Sex Differences in Fatty Acid Metabolism and Blood Pressure Response to Dietary Salt in Humans
Abstract
1. Introduction
2. Materials and Methods
3. Results
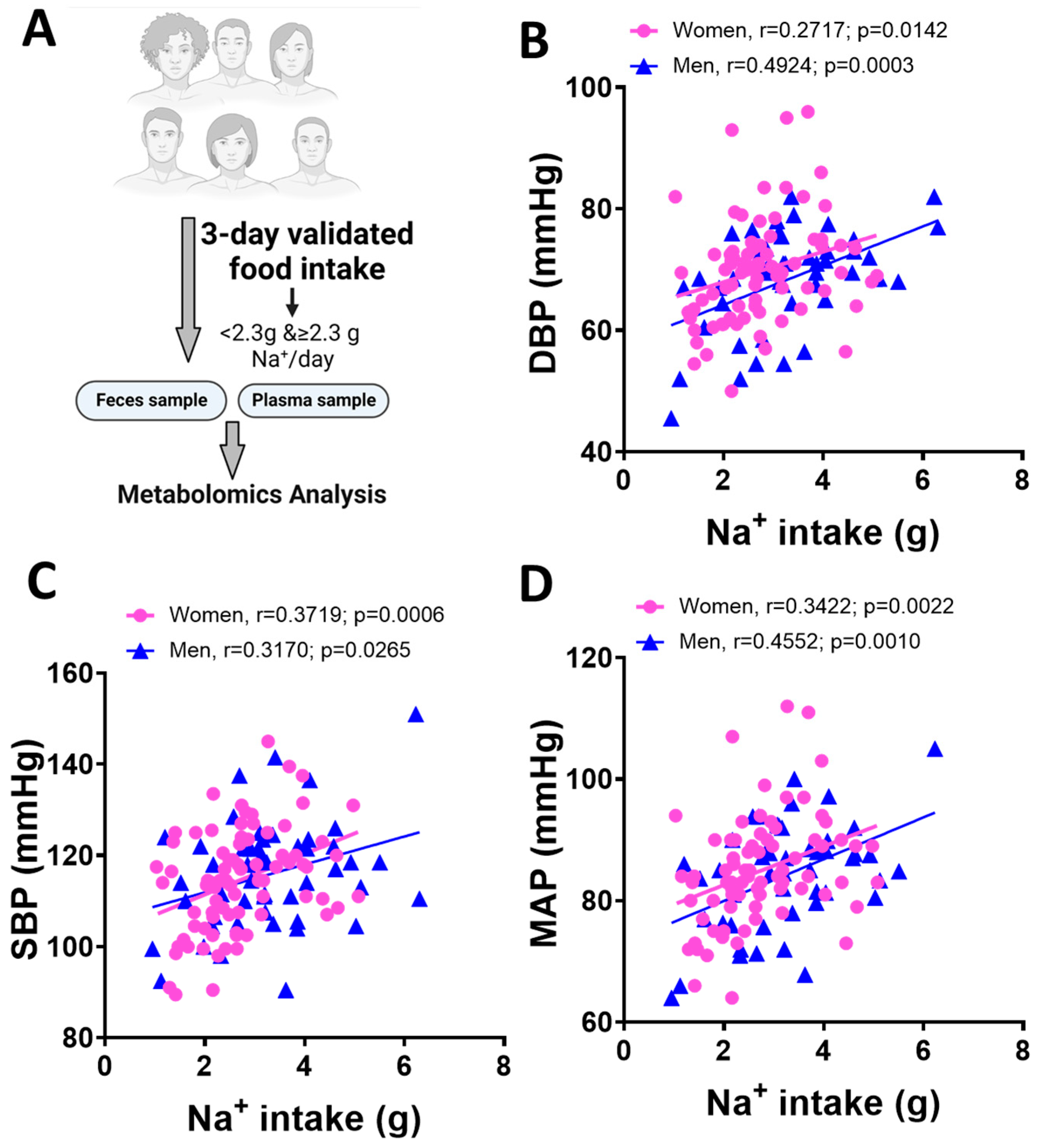
3.1. Blood Pressure and Dietary Na+ Intake in Men and Women
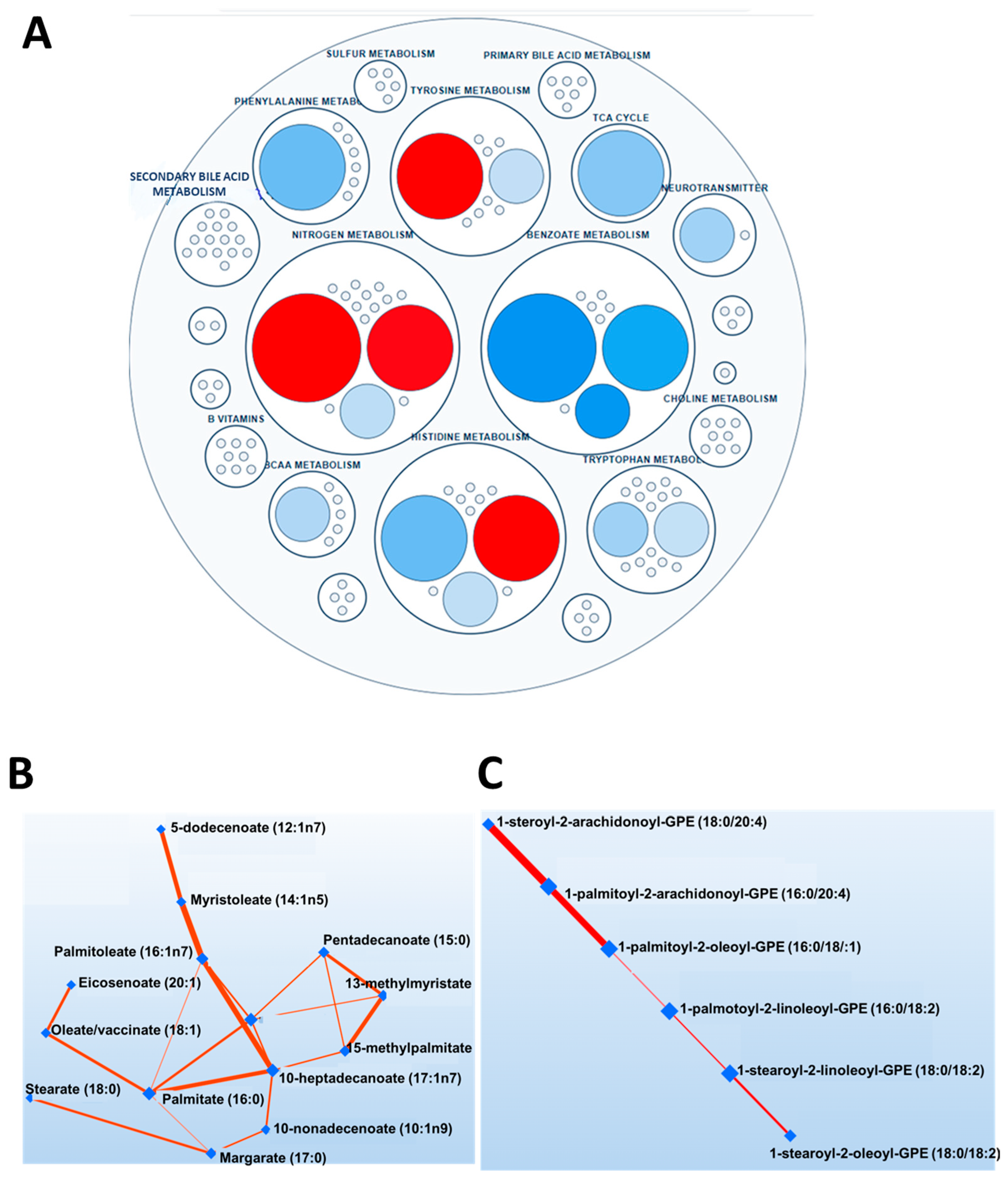
3.2. Metabolomics Analysis Results
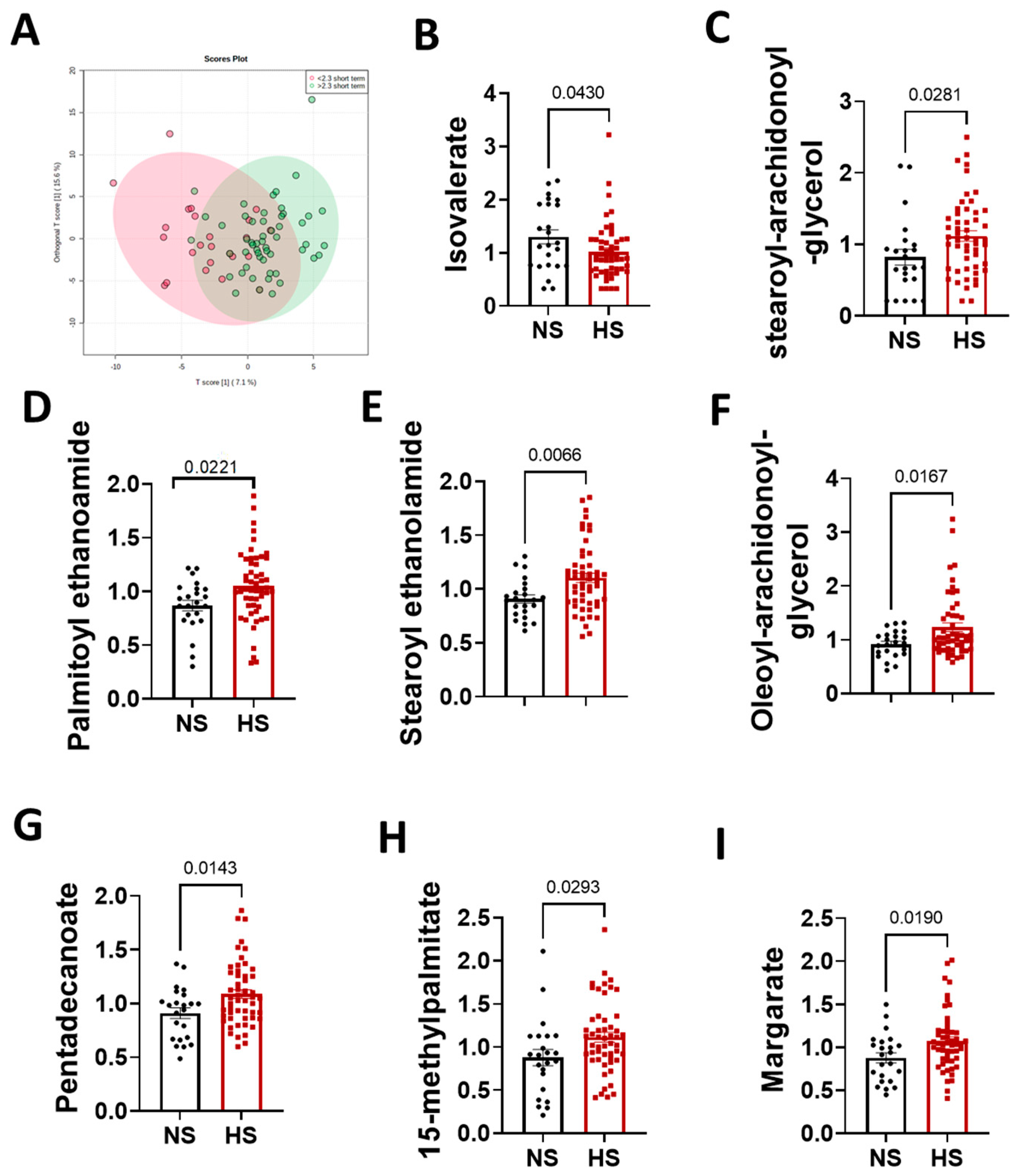
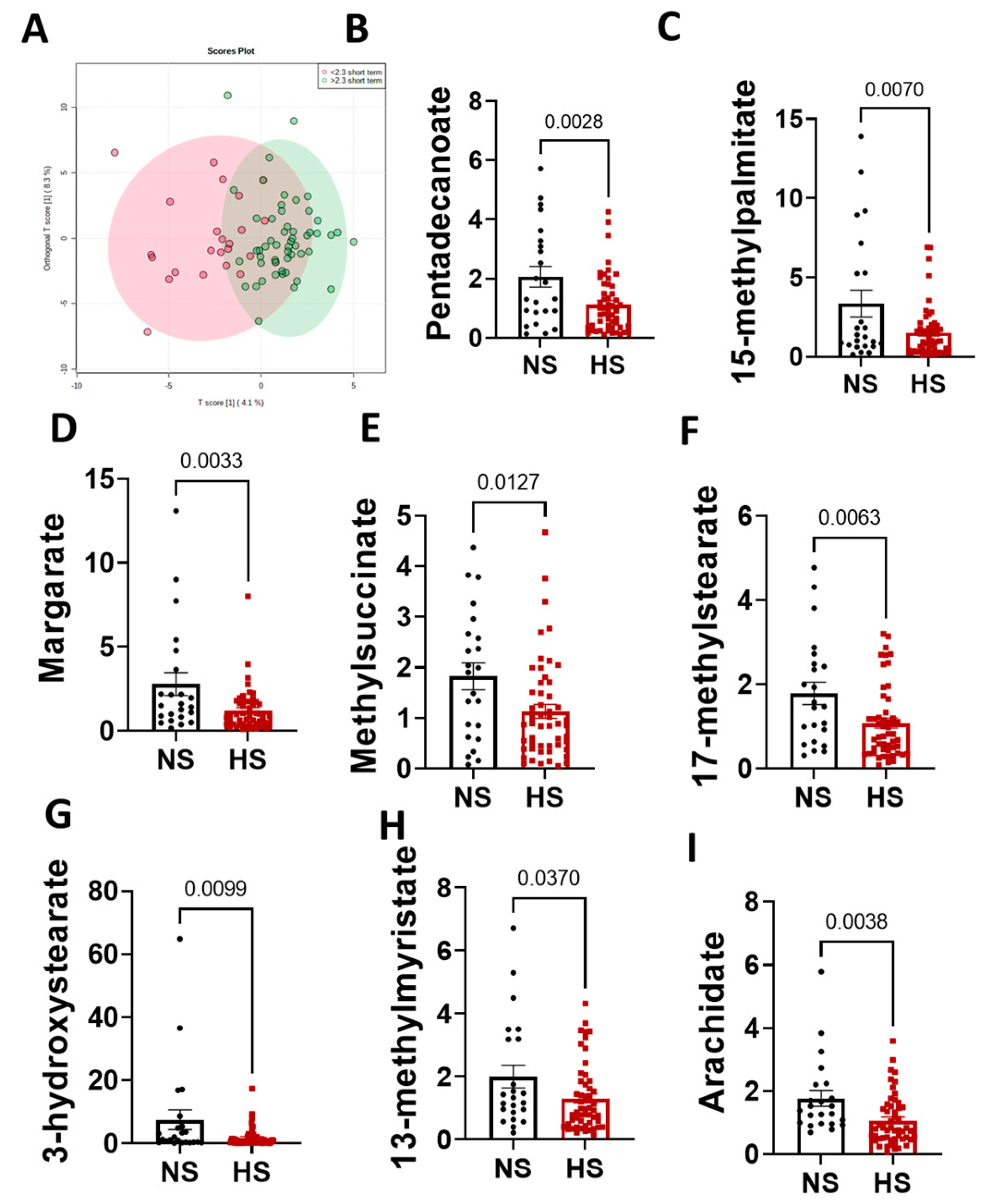
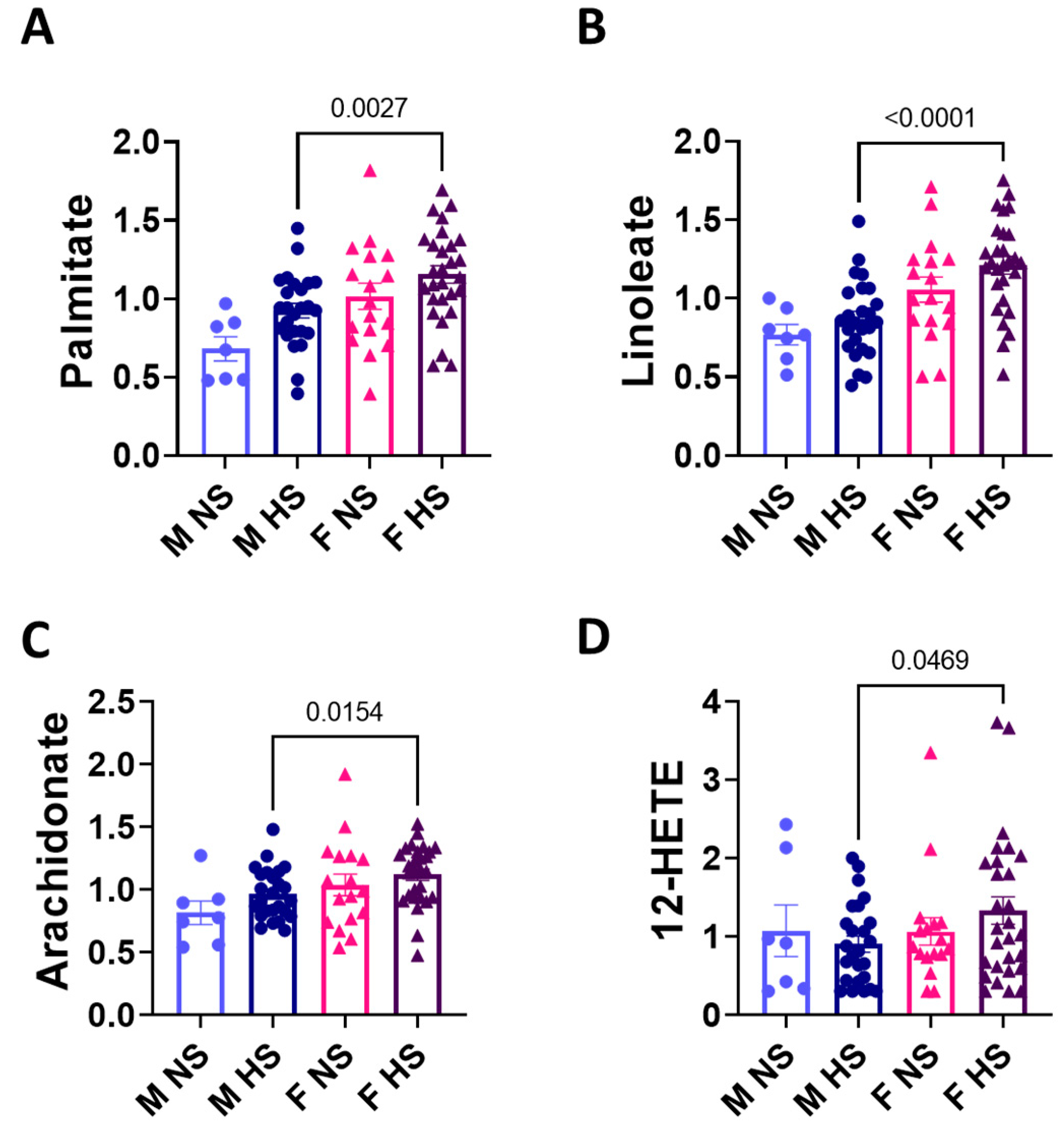
3.3. Sex Differences in the Arachidonic Acid Pathway
3.4. Elevated Sodium-Induced Changes in Genes Related to Fatty Acids Signaling in Monocytes Isolated from Women
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tsao, C.W.; Aday, A.W.; Almarzooq, Z.I.; Alonso, A.; Beaton, A.Z.; Bittencourt, M.S.; Boehme, A.K.; Buxton, A.E.; Carson, A.P.; Commodore-Mensah, Y.; et al. Heart Disease and Stroke Statistics-2022 Update: A Report From the American Heart Association. Circulation 2022, 145, e153–e639. [Google Scholar] [CrossRef]
- Murray, C.J.; Lopez, A.D. Measuring the global burden of disease. N. Engl. J. Med. 2013, 369, 448–457. [Google Scholar] [CrossRef]
- He, F.J.; Li, J.; Macgregor, G.A. Effect of longer-term modest salt reduction on blood pressure. Cochrane Database Syst. Rev. 2003, 3, CD004937. [Google Scholar]
- Connelly, P.J.; Currie, G.; Delles, C. Sex Differences in the Prevalence, Outcomes and Management of Hypertension. Curr. Hypertens. Rep. 2022, 24, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Kelly, T.N.; Rebholz, C.M.; Gu, D.; Hixson, J.E.; Rice, T.K.; Cao, J.; Chen, J.; Li, J.; Lu, F.; Ma, J.; et al. Analysis of sex hormone genes reveals gender differences in the genetic etiology of blood pressure salt sensitivity: The GenSalt study. Am. J. Hypertens. 2013, 26, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Deger, M.S.; Kang, H.; Ikizler, T.A.; Titze, J.; Gore, J.C. Sex differences in sodium deposition in human muscle and skin. Magn. Reson. Imaging 2017, 36, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri Barbaro, N.; Van Beusecum, J.; Xiao, L.; do Carmo, L.; Pitzer, A.; Loperena, R.; Foss, J.D.; Elijovich, F.; Laffer, C.L.; Montaniel, K.R.; et al. Sodium activates human monocytes via the NADPH oxidase and isolevuglandin formation. Cardiovasc. Res. 2021, 117, 1358–1371. