Melanin Pathway Determination in Sclerotium cepivorum Berk Using Spectrophotometric Assays, Inhibition Compound, and Protein Validation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Micro-Organisms and Culture Media
2.2. Sclerotia Melanin Extraction
2.3. Analysis of the Melanin Composition
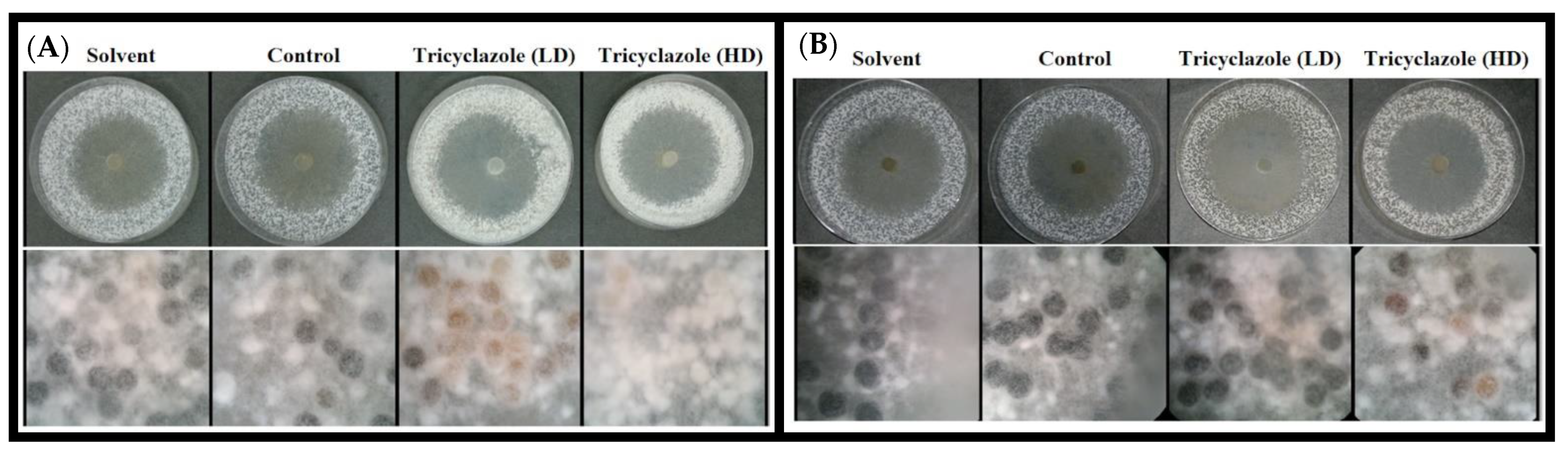
2.4. Susceptibility of S. cepivorum to Tricyclazole
2.5. Isolation of S. cepivorum DNA, RNA, and cDNA
2.6. Expression of S. cepivorum SDH Protein in P. pastoris (X-33) Cells
2.7. Zymogram Assay for Recombinant SDH Activity
2.8. Cell extract Obtention from Magnaporthe grisea
2.9. Microscopy Images
3. Results
3.1. Spectroscopy-Based Analysis of Pigment Extracted from S. cepivorum
3.2. S. cepivorum Melanization Is Affected by Treatment with Tricyclazole
3.3. Identification and Obtainment of a S. cepivorum Scytalone Dehydratase Gene
3.4. S. cepivorum Scytalone Dehydratase Recombinant Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Coley-Smith, J.; King, J. The production by species of Allium of alkyl sulphides and their effect on germination of sclerotia of Sclerotium cepivorum Berk. Ann. Appl. Biol. 1969, 64, 289–301. [Google Scholar] [CrossRef]
- Coley-Smith, J. White rot disease of Allium: Problems of soil-borne diseases in microcosm. Plant Pathol. 1990, 39, 214–222. [Google Scholar] [CrossRef]
- Willetts, H.; Bullock, S. Developmental biology of sclerotia. Mycol. Res. 1992, 96, 801–816. [Google Scholar] [CrossRef]
- Coley-Smith, J.R.; Parfitt, D. Some effects of diallyl bisulphide on sclerotia of Sclerotium cepivorum: Possible novel control method for white rot disease of onions. Pestic. Sci. 1986, 17, 587–594. [Google Scholar] [CrossRef]
- Fullerton, R.; Stewart, A. Chemical control of onion white rot (Sclerotium cepivorum Berk.) in the Pukekohe district of New Zealand. N. Z. J. Crop Hortic. Sci. 1991, 19, 121–127. [Google Scholar] [CrossRef]
- Entwistle, A. Allium white rot and its control. Soil Use Manag. 1990, 6, 201–208. [Google Scholar] [CrossRef]
- Butler, M.J.; Gardiner, R.B.; Day, A.W. Melanin synthesis by Sclerotinia sclerotiorum. Mycologia 2009, 101, 296–304. [Google Scholar] [CrossRef]
- Cordero, R.J.; Casadevall, A. Functions of fungal melanin beyond virulence. Fungal Biol. Rev. 2017, 31, 99–112. [Google Scholar] [CrossRef]
