactive oxygen species and neutrophil function. Annu. Rev. Biochem. 2016, 85, 765–792. [Google Scholar] [CrossRef]
- Imlay, J.A. Pathways of oxidative damage. Annu. Rev. Microbiol. 2003, 57, 395–418. [Google Scholar] [CrossRef]
- Ghssein, G.; Ezzeddine, Z. A Review of Pseudomonas aeruginosa Metallophores: Pyoverdine, Pyochelin and Pseudopaline. Biology 2022, 11, 1711. [Google Scholar] [CrossRef]
- Saha, P.; Yeoh, B.S.; Xiao, X.; Golonka, R.M.; Abokor, A.A.; Wenceslau, C.F.; Shah, Y.M.; Joe, B.; Vijay-Kumar, M. Enterobactin induces the chemokine, interleukin-8, from intestinal epithelia by chelating intracellular iron. Gut Microbes 2020, 12, 1841548. [Google Scholar] [CrossRef]
- Patzer, S.I.; Hantke, K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 1998, 28, 1199–1210. [Google Scholar] [CrossRef]
- Panina, E.M.; Mironov, A.A.; Gelfand, M.S. Comparative genomics of bacterial zinc regulons: Enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. USA 2003, 100, 9912–9917. [Google Scholar] [CrossRef]
- Lewis, D.A.; Klesney-Tait, J.; Lumbley, S.R.; Ward, C.K.; Latimer, J.L.; Ison, C.A.; Hansen, E.J. Identification of the znuA-encoded periplasmic zinc transport protein of Haemophilus ducreyi. Infect. Immun. 1999, 67, 5060–5068. [Google Scholar] [CrossRef]
- Campoy, S.; Jara, M.; Busquets, N.; Pérez De Rozas, A.M.; Badiola, I.; Barbé, J. Role of the high-affinity zinc uptake znuABC system in Salmonella enterica serovar typhimurium virulence. Infect. Immun. 2002, 70, 4721–4725. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Randich, A.M.; Bhattacharyya-Pakrasi, M.; Pakrasi, H.B.; Smith, T.J. Possible regulatory role for the histidine-rich loop in the zinc transport protein, ZnuA. Biochemistry 2007, 46, 8734–8743. [Google Scholar] [CrossRef] [PubMed]
- Yatsunyk, L.A.; Easton, J.A.; Kim, L.R.; Sugarbaker, S.A.; Bennett, B.; Breece, R.M.; Vorontsov, I.I.; Tierney, D.L.; Crowder, M.W.; Rosenzweig, A.C. Structure and metalbinding properties of ZnuA, a periplasmic zinc transporterfrom Escherichia coli. J. Biol. Inorg. Chem. 2008, 13, 271–288. [Google Scholar] [CrossRef]
- Ilari, A.; Alaleona, F.; Tria, G.; Petrarca, P.; Battistoni, A.; Zamparelli, C.; Verzili, D.; Falconi, M.; Chiancone, E. The Salmonella enterica ZinT structure, zinc affinity and interactionwith the high-affinity uptake protein ZnuA provideinsight into the management of periplasmic zinc. Biochim. Biophys. Acta 2014, 1840, 535–544. [Google Scholar] [CrossRef][Green Version]
- Bellotti, D.; Rowińska-Żyrek, M.; Remelli, M. Novel insights into the metal binding ability of ZinT periplasmic protein from Escherichia coli and Salmonella enterica. Dalton Trans. 2020, 49, 9393–9403. [Google Scholar] [CrossRef] [PubMed]
- Petrarca, P.; Ammendola, S.; Pasquali, P.; Battistoni, A. The Zur-regulated ZinT protein is an auxiliary component of the high-affinity ZnuABC zinc transporter that facilitates metal recruitment during severe zinc shortage. J. Bacteriol. 2010, 192, 1553–1564. [Google Scholar] [CrossRef] [PubMed]
- Pesciaroli, M.; Aloisio, F.; Ammendola, S.; Pistoia, C.; Petrucci, P.; Tarantino, M.; Francia, M.; Battistoni, A.; Pasquali, P. An attenuated Salmonella enterica serovar Typhimurium strain lacking the ZnuABC transporter induces protection in a mouse intestinal model of Salmonella infection. Vaccine 2011, 29, 1783–1790. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Z.; Jellbauer, S.; Poe, A.J.; Ton, V.; Pesciaroli, M.; Kehl-Fie, T.E.; Restrepo, N.A.; Hosking, M.P.; Edwards, R.A.; Battistoni, A.; et al. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 2012, 11, 227–239. [Google Scholar] [CrossRef]
- Mey, A.R.; Gómez-Garzón, C.; Payne, S.M. Iron Transport and Metabolism in Escherichia, Shigella, and Salmonella. EcoSal Plus 2021, 9, eESP00342020. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohsen, Y.; Tarchichi, N.; Barakat, R.; Kawtharani, I.; Ghandour, R.; Ezzeddine, Z.; Ghssein, G. The Different Types of Metallophores Produced by Salmonella enterica: A Review. Microbiol. Res. 2023, 14, 1457-1469. https://doi.org/10.3390/microbiolres14030099
Mohsen Y, Tarchichi N, Barakat R, Kawtharani I, Ghandour R, Ezzeddine Z, Ghssein G. The Different Types of Metallophores Produced by Salmonella enterica: A Review. Microbiology Research. 2023; 14(3):1457-1469. https://doi.org/10.3390/microbiolres14030099
Chicago/Turabian StyleMohsen, Yehya, Nathalie Tarchichi, Rana Barakat, Inas Kawtharani, Rayane Ghandour, Zeinab Ezzeddine, and Ghassan Ghssein. 2023. "The Different Types of Metallophores Produced by Salmonella enterica: A Review" Microbiology Research 14, no. 3: 1457-1469. https://doi.org/10.3390/microbiolres14030099
APA StyleMohsen, Y., Tarchichi, N., Barakat, R., Kawtharani, I., Ghandour, R., Ezzeddine, Z., & Ghssein, G. (2023). The Different Types of Metallophores Produced by Salmonella enterica: A Review. Microbiology Research, 14(3), 1457-1469. https://doi.org/10.3390/microbiolres14030099