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, J.F.; Aden, L.A.; Barbaro, N.R.; Van Beusecum, J.P.; Xiao, L.; Simmons, A.J.; Warden, C.; Pasic, L.; Himmel, L.E.; Washington, M.K.; et al. High dietary salt-induced dendritic cell activation underlies microbial dysbiosis-associated hypertension. JCI Insight 2019, 5, e126241. [Google Scholar] [CrossRef] [PubMed]
- Kirabo, A. A new paradigm of sodium regulation in inflammation and hypertension. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2017, 313, R706–R710. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.G.; Guzik, T.J.; Lob, H.E.; Madhur, M.S.; Marvar, P.J.; Thabet, S.R.; Vinh, A.; Weyand, C.M. Inflammation, immunity, and hypertension. Hypertension 2011, 57, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Pitzer, A.; Elijovich, F.; Laffer, C.L.; Ertuglu, L.A.; Sahinoz, M.; Saleem, M.; Krishnan, J.; Dola, T.; Aden, L.A.; Sheng, Q.; et al. DC ENaC-Dependent Inflammasome Activation Contributes to Salt-Sensitive Hypertension. Circ. Res. 2022, 131, 328–344. [Google Scholar] [CrossRef]
- Wu, J.; Saleh, M.A.; Kirabo, A.; Itani, H.A.; Montaniel, K.R.; Xiao, L.; Chen, W.; Mernaugh, R.L.; Cai, H.; Bernstein, K.E.; et al. Immune activation caused by vascular oxidation promotes fibrosis and hypertension. J. Clin. Investig. 2016, 126, 50–67. [Google Scholar] [CrossRef]
- Kleinewietfeld, M.; Manzel, A.; Titze, J.; Kvakan, H.; Yosef, N.; Linker, R.A.; Muller, D.N.; Hafler, D.A. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature 2013, 496, 518–522. [Google Scholar] [CrossRef]
- Safa, K.; Ohori, S.; Borges, T.J.; Uehara, M.; Batal, I.; Shimizu, T.; Magee, C.N.; Belizaire, R.; Abdi, R.; Wu, C.; et al. Salt Accelerates Allograft Rejection through Serum- and Glucocorticoid-Regulated Kinase-1-Dependent Inhibition of Regulatory T Cells. J. Am. Soc. Nephrol. 2015, 26, 2341–2347. [Google Scholar] [CrossRef]
- McMaster, W.G.; Kirabo, A.; Madhur, M.S.; Harrison, D.G. Inflammation, immunity, and hypertensive end-organ damage. Circ. Res. 2015, 116, 1022–1033. [Google Scholar] [CrossRef]