- Arazoe, T.; Miyoshi, K.; Yamato, T.; Ogawa, T.; Ohsato, S.; Arie, T.; Kuwata, S. Tailor-made CRISPR/Cas system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol. Bioeng. 2015, 112, 2543–2549. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. Ribosomal DNA sequence divergence within internal transcribed spacer 1 of the Sclerotiniaceae. Mycologia 1993, 85, 415–427. [Google Scholar] [CrossRef]
- Xu, Z.; Harrington, T.C.; Gleason, M.L.; Batzer, J.C. Phylogenetic placement of plant pathogenic Sclerotium species among teleomorph genera. Mycologia 2010, 102, 337–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.; Xiong, W.; Steinkellner, S.; Feng, J. Deficiency of the melanin biosynthesis genes SCD1 and THR1 affects sclerotial development and vegetative growth, but not pathogenicity, in Sclerotinia sclerotiorum. Mol. Plant Pathol. 2018, 19, 1444–1453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenman, H.C.; Casadevall, A. Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 2012, 93, 931–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattoon, E.R.; Cordero, R.J.; Casadevall, A. Fungal Melanins and Applications in Healthcare, Bioremediation and Industry. J. Fungi 2021, 7, 488. [Google Scholar] [CrossRef]
- Langfelder, K.; Streibel, M.; Jahn, B.; Haase, G.; Brakhage, A.A. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet. Biol. 2003, 38, 143–158. [Google Scholar] [CrossRef]
- Miñambres, G.G.; Conles, M.Y.; Lucini, E.I.; Verdenelli, R.A.; Meriles, J.M.; Zygadlo, J.A. Application of thymol and iprodione to control garlic white rot (Sclerotium cepivorum) and its effect on soil microbial communities. World J. Microbiol. Biotechnol. 2010, 26, 161. [Google Scholar] [CrossRef]
- Wheeler, M.H.; Bell, A.A. Melanins and their importance in pathogenic fungi. In Current Topics in Medical Mycology; Springer: Berlin/Heidelberg, Germany, 1988; pp. 338–387. [Google Scholar]
- Saitoh, Y.; Izumitsu, K.; Morita, A.; Tanaka, C.; Shimizu, K. Cloning of Sal1, a scytalone dehydratase gene involved in melanin biosynthesis in Cochliobolus heterostrophus. Mycoscience 2012, 53, 330–334. [Google Scholar] [CrossRef]
- Sezonov, G.; Joseleau-Petit, D.; d’Ari, R. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef] [Green Version]
