Specific Deletion of the FHA Domain Containing SLMAP3 Isoform in Postnatal Myocardium Has No Impact on Structure or Function
Abstract
:1. Introduction
2. Materials and Methods
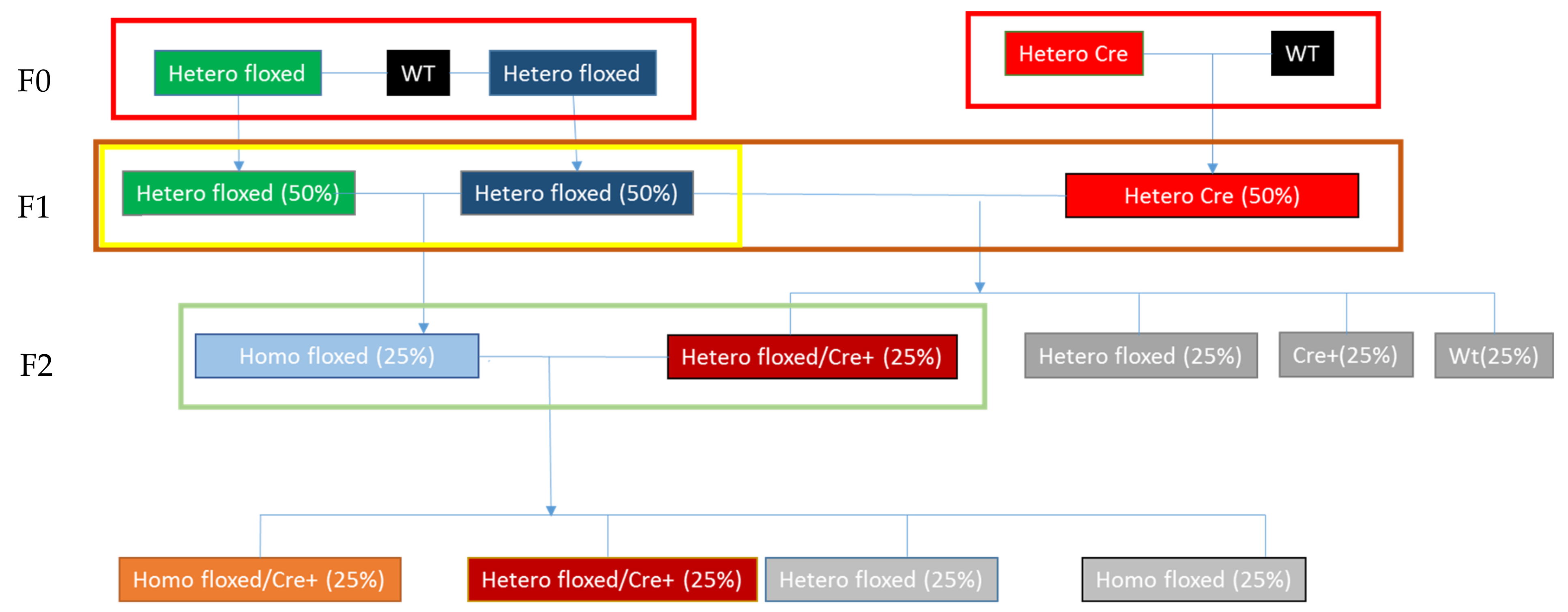
2.1. Generating Transgenic Flox-SLMAP Mouse Lines
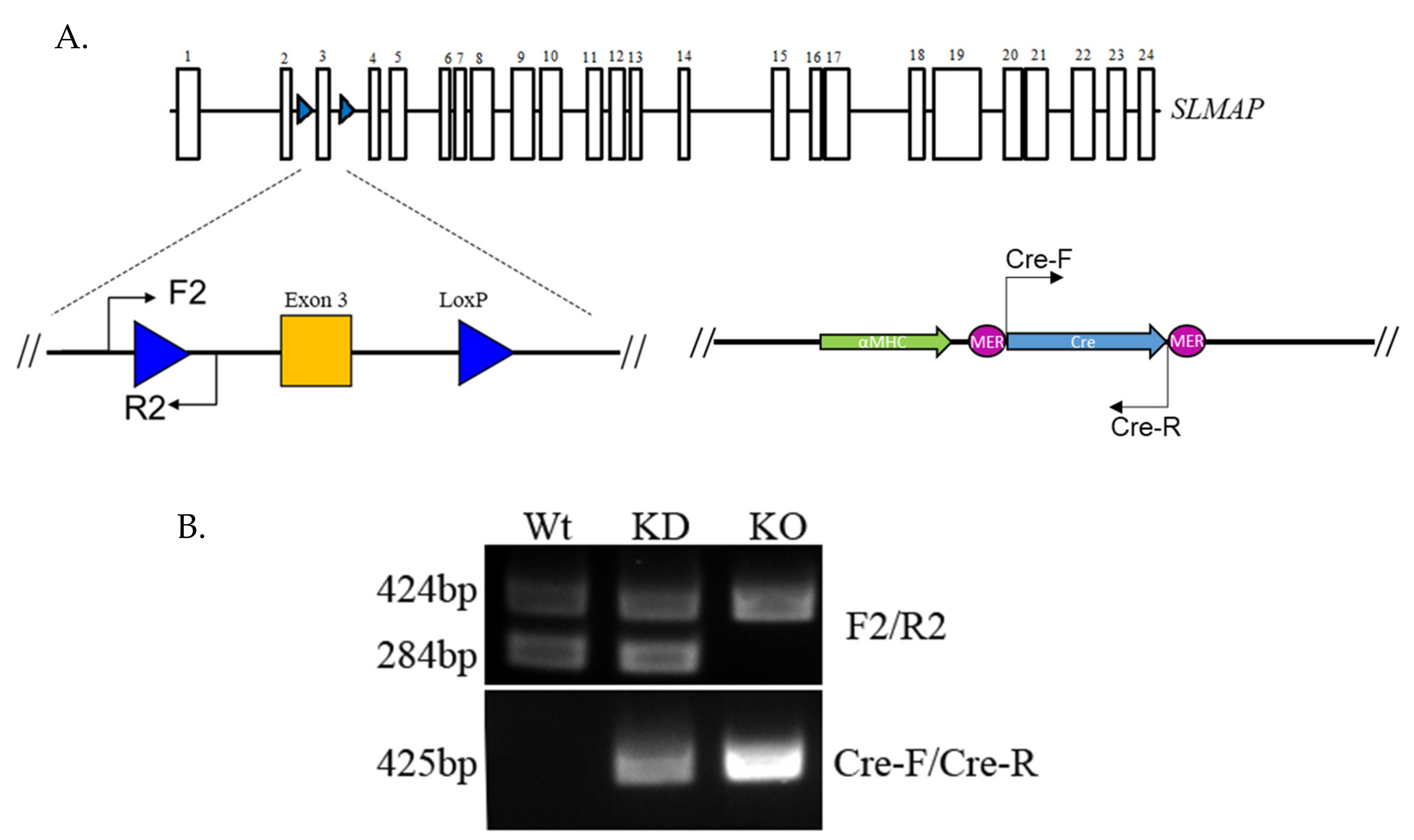
2.2. Generating and Genotyping the SLMAP Knockout Mice
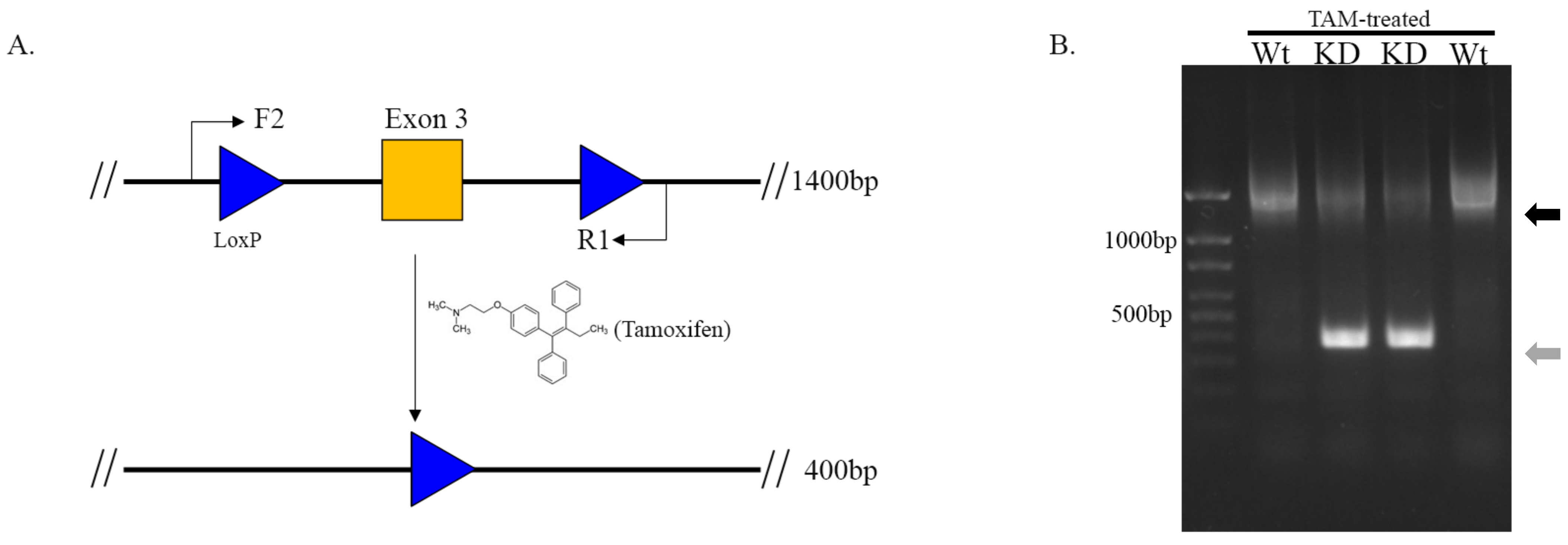
2.3. Activation of Cre and Assessing Recombination of Floxed SLMAP
2.4. Protein Isolation from Mouse Heart
2.5. SDS-PAGE and Western Blotting
2.6. Quantitative PCR
2.7. Echocardiography
2.8. Isoproterenol Delivery
2.9. Histological Analysis
2.10. Electrocardiography
2.11. Statistical Analyses
3. Results
3.1. Producing the αMHC-MerCreMer-Flox-SLMAP Mice
3.2. Cleavage of Floxed-SLMAP by Activating αMHC-MerCreMer
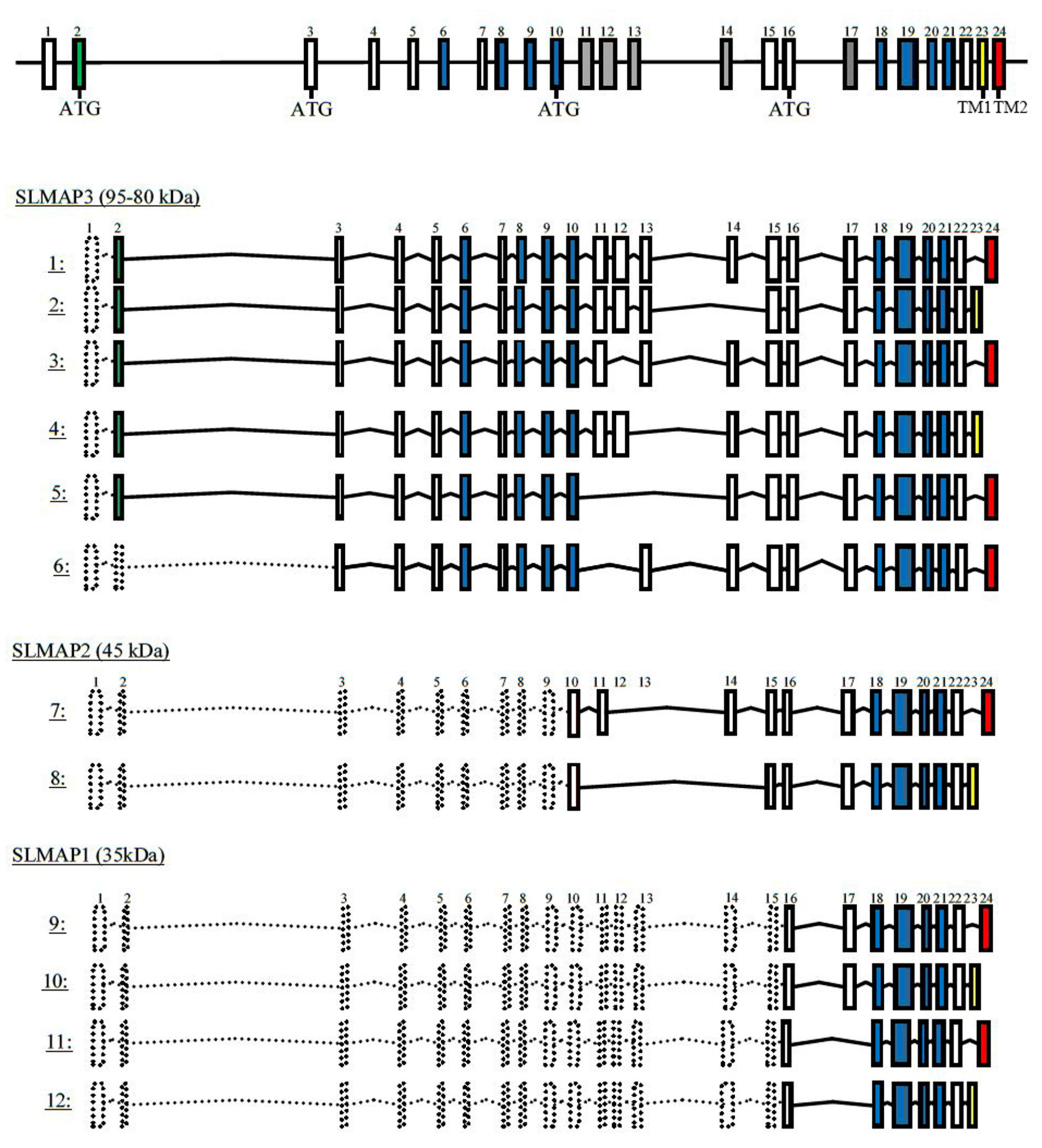
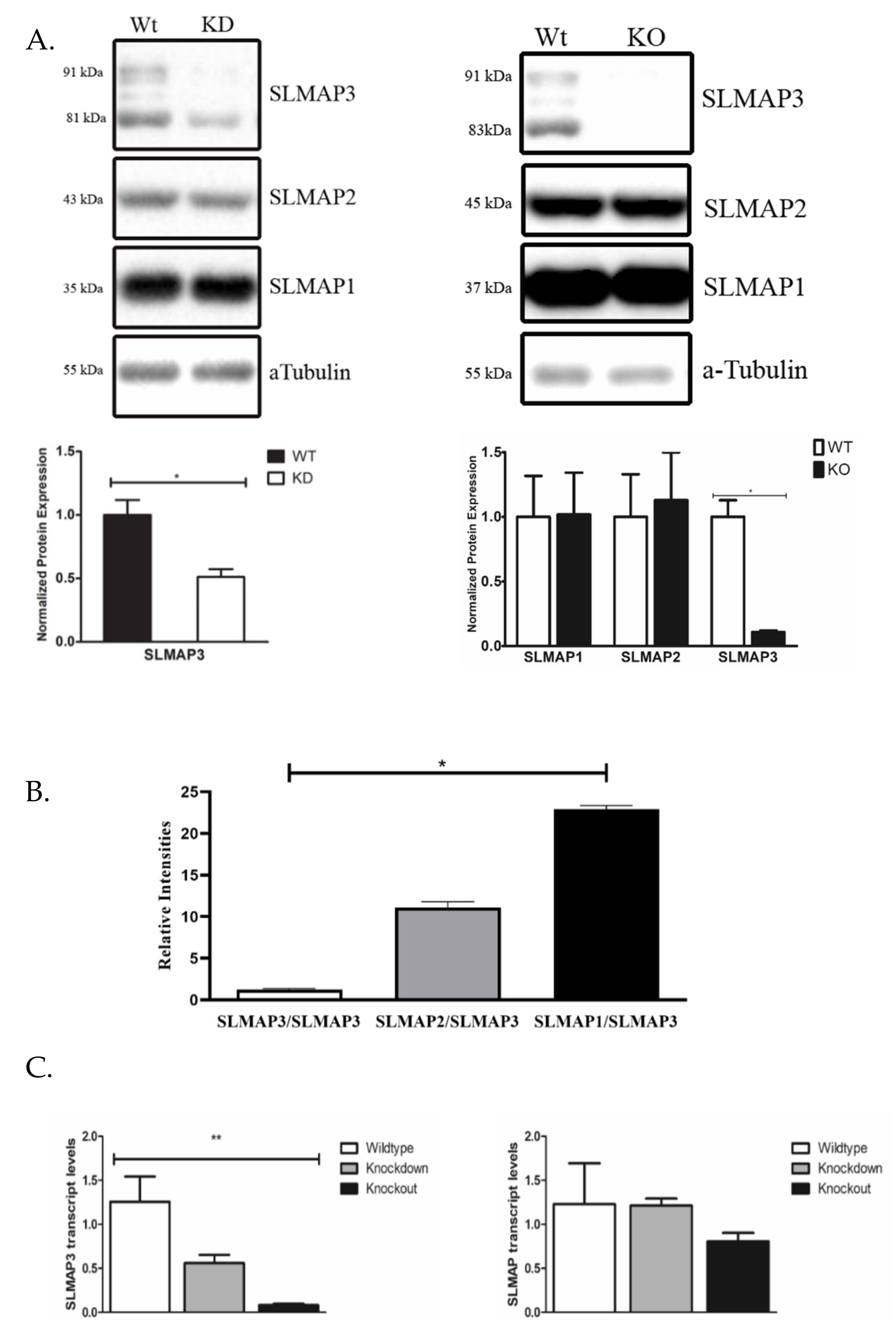
3.3. Protein and Transcript Expression of SLMAPs in Wt, KD and KO Mice
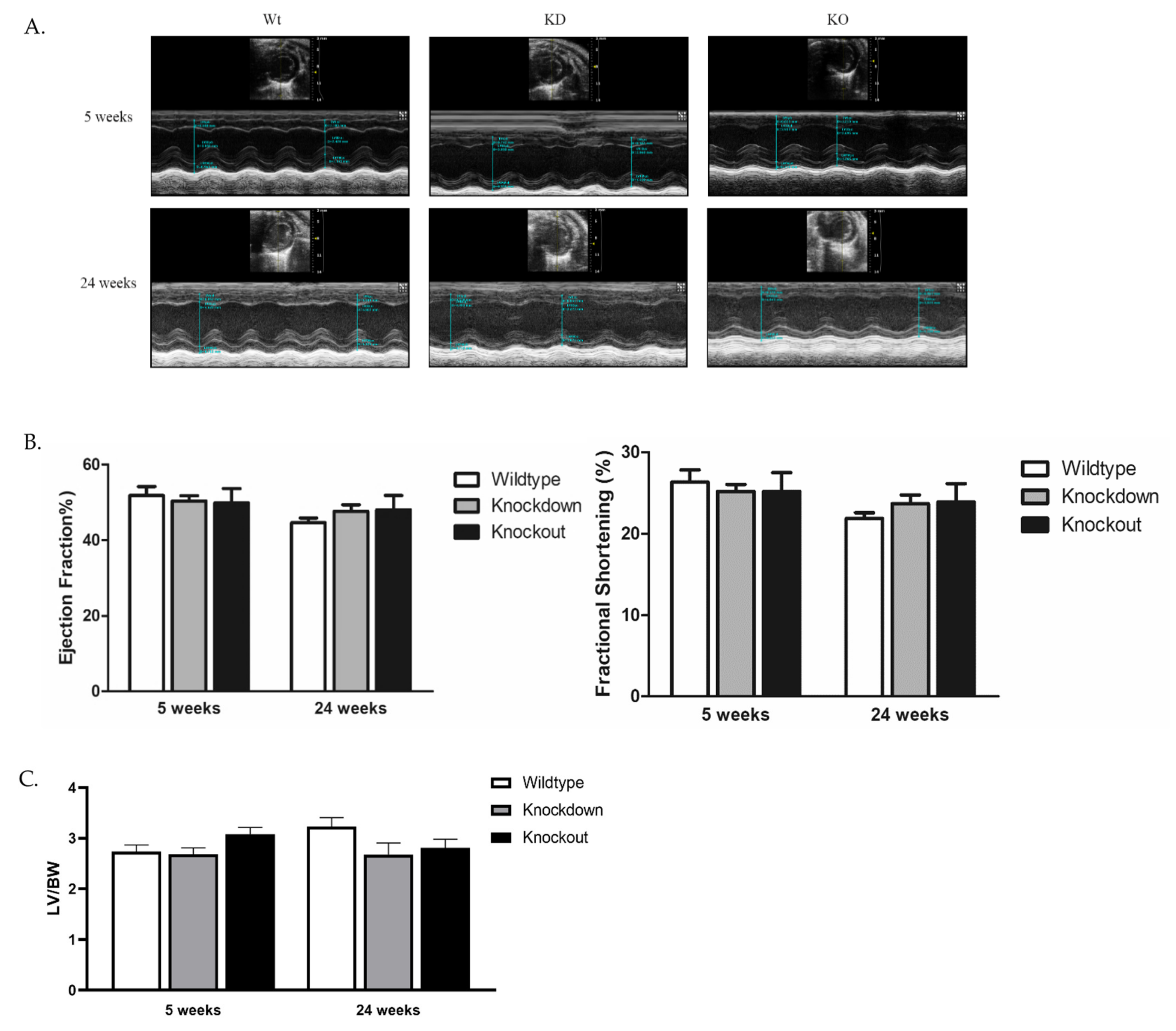
3.4. Cardiac Structure and Function of SLMAP3-Deficient Hearts
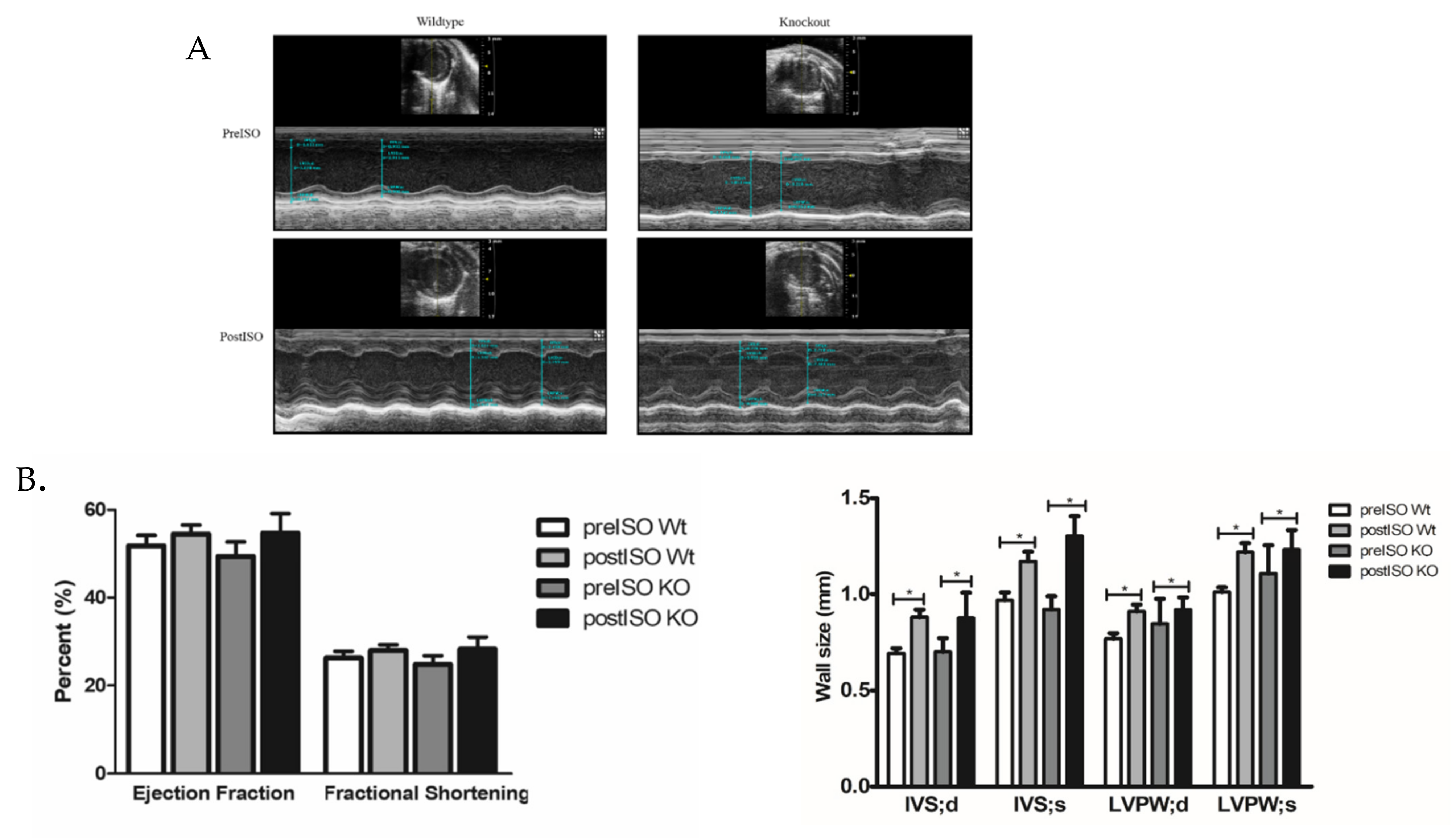
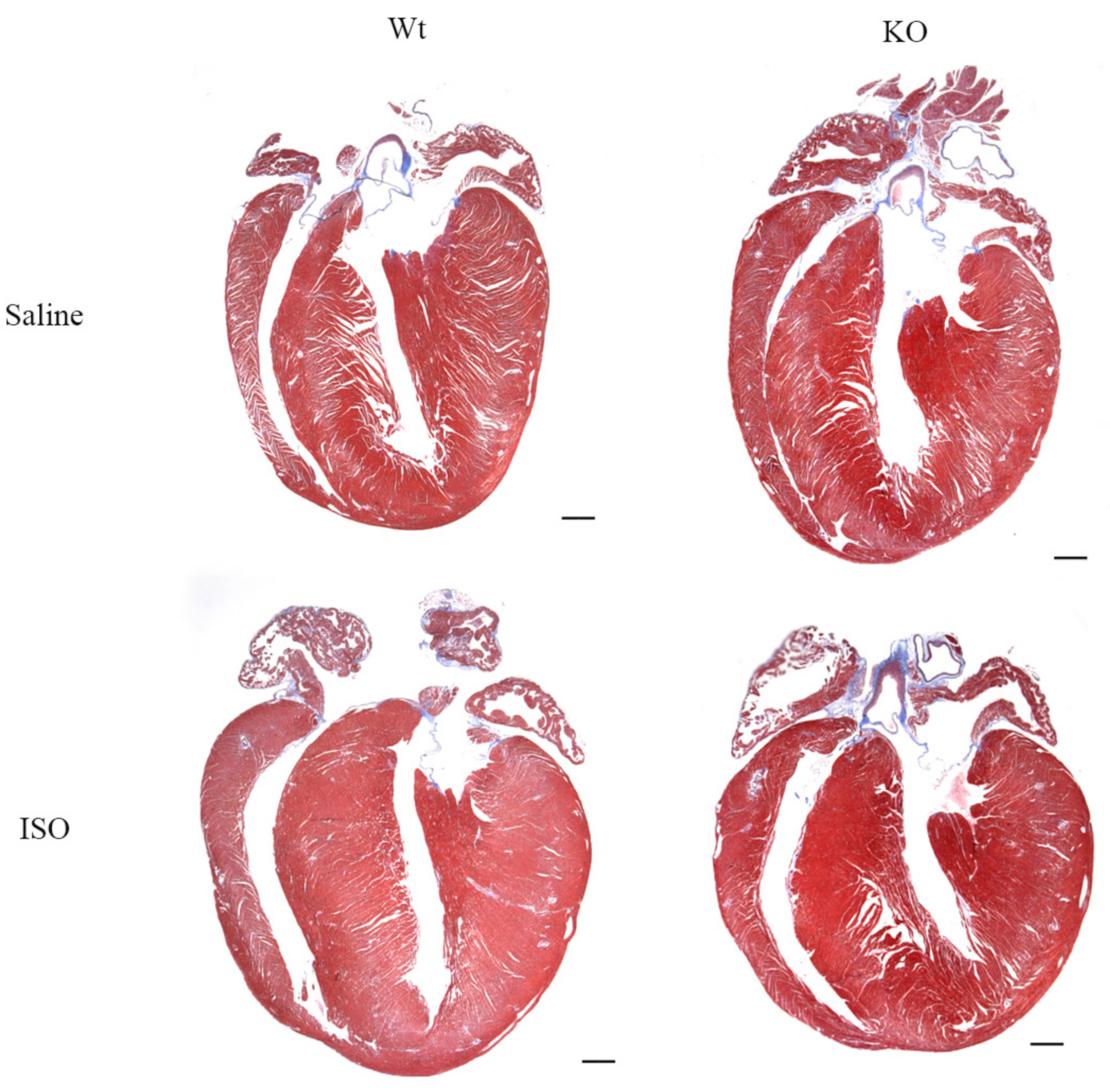
3.5. Impact of Isoproterenol Induced Stress on SLMAP3-KO Hearts
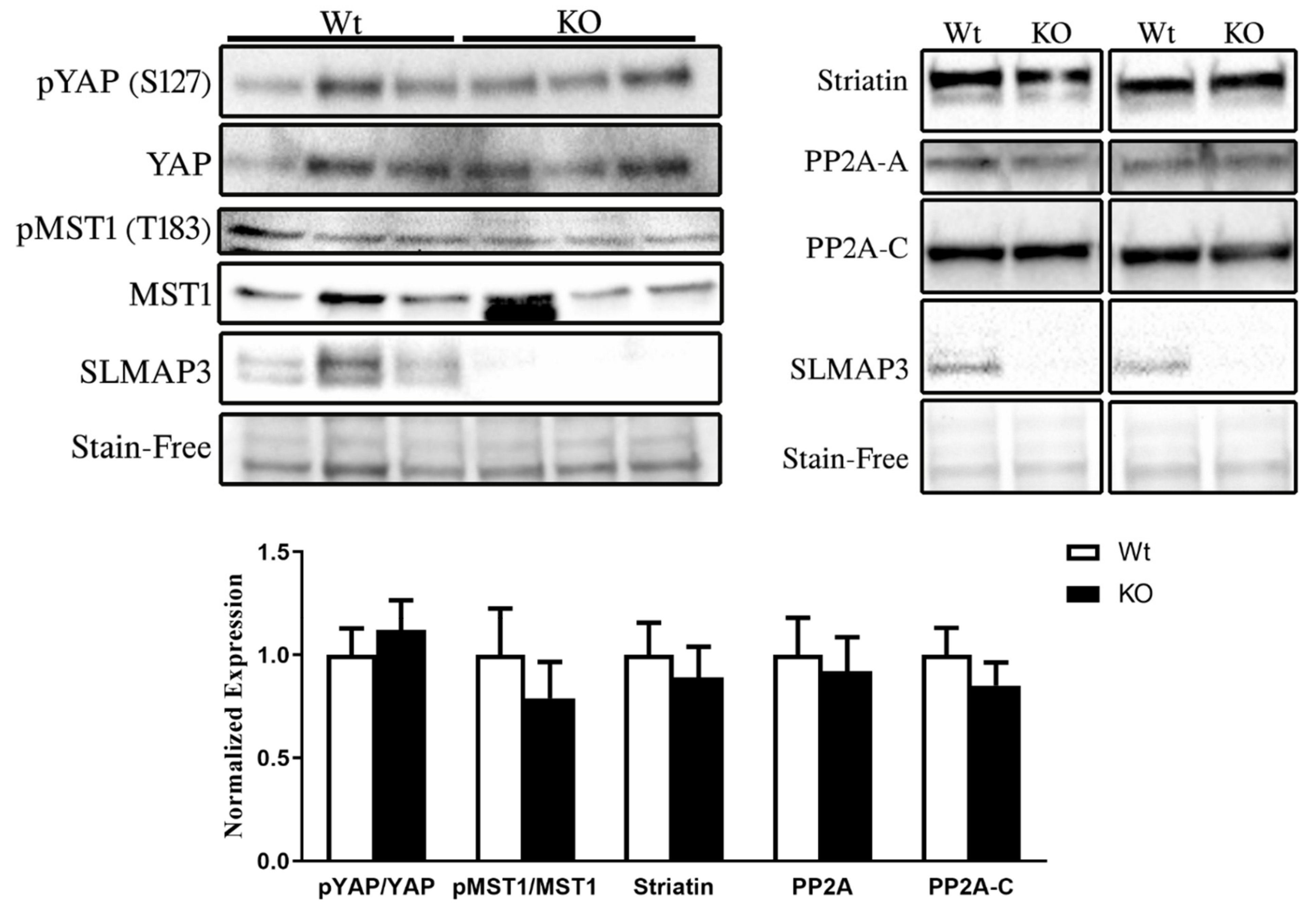
3.6. STRIPAK Complex and Hippo Signaling in SLMAP3-KO Myocardium
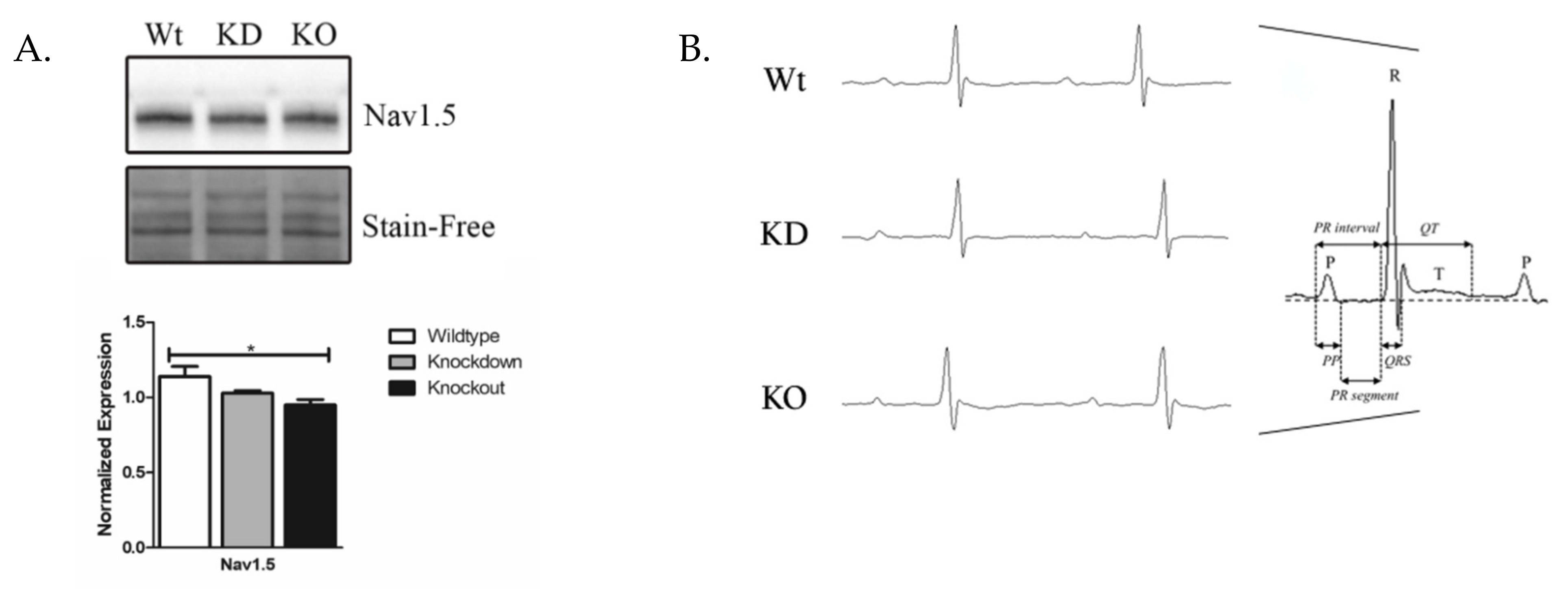
3.7. Nav1.5 Expression and Electrophysiology in SLMAP3-Deficient Myocardium
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wigle, J.; Demchyshyn, L.; Pratt, M.; Staines, W.; Salih, M.; Tuana, B. Molecular cloning, expression, and chromosomal assignment of sarcolemmal-associated proteins. A family of acidic amphipathic alpha-helical proteins associated with the membrane. J. Biol. Chem. 1997, 272, 32384–32394. [Google Scholar] [CrossRef] [Green Version]
- Wielowieyski, P.; Sevinc, S.; Guzzo, R.; Salih, M.; Wigle, J.; Tuana, B. Alternative splicing, expression, and genomic structure of the 3’ region of the gene encoding the sarcolemmal-associated proteins (SLAPs) defines a novel class of coiled-coil tail-anchored membrane proteins. J. Biol. Chem. 2000, 275, 38474–38481. [Google Scholar] [CrossRef] [Green Version]