- Masenga, S.K.; Hamooya, B.; Hangoma, J.; Hayumbu, V.; Ertuglu, L.A.; Ishimwe, J.; Rahman, S.; Saleem, M.; Laffer, C.L.; Elijovich, F.; et al. Recent advances in modulation of cardiovascular diseases by the gut microbiota. J. Hum. Hypertens. 2022, 36, 952–959. [Google Scholar] [CrossRef]
- Tang, Z.Z.; Chen, G.; Hong, Q.; Huang, S.; Smith, H.M.; Shah, R.D.; Scholz, M.; Ferguson, J.F. Multi-Omic Analysis of the Microbiome and Metabolome in Healthy Subjects Reveals Microbiome-Dependent Relationships Between Diet and Metabolites. Front. Genet. 2019, 10, 454. [Google Scholar] [CrossRef] [PubMed]
- Telleria, O.; Alboniga, O.E.; Clos-Garcia, M.; Nafria-Jimenez, B.; Cubiella, J.; Bujanda, L.; Falcon-Perez, J.M. A Comprehensive Metabolomics Analysis of Fecal Samples from Advanced Adenoma and Colorectal Cancer Patients. Metabolites 2022, 12, 550. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.E.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.C.; Conklin, S.M.; Manuck, S.B.; Yao, J.K.; Muldoon, M.F. Long-chain omega-3 fatty acids and blood pressure. Am. J. Hypertens. 2011, 24, 1121–1126. [Google Scholar] [CrossRef]
- Simon, J.A.; Fong, J.; Bernert, J.T., Jr. Serum fatty acids and blood pressure. Hypertension 1996, 27, 303–307. [Google Scholar] [CrossRef]
- Zhang, X.; Ritonja, J.A.; Zhou, N.; Chen, B.E.; Li, X. Omega-3 Polyunsaturated Fatty Acids Intake and Blood Pressure: A Dose-Response Meta-Analysis of Randomized Controlled Trials. J. Am. Heart Assoc. 2022, 11, e025071. [Google Scholar] [CrossRef]
- Koliada, A.; Moseiko, V.; Romanenko, M.; Lushchak, O.; Kryzhanovska, N.; Guryanov, V.; Vaiserman, A. Sex differences in the phylum-level human gut microbiota composition. BMC Microbiol. 2021, 21, 131. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, M.; Xue, J.; Huang, J.; Zhuang, R.; Zhou, X.; Zhang, H.; Fu, Q.; Hao, Y. Body Mass Index Differences in the Gut Microbiota Are Gender Specific. Front. Microbiol. 2018, 9, 1250. [Google Scholar] [CrossRef]