- Gadd, G. Effects of media composition and light on colony differentiation and melanin synthesis in Microdochium bolleyi. Trans. Br. Mycol. Soc. 1982, 78, 115–122. [Google Scholar] [CrossRef]
- Froyd, J.; Paget, C.; Guse, L.; Dreikorn, B.; Pafford, J. Tricyclazole: A new systemic fungicide for control of Pyricularia oryzae on rice. Phytopathology 1976, 66, 1135–1139. [Google Scholar] [CrossRef]
- Raeder, U.; Broda, P. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1985, 1, 17–20. [Google Scholar] [CrossRef]
- Trujillo-Esquivel, E.; Franco, B.; Flores-Martínez, A.; Ponce-Noyola, P.; Mora-Montes, H.M. Purification of single-stranded cDNA based on RNA degradation treatment and adsorption chromatography. Nucleosides Nucleotides Nucleic Acids 2016, 35, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, M.H. Comparisons of fungal melanin biosynthesis in ascomycetous, imperfect and basidiomycetous fungi. Trans. Br. Mycol. Soc. 1983, 81, 29–36. [Google Scholar] [CrossRef]
- Simon, W.; Clerc, T. Elucidación Estructural de Compuestos Orgánicos por Métodos Espectroscópicos; Alhambra: Madrid, Spain, 1970. [Google Scholar]
- Rajagopal, K.; Kathiravan, G.; Karthikeyan, S. Extraction and characterization of melanin from Phomopsis: A phellophytic fungi isolated from Azadirachta indica A. Juss. Afr. J. Microbiol. Res. 2011, 5, 762–766. [Google Scholar]
- Pretsch, E.; Clerc, T.; Seibl, J.; Simon, W. Tables of Spectral Data for Structure Determination of Organic Compounds; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Raman, N.M.; Ramasamy, S. Genetic validation and spectroscopic detailing of DHN-melanin extracted from an environmental fungus. Biochem. Biophys. Rep. 2017, 12, 98–107. [Google Scholar] [CrossRef]
- Manini, P.; Lino, V.; D’Errico, G.; Reale, S.; Napolitano, A.; De Angelis, F.; d’Ischia, M. “Blackness” is an index of redox complexity in melanin polymers. Polym. Chem. 2020, 11, 5005–5010. [Google Scholar] [CrossRef]
- Chatterjee, S.; Prados-Rosales, R.; Itin, B.; Casadevall, A.; Stark, R.E. Solid-state NMR reveals the carbon-based molecular architecture of Cryptococcus neoformans fungal eumelanins in the cell wall. J. Biol. Chem. 2015, 290, 13779–13790. [Google Scholar] [CrossRef] [Green Version]