- Guzzo, R.; Wigle, J.; Salih, M.; Moore, E.; Tuana, B. Regulated expression and temporal induction of the tail-anchored sarcolemmal-membrane-associated protein is critical for myoblast fusion. Biochem. J. 2004, 381, 599–608. [Google Scholar] [CrossRef] [Green Version]
- Byers, J.; Guzzo, R.; Salih, M.; Tuana, B. Hydrophobic profiles of the tail anchors in SLMAP dictate subcellular targeting. BMC Cell Biol. 2009, 10, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzzo, R.; Sevinc, S.; Salih, M.; Tuana, B. A novel isoform of sarcolemmal membrane-associated protein (SLMAP) is a component of the microtubule organizing center. J. Cell Sci. 2004, 117 Pt 11, 2271–2281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, A.; Yuan, C.; Lee, H.; Chen, E.; Wu, P.; Tsai, M. Structure and function of the phosphothreonine-specific FHA domain. Sci. Signal. 2008, 1, re12. [Google Scholar] [CrossRef] [PubMed]
- Couzens, A.; Knight, J.; Kean, M.; Teo, G.; Weiss, A.; Dunham, W.; Lin, Z.; Bagshaw, R.; Sicheri, F.; Pawson, T.; et al. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci. Signal. 2013, 6, rs15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goudreault, M.; D’Ambrosio, L.; Kean, M.; Mullin, M.; Larsen, B.; Sanchez, A.; Chaudhry, S.; Chen, G.; Sicheri, F.; Nesvizhskii, A.; et al. A PP2A Phosphatase High Density Interaction Network Identifies a Novel Striatin-interacting Phosphatase and Kinase Complex Linked to the Cerebral Cavernous Malformation 3 (CCM3) Protein. Mol. Cell Proteom. 2009, 8, 157–171. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Pan, D. The Hippo Signaling Pathway in Development and Disease. Dev. Cell 2019, 50, 264–282. [Google Scholar] [CrossRef]
- Wang, J.; Liu, S.; Heallen, T.; Martin, J. The Hippo pathway in the heart: Pivotal roles in development, disease, and regeneration. Nat. Rev. Cardiol. 2018, 15, 672–684. [Google Scholar] [CrossRef] [PubMed]