- Haro, C.; Rangel-Zuniga, O.A.; Alcala-Diaz, J.F.; Gomez-Delgado, F.; Perez-Martinez, P.; Delgado-Lista, J.; Quintana-Navarro, G.M.; Landa, B.B.; Navas-Cortes, J.A.; Tena-Sempere, M.; et al. Intestinal Microbiota Is Influenced by Gender and Body Mass Index. PLoS ONE 2016, 11, e0154090. [Google Scholar] [CrossRef]
- Karastergiou, K.; Smith, S.R.; Greenberg, A.S.; Fried, S.K. Sex differences in human adipose tissues—The biology of pear shape. Biol. Sex Differ. 2012, 3, 13. [Google Scholar] [CrossRef]
- Hall, M.E.; Hall, J.E. Sex Differences in Hypertension: Related to Genes, Jean Sizes, and Salt Sensitivity? Hypertension 2022, 79, 47–49. [Google Scholar] [CrossRef]
- Sabbatini, A.R.; Kararigas, G. Estrogen-related mechanisms in sex differences of hypertension and target organ damage. Biol. Sex Differ. 2020, 11, 31. [Google Scholar] [CrossRef]
- Ji, H.; Kim, A.; Ebinger, J.E.; Niiranen, T.J.; Claggett, B.L.; Bairey Merz, C.N.; Cheng, S. Sex Differences in Blood Pressure Trajectories Over the Life Course. JAMA Cardiol. 2020, 5, 19–26. [Google Scholar] [CrossRef]
- Hinojosa-Laborde, C.; Lange, D.L.; Haywood, J.R. Role of female sex hormones in the development and reversal of dahl hypertension. Hypertension 2000, 35, 484–489. [Google Scholar] [CrossRef]
- Tominaga, T.; Suzuki, H.; Ogata, Y.; Matsukawa, S.; Saruta, T. The role of sex hormones and sodium intake in postmenopausal hypertension. J. Hum. Hypertens. 1991, 5, 495–500. [Google Scholar]
- Ou-Yang, Y.N.; Yuan, M.D.; Yang, Z.M.; Min, Z.; Jin, Y.X.; Tian, Z.M. Revealing the Pathogenesis of Salt-Sensitive Hypertension in Dahl Salt-Sensitive Rats through Integrated Multi-Omics Analysis. Metabolites 2022, 12, 1076. [Google Scholar] [CrossRef]
- He, W.J.; Li, C.; Mi, X.; Shi, M.; Gu, X.; Bazzano, L.A.; Razavi, A.C.; Nierenberg, J.L.; Dorans, K.; He, H.; et al. An untargeted metabolomics study of blood pressure: Findings from the Bogalusa Heart Study. J. Hypertens. 2020, 38, 1302–1311. [Google Scholar] [CrossRef]
- Lees, H.J.; Swann, J.R.; Wilson, I.D.; Nicholson, J.K.; Holmes, E. Hippurate: The natural history of a mammalian-microbial cometabolite. J. Proteome Res. 2013, 12, 1527–1546. [Google Scholar] [CrossRef]