- Kang, X.; Kirui, A.; Muszyński, A.; Widanage, M.C.D.; Chen, A.; Azadi, P.; Wang, P.; Mentink-Vigier, F.; Wang, T. Molecular architecture of fungal cell walls revealed by solid-state NMR. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Di Pietro, M.E.; Mannu, A.; Mele, A. NMR determination of free fatty acids in vegetable oils. Processes 2020, 8, 410. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.-Q.; Huang, X.; Wang, J.-X.; Zhou, M.-G. Resistance development in rice blast disease caused by Magnaporthe grisea to tricyclazole. Pestic. Biochem. Physiol. 2009, 94, 43–47. [Google Scholar] [CrossRef]
- Butler, M.; Day, A. Fungal melanins: A review. Can. J. Microbiol. 1998, 44, 1115–1136. [Google Scholar] [CrossRef]
- Amselem, J.; Cuomo, C.A.; van Kan, J.A.; Viaud, M.; Benito, E.P.; Couloux, A.; Coutinho, P.M.; de Vries, R.P.; Dyer, P.S.; Fillinger, S. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011, 7, e1002230. [Google Scholar] [CrossRef] [Green Version]
- Jordan, D.B.; Basarab, G.S.; Steffens, J.J.; Lundqvist, T.; Pfrogner, B.R.; Schwartz, R.S.; Wawrzak, Z. Catalytic mechanism of scytalone dehydratase from Magnaporthe grisea. Pestic. Sci. 1999, 55, 277–280. [Google Scholar] [CrossRef]
- Tsuji, G.; Takeda, T.; Furusawa, I.; Horino, O.; Kubo, Y. Carpropamid, an Anti-Rice Blast Fungicide, Inhibits Scytalone Dehydratase Activity and Appressorial Penetration in Colletotrichum lagenarium. Pestic. Biochem. Physiol. 1997, 57, 211–219. [Google Scholar] [CrossRef]
- Liachko, I.; Youngblood, R.A.; Tsui, K.; Bubb, K.L.; Queitsch, C.; Raghuraman, M.; Nislow, C.; Brewer, B.J.; Dunham, M.J. GC-rich DNA elements enable replication origin activity in the methylotrophic yeast Pichia pastoris. PLoS Genet. 2014, 10, e1004169. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, T.; Rice, J.; Hodge, C.N.; Basarab, G.S.; Pierce, J.; Lindqvist, Y. Crystal structure of scytalone dehydratase—A disease determinant of the rice pathogen, Magnaporthe grisea. Structure 1994, 2, 937–944. [Google Scholar] [CrossRef] [Green Version]
- Allam, N.G.; El-Zaher, E.A. Protective role of Aspergillus fumigatus melanin against ultraviolet (UV) irradiation and Bjerkandera adusta melanin as a candidate vaccine against systemic candidiasis. Afr. J. Biotechnol. 2012, 11, 6566–6577. [Google Scholar]
- Potgieter, H.; Alexander, M. Susceptibility and resistance of several fungi to microbial lysis. J. Bacteriol. 1966, 91, 1526–1532. [Google Scholar] [CrossRef] [Green Version]