- Misra, J.; Irvine, K. The Hippo Signaling Network and Its Biological Functions. Annu. Rev. Genet. 2018, 52, 65–87. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Li, L.; Zhao, B.; Guan, K.L. The Hippo pathway in heart development, regeneration, and diseases. Circ. Res. 2015, 116, 1431–1447. [Google Scholar] [CrossRef] [PubMed]
- Von Gise, A.; Lin, Z.; Schlegelmilch, K.; Honor, L.; Pan, G.; Buck, J.; Ma, Q.; Ishiwata, T.; Zhou, B.; Camargo, F.; et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc. Natl. Acad. Sci. USA 2012, 109, 2394–2399. [Google Scholar] [CrossRef] [Green Version]
- Xin, M.; Kim, Y.; Sutherland, L.; Murakami, M.; Qi, X.; McAnally, J.; Porrello, E.; Mahmoud, A.; Tan, W.; Shelton, J.; et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 13839–13844. [Google Scholar] [CrossRef] [Green Version]
- Heallen, T.; Zhang, M.; Wang, J.; Bonilla-Claudio, M.; Klysik, E.; Johnson, R.; Martin, J. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 2011, 332, 458–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byun, J.; Del Re, D.; Zhai, P.; Ikeda, S.; Shirakabe, A.; Mizushima, W.; Miyamoto, S.; Brown, J.; Sadoshima, J. Yes-associated protein (YAP) mediates adaptive cardiac hypertrophy in response to pressure overload. J. Biol. Chem. 2019, 294, 3603–3617. [Google Scholar] [CrossRef] [Green Version]
- Jeong, B.; Bae, S.; Ni, L.; Zhang, X.; Bai, X.; Luo, X. Cryo-EM structure of the Hippo signaling integrator human STRIPAK. Nat. Struct. Mol. Biol. 2021, 28, 290–299. [Google Scholar] [CrossRef]
- Hauri, S.; Wepf, A.; van Drogen, A.; Varjosalo, M.; Tapon, N.; Aebersold, R.; Gstaiger, M. Interaction proteome of human Hippo signaling: Modular control of the co-activator YAP1. Mol. Syst. Biol. 2013, 9, 713. [Google Scholar] [CrossRef] [Green Version]
- Bae, S.; Ni, L.; Osinski, A.; Tomchick, D.; Brautigam, C.; Luo, X. SAV1 promotes Hippo kinase activation through antagonizing the PP2A phosphatase STRIPAK. eLife 2017, 6, e30278. [Google Scholar] [CrossRef]
- Zheng, Y.; Liu, B.; Wang, L.; Lei, H.; Pulgar Prieto, K.; Pan, D. Homeostatic Control of Hpo/MST Kinase Activity through Autophosphorylation-Dependent Recruitment of the STRIPAK PP2A Phosphatase Complex. Cell Rep. 2017, 21, 3612–3623. [Google Scholar] [CrossRef] [Green Version]
