The Flavonoid Biosynthesis and Regulation in Brassica napus: A Review
Abstract
1. Introduction
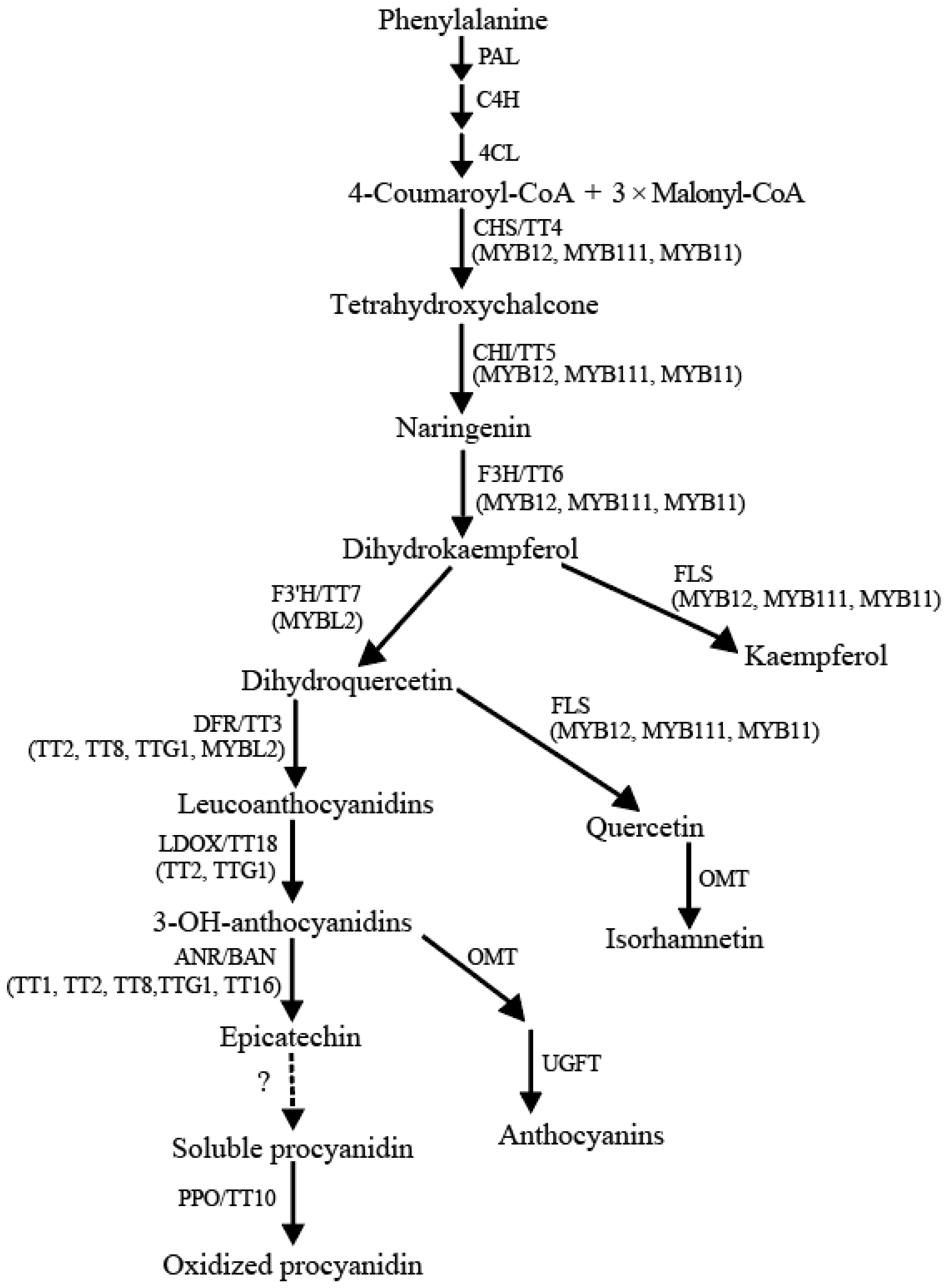
2. Flavonoid Biosynthesis in Arabidopsis
3. Flavonoid Profiles Tentatively Identified in Rapeseed
4. Regulation of Flavonoid Biosynthesis in Seeds of B. napus
4.1. Gene Expressional Changes in Yellow Seed of B. napus
4.2. Quantitative Trait Loci of Yellow Seed Trait in B. napus
4.3. Functionally Characterized Genes Regulating Yellow Seed Trait of B. napus
5. Regulation of Anthocyanin Biosynthesis in B. napus Flowers
6. Flavonoid Regulation in Rapeseed Leaves
7. Flavonoid Regulation Related to Abiotic and Biotic Stress Responses in Rapeseed
7.1. Flavonoid Regulation in Response to Abiotic Stresses
7.2. Flavonoid Regulation in Response to Light Conditions
7.3. Flavonoid Regulation in Response to Biotic Stresses
8. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food Chem. 2022, 383, 132531. [Google Scholar] [CrossRef] [PubMed]
- Teka, T.; Zhang, L.; Ge, X.; Li, Y.; Han, L.; Yan, X. Stilbenes: Source plants, chemistry, biosynthesis, pharmacology, application and problems related to their clinical Application-A comprehensive review. Phytochemistry 2022, 197, 113128. [Google Scholar] [CrossRef] [PubMed]
- Andersen, O.M.; Markham, K.R. The Flavonoids: Chemistry, Biochemistry and Applications; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Alappat, B.; Alappat, J. Anthocyanin pigments: Beyond aesthetics. Molecules 2020, 25, 5500. [Google Scholar] [CrossRef]
- Laoue, J.; Fernandez, C.; Ormeno, E. Plant flavonoids in Mediterranean species: A focus on flavonols as protective metabolites under climate stress. Plants 2022, 11, 172. [Google Scholar] [CrossRef]
- Morales-Quintana, L.; Mino, R.; Mendez-Yanez, A.; Gundel, P.E.; Ramos, P. Do fungal-endosymbionts improve crop nutritional quality and tolerance to stress by boosting flavonoid-mediated responses? Food Res. Int. 2022, 161, 111850. [Google Scholar] [CrossRef]
- Hsieh, K.; Huang, A.H. Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 2007, 19, 582–596. [Google Scholar] [CrossRef]
- Cushnie, T.P.; Lamb, A.J. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 2005, 26, 343–356. [Google Scholar] [CrossRef]
- Rousserie, P.; Rabot, A.; Geny-Denis, L. From flavanols biosynthesis to wine tannins: What place for grape seeds? J. Agric. Food Chem. 2019, 67, 1325–1343. [Google Scholar] [CrossRef]
- Maleki, S.J.; Crespo, J.F.; Cabanillas, B. Anti-inflammatory effects of flavonoids. Food Chem. 2019, 299, 125124. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Farias, M.; Carrasco-Pozo, C. The anti-cancer effect of quercetin: Molecular implications in cancer metabolism. Int. J. Mol. Sci. 2019, 20, 3177. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Plant genetics. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ye, S.; Yuan, G.; Ma, X.; Heng, S.; Yi, B.; Ma, C.; Shen, J.; Tu, J.; Fu, T.; et al. Gene silencing of BnaA09.ZEP and BnaC09.ZEP confers orange color in Brassica napus flowers. Plant J. 2020, 104, 932–949. [Google Scholar] [CrossRef]