- Jonker, G.J.; Visscher, C.A.; de Zeeuw, D.; Huisman, R.M.; Piers, D.A.; Beekhuis, H.; van der Hem, G.K. Changes in renal function induced by ACE-inhibition in the conscious two-kidney, one-clip Goldblatt hypertensive dog. Nephron 1992, 60, 226–231. [Google Scholar] [CrossRef]
- Dias, P.; Pourova, J.; Voprsalova, M.; Nejmanova, I.; Mladenka, P. 3-Hydroxyphenylacetic Acid: A Blood Pressure-Reducing Flavonoid Metabolite. Nutrients 2022, 14, 328. [Google Scholar] [CrossRef]
- Shi, M.; He, J.; Li, C.; Lu, X.; He, W.J.; Cao, J.; Chen, J.; Chen, J.C.; Bazzano, L.A.; Li, J.X.; et al. Metabolomics study of blood pressure salt-sensitivity and hypertension. Nutr. Metab. Cardiovasc. Dis. 2022, 32, 1681–1692. [Google Scholar] [CrossRef]
- Yang, P.; Zhao, X.; Zhou, L.; Jin, Y.; Zheng, X.; Ouyang, Y.; Chen, M.; Zeng, L.; Chen, S.; Chen, X.; et al. Protective effect of oral histidine on hypertension in Dahl salt-sensitive rats induced by high-salt diet. Life Sci. 2021, 270, 119134. [Google Scholar] [CrossRef]
- Bertero, T.; Perk, D.; Chan, S.Y. The molecular rationale for therapeutic targeting of glutamine metabolism in pulmonary hypertension. Expert. Opin. Ther. Targets 2019, 23, 511–524. [Google Scholar] [CrossRef]
- Barbaro, N.R.; Foss, J.D.; Kryshtal, D.O.; Tsyba, N.; Kumaresan, S.; Xiao, L.; Mernaugh, R.L.; Itani, H.A.; Loperena, R.; Chen, W.; et al. Dendritic Cell Amiloride-Sensitive Channels Mediate Sodium-Induced Inflammation and Hypertension. Cell Rep. 2017, 21, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Huang, C.; Su, J.; Pan, C.W.; Ke, C. Identification of biomarkers for essential hypertension based on metabolomics. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 382–395. [Google Scholar] [CrossRef]
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B.; et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017, 5, 14. [Google Scholar] [CrossRef]
- Derkach, A.; Sampson, J.; Joseph, J.; Playdon, M.C.; Stolzenberg-Solomon, R.Z. Effects of dietary sodium on metabolites: The Dietary Approaches to Stop Hypertension (DASH)-Sodium Feeding Study. Am. J. Clin. Nutr. 2017, 106, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Rebholz, C.M.; Lichtenstein, A.H.; Zheng, Z.; Appel, L.J.; Coresh, J. Serum