- Guzzo, R.M.; Salih, M.; Moore, E.D.; Tuana, B. Molecular properties of cardiac tail-anchored membrane protein SLMAP are consistent with structural role in arrangement of excitation-contraction coupling apparatus. Am. J. Physiol. Heart Circ. Physiol. 2005, 288, H1810–H1819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nader, M.; Westendorp, B.; Hawari, O.; Salih, M.; Stewart, A.F.; Leenen, F.H.; Tuana, B.S. Tail-anchored membrane protein SLMAP is a novel regulator of cardiac function at the sarcoplasmic reticulum. Am. J. Physiol. Heart Circ. Physiol. 2012, 302, 1138–1145. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, T.; Sato, A.; Marcou, C.; Tester, D.; Ackerman, M.; Crotti, L.; Schwartz, P.; On, Y.; Park, J.; Nakamura, K.; et al. A novel disease gene for Brugada syndrome: Sarcolemmal membrane-associated protein gene mutations impair intracellular trafficking of hNav1.5. Circ. Arrhythm Electrophysiol. 2012, 5, 1098–1107. [Google Scholar] [CrossRef] [Green Version]
- Brugada, R.; Campuzano, O.; Sarquella-Brugada, G.; Brugada, J.; Brugada, P. Brugada syndrome. Methodist Debakey Cardiovasc. J. 2014, 10, 25–28. [Google Scholar] [CrossRef]
- Mlynarova, J.; Trentin-Sonoda, M.; Gaisler da Silva, F.; Major, J.; Salih, M.; Carneiro-Ramos, M.; Tuana, B. SLMAP3 isoform modulates cardiac gene expression and function. PLoS ONE 2019, 14, e0214669. [Google Scholar] [CrossRef]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, G.; Jeron, A.; Koren, G. Measurement of heart rate and Q-T interval in the conscious mouse. Am. J. Physiol. 1998, 274, H747–H751. [Google Scholar] [CrossRef] [PubMed]
- Aken, B.; Ayling, S.; Barrell, D.; Clarke, L.; Curwen, V.; Fairley, S.; Fernandez, B.J.; Billis, K.; García, G.C.; Hourlier, T.; et al. The Ensembl gene annotation system. Database 2016, 2016, baw093. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, T.; Samaras, P.; Frejno, M.; Gessulat, S.; Barnert, M.; Kienegger, H.; Krcmar, H.; Schlegl, J.; Ehrlich, H.C.; Aiche, S.; et al. ProteomicsDB. Nucleic Acids Res. 2018, 4, D1271–D1281. [Google Scholar] [CrossRef]