- Chumley, F.G.; Valent, B. Genetic analysis of melanin-deficient, nonpathogenic mutants of Magnaporthe grisea. Mol. Plant-Microbe Interact 1990, 3, 135–143. [Google Scholar] [CrossRef]
- Mahmoud, Y.A.; Sammour, R.H.; Mustafa, A.-Z.M.; Alhozeim, R.A. Role of sclerotia in the aggressiveness and pathogenicity of Sclerotium cepivorum. Asian J. Microbiol. Biotechnol. Environ. Sci. 2021, 23, 16–23. [Google Scholar]
- Moriwaki, A.; Kihara, J.; Kobayashi, T.; Tokunaga, T.; Arase, S.; Honda, Y. Insertional mutagenesis and characterization of a polyketide synthase gene (PKS1) required for melanin biosynthesis in Bipolaris oryzae. FEMS Microbiol. Lett. 2004, 238, 1–8. [Google Scholar] [PubMed]
- Rehnstrom, A.; Free, S. The isolation and characterization of melanin-deficient mutants of Monilinia fructicola. Physiol. Mol. Plant Pathol. 1996, 49, 321–330. [Google Scholar] [CrossRef]
- Thines, E.; Heidrun, A.; Weber, R.W. Fungal secondary metabolites as inhibitors of infection-related morphogenesis in phytopathogenic fungi. Mycol. Res. 2004, 108, 14–25. [Google Scholar] [CrossRef]
- Bell, A.A.; Wheeler, M.H. Biosynthesis and functions of fungal melanins. Annu. Rev. Phytopathol. 1986, 24, 411–451. [Google Scholar] [CrossRef]
- De la Rosa, J.M.; Martin-Sanchez, P.M.; Sanchez-Cortes, S.; Hermosin, B.; Knicker, H.; Saiz-Jimenez, C. Structure of melanins from the fungi Ochroconis lascauxensis and Ochroconis anomala contaminating rock art in the Lascaux Cave. Sci. Rep. 2017, 7, 13441. [Google Scholar] [CrossRef] [Green Version]
- Pal, A.K.; Gajjar, D.U.; Vasavada, A.R. DOPA and DHN pathway orchestrate melanin synthesis in Aspergillus species. Med. Mycol. 2014, 52, 10–18. [Google Scholar]
- Suryanarayanan, T.S.; Ravishankar, J.P.; Venkatesan, G.; Murali, T.S. Characterization of the melanin pigment of a cosmopolitan fungal endophyte. Mycol. Res. 2004, 108, 974–978. [Google Scholar] [CrossRef]
- Solano, F. Melanins: Skin pigments and much more—Types, structural models, biological functions, and formation routes. New J. Sci. 2014, 2014, 498276. [Google Scholar] [CrossRef] [Green Version]
- Correa, N.; Covarrubias, C.; Rodas, P.I.; Hermosilla, G.; Olate, V.R.; Valdés, C.; Meyer, W.; Magne, F.; Tapia, C.V. Differential antifungal activity of human and cryptococcal melanins with structural discrepancies. Front. Microbiol. 2017, 8, 1292. [Google Scholar] [CrossRef]