untargeted metabolomic profile of the Dietary Approaches to Stop Hypertension (DASH) dietary pattern. Am. J. Clin. Nutr. 2018, 108, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Swietlik, E.M.; Ghataorhe, P.; Zalewska, K.I.; Wharton, J.; Howard, L.S.; Taboada, D.; Cannon, J.E.; UK National Cohort Study of PAH; Morrell, N.W.; Wilkins, M.R.; et al. Plasma metabolomics exhibit response to therapy in chronic thromboembolic pulmonary hypertension. Eur. Respir. J. 2021, 57, 2003201. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, M.H.; Fineberg, N.S.; Fineberg, S.E.; Weinberger, M. Salt sensitivity, pulse pressure, and death in normal and hypertensive humans. Hypertension 2001, 37, 429–432. [Google Scholar] [CrossRef]
- Ingersoll, M.A.; Platt, A.M.; Potteaux, S.; Randolph, G.J. Monocyte trafficking in acute and chronic inflammation. Trends Immunol. 2011, 32, 470–477. [Google Scholar] [CrossRef]
- Wenzel, P.; Knorr, M.; Kossmann, S.; Stratmann, J.; Hausding, M.; Schuhmacher, S.; Karbach, S.H.; Schwenk, M.; Yogev, N.; Schulz, E.; et al. Lysozyme M-positive monocytes mediate angiotensin II-induced arterial hypertension and vascular dysfunction. Circulation 2011, 124, 1370–1381. [Google Scholar] [CrossRef]
- Kopp, C.; Linz, P.; Dahlmann, A.; Hammon, M.; Jantsch, J.; Muller, D.N.; Schmieder, R.E.; Cavallaro, A.; Eckardt, K.U.; Uder, M.; et al. 23Na magnetic resonance imaging-determined tissue sodium in healthy subjects and hypertensive patients. Hypertension 2013, 61, 635–640. [Google Scholar] [CrossRef]
- Gagnon, L.; Leduc, M.; Thibodeau, J.F.; Zhang, M.Z.; Grouix, B.; Sarra-Bournet, F.; Gagnon, W.; Hince, K.; Tremblay, M.; Geerts, L.; et al. A Newly Discovered Antifibrotic Pathway Regulated by Two Fatty Acid Receptors: GPR40 and GPR84. Am. J. Pathol. 2018, 188, 1132–1148. [Google Scholar] [CrossRef]
- Sartorius, T.; Drescher, A.; Panse, M.; Lastovicka, P.; Peter, A.; Weigert, C.; Kostenis, E.; Ullrich, S.; Haring, H.U. Mice Lacking Free Fatty Acid Receptor 1 (GPR40/FFAR1) are Protected Against Conjugated Linoleic Acid-Induced Fatty Liver but Develop Inflammation and Insulin Resistance in the Brain. Cell Physiol. Biochem. 2015, 35, 2272–2284. [Google Scholar] [CrossRef]