- Davis, J.; Maillet, M.; Miano, J.; Molkentin, J. Lost in transgenesis: A user’s guide for genetically manipulating the mouse in cardiac research. Circ. Res. 2012, 111, 761–777. [Google Scholar] [CrossRef] [PubMed]
- Sohal, D.S.; Nghiem, M.; Crackower, M.A.; Witt, S.A.; Kimball, T.R.; Tymitz, K.M.; Penninger, J.M.; Molkentin, J.D. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ. Res. 2001, 89, 20–25. [Google Scholar] [CrossRef]
- Demayo, J.; Wang, J.; Liang, D.; Zhang, R.; Demayo, F. Genetically Engineered Mice by Pronuclear DNA microinjection. Curr. Protoc. Mouse Biol. 2012, 2, 245–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayadi, A.; Ferrand, G.; Cruz, I.; Warot, X. Mouse Breeding and Colony Management. Curr. Protoc. Mouse Biol. 2011, 1, 239–264. [Google Scholar]
- Kuhn, R.; Torres, R.M. Cre/LoxP Recombination System and Gene Targeting. Transgenesis Tech. 2002, 180, 175–204. [Google Scholar]
- Bersell, K.; Choudhury, S.; Mollova, M.; Polizzotti, B.D.; Ganapathy, B.; Walsh, S.; Wadugu, B.; Arab, S.; Kühn, B. Moderate and high amounts of tamoxifen in αMHC-MerCreMer mice induce a DNA damage response, leading to heart failure and death. Dis. Models Mech. 2013, 6, 1459–1469. [Google Scholar]
- Yang, X.; Liu, Y.; Rhaleb, N.; Kurihara, N.; Kim, H.; Carretero, O. Echocardiographic assessment of cardiac function in conscious and anesthetized mice. Am. J. Physiol. 1999, 277, 1967–1974. [Google Scholar] [CrossRef]
- Zhang, G.; Kimura, S.; Nishiyama, A.; Shokoji, T.; Rahman, M.; Yao, L.; Nagai, Y.; Fujisawa, Y.; Miyatake, A.; Abe, Y. Cardiac oxidative stress in acute and chronic isoproterenol-infused rats. Cardiovasc. Res. 2005, 65, 230–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Major, J.; Salih, M.; Tuana, B. Interplay between the E2F pathway and β-adrenergic signaling in the pathological hypertrophic response of myocardium. J. Mol. Cell Cardiol. 2015, 84, 179–190. [Google Scholar] [CrossRef]
- Dong, C.; Wang, Y.; Ma, A.; Wang, T. Life Cycle of the Cardiac Voltage-Gated Sodium Channel NaV1.5. Front. Physiol. 2020, 11, 609733. [Google Scholar] [CrossRef]