- Tokousbalides, M.C.; Sisler, H. Site of inhibition by tricyclazole in the melanin biosynthetic pathway of Verticillium dahliae. Pestic. Biochem. Physiol. 1979, 11, 64–73. [Google Scholar] [CrossRef]
- Andersson, A.; Jordan, D.; Schneider, G.; Lindqvist, Y. Crystal structure of the ternary complex of 1, 3, 8-trihydroxynaphthalene reductase from Magnaporthe grisea with NADPH and an active-site inhibitor. Structure 1996, 4, 1161–1170. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Min, H.; Ye, L. The negative interaction between the degradation of phenanthrene and tricyclazole in medium, soil and soil/compost mixture. Biodegradation 2008, 19, 695–703. [Google Scholar] [CrossRef]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.-L.; Chi, Z.; Chen, L.; Liu, G.-L.; Hu, Z.; Chi, Z.-M. Molecular evolution and regulation of DHN melanin-related gene clusters are closely related to adaptation of different melanin-producing fungi. Genomics 2021, 113, 1962–1975. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Cuevas, R.I. Clonación y Expresón Heteróloga de la Scitalona Deshidratasa de Sclerotium Cepivorum Berk. Master’s Thesis, Department of Biology, University of Guanajuato, Guanajuato, Mexico, 2015. [Google Scholar]
- Yamada, N.; Motoyama, T.; Nakasako, M.; Kagabu, S.; Kudo, T.; Yamaguchi, I. Enzymatic characterization of scytalone dehydratase Val75Met variant found in melanin biosynthesis dehydratase inhibitor (MBI-D) resistant strains of the rice blast fungus. Biosci. Biotechnol. Biochem. 2004, 68, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Chizhov, D.L.; Sosnovskikh, V.Y.; Pryadeina, M.V.; Burgart, Y.V.; Saloutin, V.I.; Charushin, V.N. The first synthesis of 4-unsubstituted 3-(trifluoroacetyl) coumarins by the Knoevenagel condensation of salicylaldehydes with ethyl trifluoroacetoacetate followed by chromene-coumarin recyclization. Synlett 2008, 19, 281–285. [Google Scholar] [CrossRef]
- Xu, C.; Yang, G.; Wang, C.; Fan, S.; Xie, L.; Gao, Y. An efficient solvent-free synthesis of 2-hydroxy-2-(trifluoromethyl)-2H-chromenes using silica-immobilized L-proline. Molecules 2013, 18, 11964–11977. [Google Scholar] [CrossRef] [Green Version]