- Burns, J.L.; Nakamura, M.T.; Ma, D.W.L. Differentiating the biological effects of linoleic acid from arachidonic acid in health and disease. Prostaglandins Leukot Essent Fatty Acids 2018, 135, 1–4. [Google Scholar] [CrossRef]
- Shureiqi, I.; Chen, D.; Day, R.S.; Zuo, X.; Hochman, F.L.; Ross, W.A.; Cole, R.A.; Moy, O.; Morris, J.S.; Xiao, L.; et al. Profiling lipoxygenase metabolism in specific steps of colorectal tumorigenesis. Cancer Prev. Res. 2010, 3, 829–838. [Google Scholar] [CrossRef]
- Zuo, X.; Peng, Z.; Wu, Y.; Moussalli, M.J.; Yang, X.L.; Wang, Y.; Parker-Thornburg, J.; Morris, J.S.; Broaddus, R.R.; Fischer, S.M.; et al. Effects of gut-targeted 15-LOX-1 transgene expression on colonic tumorigenesis in mice. J. Natl. Cancer Inst. 2012, 104, 709–716. [Google Scholar] [CrossRef]
- Bonyek-Silva, I.; Machado, A.F.A.; Cerqueira-Silva, T.; Nunes, S.; Silva Cruz, M.R.; Silva, J.; Santos, R.L.; Barral, A.; Oliveira, P.R.S.; Khouri, R.; et al. LTB4-Driven Inflammation and Increased Expression of ALOX5/ACE2 During Severe COVID-19 in Individuals With Diabetes. Diabetes 2021, 70, 2120–2130. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Li, Y.; Jin, G.; Huang, T.; Zou, M.; Duan, S. The biological role of arachidonic acid 12-lipoxygenase (ALOX12) in various human diseases. Biomed. Pharmacother. 2020, 129, 110354. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Nunez, D.; Claria, J.; Rivera, F.; Poch, E. Increased levels of 12(S)-HETE in patients with essential hypertension. Hypertension 2001, 37, 334–338. [Google Scholar] [CrossRef]
- Manega, C.M.; Fiorelli, S.; Porro, B.; Turnu, L.; Cavalca, V.; Bonomi, A.; Cosentino, N.; Di Minno, A.; Marenzi, G.; Tremoli, E.; et al. 12(S)-Hydroxyeicosatetraenoic acid downregulates monocyte-derived macrophage efferocytosis: New insights in atherosclerosis. Pharmacol. Res. 2019, 144, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, J.; Pye, C.; Al-Shabrawey, M.; Elmarakby, A.A. Inhibition of 12/15-Lipoxygenase Reduces Renal Inflammation and Injury in Streptozotocin-Induced Diabetic Mice. J. Diabetes Metab 2015, 6, 555. [Google Scholar] [PubMed]