- Ayoubi, T.; Van De Ven, W. Regulation of gene expression by alternative promoters. FASEB J. 1996, 4, 453–460. [Google Scholar] [CrossRef]
- Papadatos, G.; Wallerstein, P.; Head, C.; Ratcliff, R.; Brady, P.; Benndorf, K.; Saumarez, R.; Trezise, A.; Huang, C.; Vandenberg, J.; et al. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc. Natl. Acad. Sci. USA 2002, 99, 6210–6215. [Google Scholar] [CrossRef] [Green Version]
- Bae, S.; Ni, L.; Luo, X. STK25 suppresses Hippo signaling by regulating SAV1-STRIPAK antagonism. eLife 2020, 9, e54863. [Google Scholar] [CrossRef]
- Chen, R.; Xie, R.; Meng, Z.; Ma, S.; Guan, K. STRIPAK integrates upstream signals to initiate the Hippo kinase cascade. Nat. Cell Biol. 2019, 21, 1565–1577. [Google Scholar] [CrossRef]
- Kaya-Çopur, A.; Marchiano, F.; Hein, M.; Alpern, D.; Russeil, J.; Luis, N.; Mann, M.; Deplancke, B.; Habermann, B.; Schnorrer, F. The Hippo pathway controls myofibril assembly and muscle fiber growth by regulating sarcomeric gene expression. Elife 2021, 10, e63726. [Google Scholar] [CrossRef] [PubMed]
- Del Re, D.; Yang, Y.; Nakano, N.; Cho, J.; Zhai, P.; Yamamoto, T.; Zhang, N.; Yabuta, N.; Nojima, H.; Pan, D.; et al. Yes-associated protein isoform 1 (Yap1) promotes cardiomyocyte survival and growth to protect against myocardial ischemic injury. J. Biol. Chem. 2013, 288, 3977–3988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, S.; Yang, G.; Zablocki, D.; Liu, J.; Hong, C.; Kim, S.; Soler, S.; Odashima, M.; Thaisz, J.; Yehia, G.; et al. Activation of Mst1 causes dilated cardiomyopathy by stimulating apoptosis without compensatory ventricular myocyte hypertrophy. J. Clin. Invest. 2003, 111, 1463–1474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, S.; Mizushima, W.; Sciarretta, S.; Abdellatif, M.; Zhai, P.; Mukai, R.; Fefelova, N.; Oka, S.; Nakamura, M.; Del Re, D.; et al. Hippo Deficiency Leads to Cardiac Dysfunction Accompanied by Cardiomyocyte Dedifferentiation During Pressure Overload. Circ. Res. 2019, 124, 292–305. [Google Scholar] [CrossRef] [PubMed]