- Hao, P.; Liu, H.; Lin, B.; Ren, Y.; Huang, L.; Jiang, L.; Hua, S. BnaA03.ANS identified by metabolomics and RNA-seq partly played irreplaceable role in pigmentation of red rapeseed (Brassica napus) petal. Front. Plant Sci. 2022, 13, 940765. [Google Scholar] [CrossRef]
- Li, H.; Du, Y.; Zhang, J.; Feng, H.; Liu, J.; Yang, G.; Zhu, Y. Unraveling the mechanism of purple leaf formation in Brassica napus by integrated metabolome and transcriptome analyses. Front. Plant Sci. 2022, 13, 945553. [Google Scholar] [CrossRef]
- Qu, C.; Zhao, H.; Fu, F.; Wang, Z.; Zhang, K.; Zhou, Y.; Wang, X.; Wang, R.; Xu, X.; Tang, Z.; et al. Genome-wide survey of flavonoid biosynthesis genes and gene expression analysis between black- and yellow-seeded Brassica napus. Front. Plant Sci. 2016, 7, 1755. [Google Scholar] [CrossRef]
- Xie, T.; Chen, X.; Guo, T.; Rong, H.; Chen, Z.; Sun, Q.; Batley, J.; Jiang, J.; Wang, Y. Targeted knockout of BnTT2 homologues for yellow-seeded Brassica napus with reduced flavonoids and improved fatty acid composition. J. Agric. Food Chem. 2020, 68, 5676–5690. [Google Scholar] [CrossRef]
- Zhai, Y.; Yu, K.; Cai, S.; Hu, L.; Amoo, O.; Xu, L.; Yang, Y.; Ma, B.; Jiao, Y.; Zhang, C.; et al. Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 2020, 18, 1153–1168. [Google Scholar] [CrossRef]
- Khare, S.; Singh, N.B.; Niharika; Singh, A.; Amist, N.; Azim, Z.; Yadav, R.K. Phytochemicals mitigation of Brassica napus by IAA grown under Cd and Pb toxicity and its impact on growth responses of Anagallis arvensis. J. Biotechnol. 2022, 343, 83–95. [Google Scholar] [CrossRef]
- Koeslin-Findeklee, F.; Rizi, V.S.; Becker, M.A.; Parra-Londono, S.; Arif, M.; Balazadeh, S.; Mueller-Roeber, B.; Kunze, R.; Horst, W.J. Transcriptomic analysis of nitrogen starvation- and cultivar-specific leaf senescence in winter oilseed rape (Brassica napus L.). Plant Sci. 2015, 233, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wu, H.; Zhu, H.; Huang, C.; Liu, C.; Chang, Y.; Kong, Z.; Zhou, Z.; Wang, G.; Lin, Y.; et al. Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytol. 2019, 223, 705–721. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, J.; Xia, X.; Zhang, Z.; He, J.; Nong, B.; Luo, T.; Feng, R.; Wu, Y.; Pan, Y.; et al. OsTTG1, a WD40 repeat gene, regulates anthocyanin biosynthesis in rice. Plant J. 2021, 107, 198–214. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, F.; Zhang, C.; Zhang, M.; Wang, W.; Zhang, C.; Xi, Y. Anthocyanin biosynthesis and a regulatory network of different-colored wheat grains revealed by multiomics analysis. J. Agric. Food Chem. 2022, 70, 887–900. [Google Scholar] [CrossRef]