- Valero-Jiménez, C.A.; Steentjes, M.B.; Slot, J.C.; Shi-Kunne, X.; Scholten, O.E.; van Kan, J.A. Dynamicsin Secondary Metabolite Gene Clustersin Otherwise Highly Syntenic and Stable Genomes in the Fungal Genus. Genome Biol. Evol. 2020, 12, 2491–2507. [Google Scholar]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Dereeper, A.; Audic, S.; Claverie, J.M.; Blanc, G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 2010, 10, 8. [Google Scholar] [CrossRef] [Green Version]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anisimova, M.; Gascuel, O. Approximate likelihood ratio test for branchs: A fast, accurate and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef]
- Chevenet, F.; Brun, C.; Banuls, A.L.; Jacq, B.; Chisten, R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006, 7, 439. [Google Scholar] [CrossRef] [Green Version]
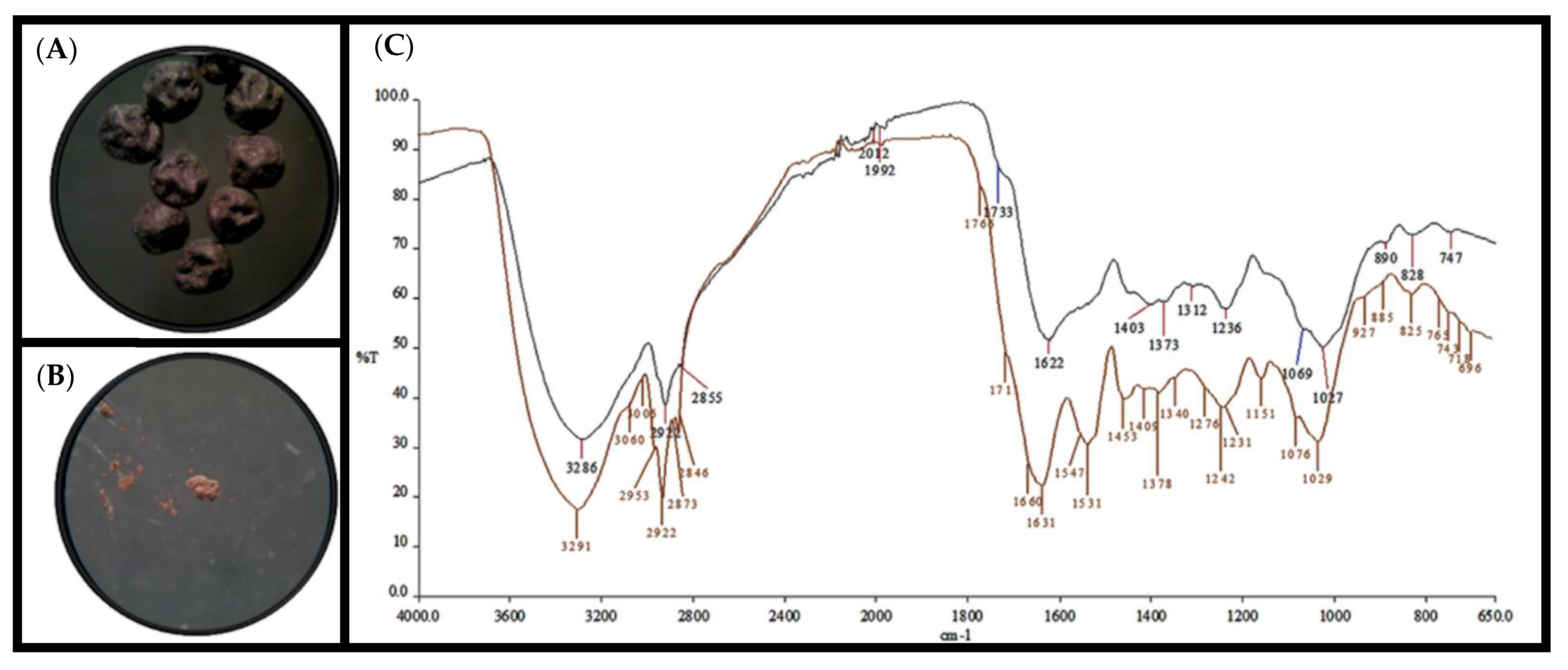

| Purified Compound | Direct Sample | ||
|---|---|---|---|
| Frequency (cm−1) | Signal Generating Group | Frequency (cm−1) | Signal Generating Group |
| 3291 | -O-H Elongation vibration. Polymeric association. | 3286 | -O-H Elongation vibration. Polymeric association. |
| 3060 3005 2953 | =C-H Elongation vibration. Aromatic compound. | 2922 | =C-H Elongation vibration. Aromatic compound. |
| 1631 1547 1531 1453 | -C=C- Deformation (in plane) aromatic compound. | 1622 | -C=C- Deformation (in plane) aromatic compound. |
| 1409 1378 | -C-O Elongation vibration. Characteristic of phenol and derivatives. | 1403 1373 | -C-O Elongation vibration. Characteristic of phenol and derivatives. |
| 1242 | -O-H Deformation (in plane). Characteristic of phenol and derivatives. | 1236 | -O-H Deformation (in plane). Characteristic of phenol and derivatives. |
| 1151 1076 1029 | -C-H Deformation (in plane) and aromatic ring substitution 1:2:3. | 1069 1027 | -C-H Deformation (in plane) and aromatic ring substitution 1:2:3. |
| 885 765 718 | -C-H Deformation (out plane) and aromatic ring substitution 1:2:3. | 890 828 747 | -C-H Deformation (out plane) and aromatic ring substitution 1:2:3. |
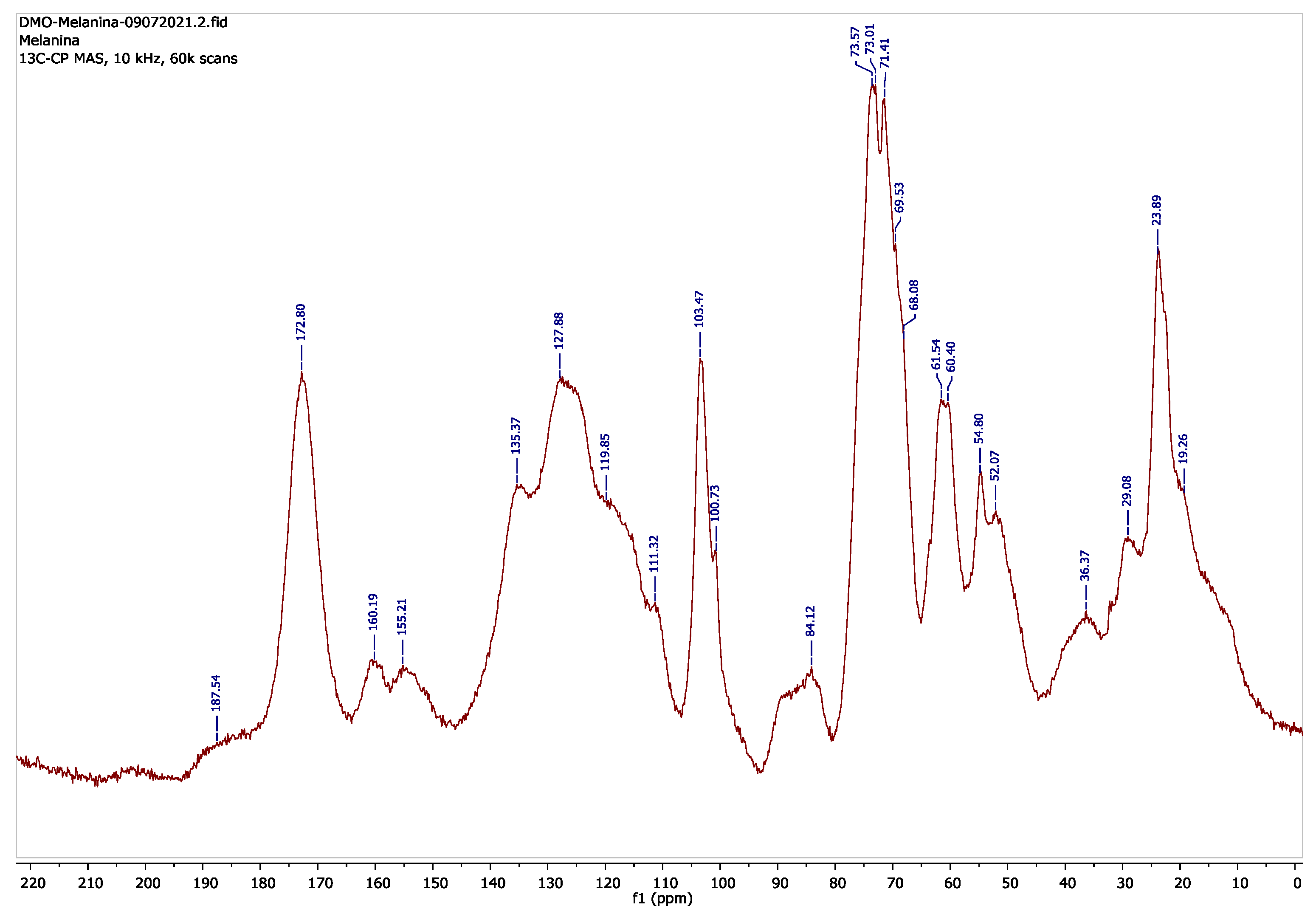
| Chemical Shift, ppm | Chemical Grouping | Reference Materials | Literature Refs. |
|---|---|---|---|
| 172.80 | -CONH | Chitin | [31,32] |
| 160.19 | Aromatic -C-, O-naphthyl | Poly 1,8 DHN | [31] |