- Leach, J.; Heallen, T.; Zhang, M.; Rahmani, M.; Morikawa, Y.; Hill, M.; Segura, A.; Willerson, J.; Martin, J. Hippo pathway deficiency reverses systolic heart failure after infarction. Nature 2017, 550, 260–264. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Mao, B.; Luo, W.; Wei, B.; Jiang, W.; Liu, D.; Song, L.; Ji, G.; Yang, Z.; Lai, Y.; et al. The alteration of Hippo/YAP signaling in the development of hypertrophic cardiomyopathy. Basic Res. Cardiol. 2014, 109, 435–438. [Google Scholar] [CrossRef]
- Heallen, T.; Morikawa, Y.; Leach, J.; Tao, G.; Willerson, J.T.; Johnson, R.L.; Martin, J.F. Hippo signaling impedes adult heart regeneration. Development 2013, 140, 4683–4690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergmann, O.; Bhardwaj, R.; Bernard, S.; Zdunek, S.; Barnabé-Heider, F.; Walsh, S.; Zupicich, J.; Alkass, K.; Buchholz, B.; Druid, H.; et al. Evidence for cardiomyocyte renewal in humans. Science 2009, 324, 98–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alkass, K.; Panula, J.; Westman, M.; Wu, T.; Guerquin-Kern, J.; Bergmann, O. No Evidence for Cardiomyocyte Number Expansion in Preadolescent Mice. Cell 2015, 163, 1026–1036. [Google Scholar] [CrossRef] [Green Version]
- Heallen, T.; Kadow, Z.; Wang, J.; Martin, J. Determinants of Cardiac Growth and Size. Cold Spring Harb. Perspect. Biol. 2020, 2, a037150. [Google Scholar] [CrossRef] [PubMed]
- Yutzey, K. Cardiomyocyte Proliferation: Teaching an Old Dogma New Tricks. Circ. Res. 2017, 120, 627–629. [Google Scholar] [CrossRef]
- Porrello, E.; Mahmoud, A.; Simpson, E.; Hill, J.; Richardson, J.; Olson, E.; Sadek, H. Transient regenerative potential of the neonatal mouse heart. Science 2011, 331, 1078–1080. [Google Scholar] [CrossRef] [Green Version]
| Antibody | Manufacturer | Dilution |
|---|---|---|
| SLMAP | Novus Biologicals (NBP1-81397) | 1:1000 |
| Nav1.5 | Alomone Labs (ASC-005) | 1:600 |
| Striatin | BD Transduction Laboratories (610838) | 1:1000 |
| PP2A-C α/β subunit | Santa Cruz Biotechnology (sc-80665) | 1:200 |
| PP2A-A α subunit | Millipore Sigma (07-250) | 1:600 |
| Phospho-YAP (S127) | Cell Signaling Technology (13008S) | 1:1000 |
| YAP | Cell Signaling Technology (14074S) | 1:1000 |
| Phospho-MST1/2 (T183/180) | Cell Signaling Technology (49332S) | 1:1000 |