- Merlo, S.; Letonja, M.S.; Vujkovac, A.C.; Delev, D.; Mozos, I.; Kruzliak, P.; Petrovic, D. Arachidonate 5-lipoxygenase (ALOX5) gene polymorphism (rs12762303) and arachidonate 5-lipoxygenase activating protein (ALOX5AP) gene polymorphism (rs3802278) and markers of carotid atherosclerosis in patients with type 2 diabetes mellitus. Int. J. Clin. Exp. Med. 2016, 9, 4509–4514. [Google Scholar]

| Female (n = 44) | Male (n = 31) | Test Statistic | |
|---|---|---|---|
| Age (years) | 28.68 ± 7.953 | 32.71± 9.324 | p =0 .0481 |
| Race | |||
| Asian | 4 | ||
| Black | 16 | 4 | |
| White | 24 | 27 | |
| Body Mass Index (kg/m2) | 24.27 ± 6.17 | 27.14 ± 5.57 | p = 0.0423 |
| Waist circumference (cm) | 76.76 ± 13.37 | 91.69 ± 15.18 | p < 0.0001 |
| Hip circumference | 98.96 ± 12.23 | 103.4 ± 10.35 | p = 0.1053 |
| Average DBP | 67.31 ± 8.075 | 72.76 ± 9.24 | p = 0.0076 |
| Average SBP | 110.80 ± 10.92 | 122.8 ± 11.50 | p < 0.0001 |
| Average MAP | 80.25 ± 14.84 | 89.44 ± 9.41 | p = 0.0033 |
| Heart Rate | 71.48 ± 7.56 | 70.35 ± 8.41 | p = 0.5476 |
| Daily Na+ intake (g) | 2.74± 1.05 | 3.25 ± 1.17 | p = 0.0582 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishimwe, J.A.; Ferguson, J.F.; Kirabo, A. Sex Differences in Fatty Acid Metabolism and Blood Pressure Response to Dietary Salt in Humans. Cardiogenetics 2023, 13, 33-46. https://doi.org/10.3390/cardiogenetics13010005
Ishimwe JA, Ferguson JF, Kirabo A. Sex Differences in Fatty Acid Metabolism and Blood Pressure Response to Dietary Salt in Humans. Cardiogenetics. 2023; 13(1):33-46. https://doi.org/10.3390/cardiogenetics13010005
Chicago/Turabian StyleIshimwe, Jeanne A., Jane F. Ferguson, and Annet Kirabo. 2023. "Sex Differences in Fatty Acid Metabolism and Blood Pressure Response to Dietary Salt in Humans" Cardiogenetics 13, no. 1: 33-46. https://doi.org/10.3390/cardiogenetics13010005
APA StyleIshimwe, J. A., Ferguson, J. F., & Kirabo, A. (2023). Sex Differences in Fatty Acid Metabolism and Blood Pressure Response to Dietary Salt in Humans. Cardiogenetics, 13(1), 33-46. https://doi.org/10.3390/cardiogenetics13010005