| 135.77–111.32 | Aromatic -C-, -C=C- | Poly 1,8 DHN, likely oleoyl | [31,33] |
| 103.47, 100.73 | Aromatic –C-, -O-CH-O- | Chitin, glucan | [32] |
| 84.12 | -CHOH | α-1.3 glucan | [32] |
| 73.57–71.41 | -CHOH, -CHO- | Chitin, α-1.3 glucan | [32] |
| 61.54 | -CH2OH | Chitin | [32] |
| 60.40 | -CH2OH | α-1.3 glucan | [32] |
| 54.80 | -CH-NH- | Chitin | [32] |
| 29.08 | -(CH2)- (C4–C7, C2–C16) | Likely oleoyl | [33] |
| 23.89 | -C(=O)-CH3, -(CH2)- (C3) | Chitin, likely oleoyl | [32,33] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salazar-García, L.M.; Ortega-Cuevas, R.I.; Martínez-Álvarez, J.A.; González-Hernández, S.E.; Martínez-Álvarez, R.A.; Mendoza-Olivares, D.; Vázquez, M.Á.; Flores-Martínez, A.; Ponce-Noyola, P. Melanin Pathway Determination in Sclerotium cepivorum Berk Using Spectrophotometric Assays, Inhibition Compound, and Protein Validation. Microbiol. Res. 2022, 13, 152-166. https://doi.org/10.3390/microbiolres13020013
Salazar-García LM, Ortega-Cuevas RI, Martínez-Álvarez JA, González-Hernández SE, Martínez-Álvarez RA, Mendoza-Olivares D, Vázquez MÁ, Flores-Martínez A, Ponce-Noyola P. Melanin Pathway Determination in Sclerotium cepivorum Berk Using Spectrophotometric Assays, Inhibition Compound, and Protein Validation. Microbiology Research. 2022; 13(2):152-166. https://doi.org/10.3390/microbiolres13020013
Chicago/Turabian StyleSalazar-García, Luis M., Rocío Ivette Ortega-Cuevas, José A. Martínez-Álvarez, Sandra E. González-Hernández, Román Antonio Martínez-Álvarez, Diana Mendoza-Olivares, Miguel Ángel Vázquez, Alberto Flores-Martínez, and Patricia Ponce-Noyola. 2022. "Melanin Pathway Determination in Sclerotium cepivorum Berk Using Spectrophotometric Assays, Inhibition Compound, and Protein Validation" Microbiology Research 13, no. 2: 152-166. https://doi.org/10.3390/microbiolres13020013
APA StyleSalazar-García, L. M., Ortega-Cuevas, R. I., Martínez-Álvarez, J. A., González-Hernández, S. E., Martínez-Álvarez, R. A., Mendoza-Olivares, D., Vázquez, M. Á., Flores-Martínez, A., & Ponce-Noyola, P. (2022). Melanin Pathway Determination in Sclerotium cepivorum Berk Using Spectrophotometric Assays, Inhibition Compound, and Protein Validation. Microbiology Research, 13(2), 152-166. https://doi.org/10.3390/microbiolres13020013