| KRS2(MST1) | Santa Cruz (sc-515051) | 1:200 |
| Electrocardiography Parameter | Wt | KD | KO | ρ-Value |
|---|---|---|---|---|
| RR (ms) | 123.370 ± 3.163 | 127.5 ± 2.644 | 123.407 ± 3.271 | 0.5738 |
| HR (bpm) | 487.4 ± 12.771 | 471.224 ± 9.847 | 487.248 ± 13.116 | 0.5787 |
| PP duration (ms) | 8.941 ± 0.177 | 8.168 ± 0.737 | 8.023 ± 0.651 | 0.5197 |
| PR segment (ms) | 29.738 ± 0.684 | 29.429 ± 4.568 | 28.781 ± 1.557 | 0.9148 |
| PR interval (ms) | 45.185 ± 0.737 | 44.685 ± 2.428 | 41.558 ± 0.883 | 0.2764 |
| QRS duration (ms) | 11.954 ± 1.289 | 10.298 ± 0.549 | 11.613 ± 0.773 | 0.4568 |
| QTc interval (ms) | 46.010 ± 2.508 | 40.969 ± 6.033 | 45.713 ± 2.166 | 0.6262 |
| P amplitude (mV) | 0.0531 ± 0.009 | 0.069 ± 0.01 | 0.067 ± 0.003 | 0.3765 |
| R amplitude (mV) | 25.670 ± 0.730 | 21.351 ± 0.789 | 0.620 ± 0.027 | 0.5823 |
| S amplitude (mV) | -0.248 ± 0.023 | -0.204 ± 0.031 | -0.219 ± 0.046 | 0.6749 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehmani, T.; Mlynarova, J.; Byers, J.; Salih, M.; Tuana, B.S. Specific Deletion of the FHA Domain Containing SLMAP3 Isoform in Postnatal Myocardium Has No Impact on Structure or Function. Cardiogenetics 2021, 11, 164-184. https://doi.org/10.3390/cardiogenetics11040018
Rehmani T, Mlynarova J, Byers J, Salih M, Tuana BS. Specific Deletion of the FHA Domain Containing SLMAP3 Isoform in Postnatal Myocardium Has No Impact on Structure or Function. Cardiogenetics. 2021; 11(4):164-184. https://doi.org/10.3390/cardiogenetics11040018
Chicago/Turabian StyleRehmani, Taha, Jana Mlynarova, Joseph Byers, Maysoon Salih, and Balwant S. Tuana. 2021. "Specific Deletion of the FHA Domain Containing SLMAP3 Isoform in Postnatal Myocardium Has No Impact on Structure or Function" Cardiogenetics 11, no. 4: 164-184. https://doi.org/10.3390/cardiogenetics11040018
APA StyleRehmani, T., Mlynarova, J., Byers, J., Salih, M., & Tuana, B. S. (2021). Specific Deletion of the FHA Domain Containing SLMAP3 Isoform in Postnatal Myocardium Has No Impact on Structure or Function. Cardiogenetics, 11(4), 164-184. https://doi.org/10.3390/cardiogenetics11040018