- Lu, N.; Rao, X.; Li, Y.; Jun, J.H.; Dixon, R.A. Dissecting the transcriptional regulation of proanthocyanidin and anthocyanin biosynthesis in soybean (Glycine max). Plant Biotechnol J. 2021, 19, 1429–1442. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Chao, H.; Zhao, X.; Wang, H.; Li, H.; Zhao, W.; Sun, T.; Li, M.; Huang, J. Anthocyanins identification and transcriptional regulation of anthocyanin biosynthesis in purple Brassica napus. Plant Mol. Biol. 2022, 110, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Mekapogu, M.; Vasamsetti, B.M.K.; Kwon, O.K.; Ahn, M.S.; Lim, S.H.; Jung, J.A. Anthocyanins in floral colors: Biosynthesis and regulation in Chrysanthemum flowers. Int. J. Mol. Sci. 2020, 21, 6537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, S.; Ma, H.; Duan, X.; Gao, S.; Zhou, X.; Cheng, Y. The R2R3-MYB gene PsMYB58 positively regulates anthocyanin biosynthesis in tree peony flowers. Plant Physiol. Biochem. 2021, 164, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Chiu, L.W.; Li, L. Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosynthesis in purple cauliflower. Planta 2012, 236, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, Y.; Sun, H.; Sun, L.; Zhang, L. Transposon-induced methylation of the RsMYB1 promoter disturbs anthocyanin accumulation in red-fleshed radish. J. Exp. Bot. 2020, 71, 2537–2550. [Google Scholar] [CrossRef]
- Jiu, S.; Guan, L.; Leng, X.; Zhang, K.; Haider, M.S.; Yu, X.; Zhu, X.; Zheng, T.; Ge, M.; Wang, C.; et al. The role of VvMYBA2r and VvMYBA2w alleles of the MYBA2 locus in the regulation of anthocyanin biosynthesis for molecular breeding of grape (Vitis spp.) skin coloration. Plant Biotechnol. J. 2021, 19, 1216–1239. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, Z.; Wu, Y.; Zheng, L.; Zhang, G. Regulatory mechanisms of anthocyanin biosynthesis in apple and pear. Int. J. Mol. Sci. 2021, 22, 8441. [Google Scholar] [CrossRef] [PubMed]
- Routaboul, J.M.; Kerhoas, L.; Debeaujon, I.; Pourcel, L.; Caboche, M.; Einhorn, J.; Lepiniec, L. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 2006, 224, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Appelhagen, I.; Thiedig, K.; Nordholt, N.; Schmidt, N.; Huep, G.; Sagasser, M.; Weisshaar, B. Update on transparent testa mutants from Arabidopsis thaliana: Characterisation of new alleles from an isogenic collection. Planta 2014, 240, 955–970. [Google Scholar] [CrossRef]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thevenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef]
- Appelhagen, I.; Jahns, O.; Bartelniewoehner, L.; Sagasser, M.; Weisshaar, B.; Stracke, R. Leucoanthocyanidin dioxygenase in Arabidopsis thaliana: Characterization of mutant alleles and regulation by MYB-BHLH-TTG1 transcription factor complexes. Gene 2011, 484, 62–69. [Google Scholar] [CrossRef]
- Farag, M.A.; Sharaf Eldin, M.G.; Kassem, H.; Abou el Fetouh, M. Metabolome classification of Brassica napus L. organs via UPLC-QTOF-PDA-MS and their anti-oxidant potential. Phytochem. Anal. 2013, 24, 277–287. [Google Scholar] [CrossRef]
- Shao, Y.; Jiang, J.; Ran, L.; Lu, C.; Wei, C.; Wang, Y. Analysis of flavonoids and hydroxycinnamic acid derivatives in rapeseeds (Brassica napus L. var. napus) by HPLC-PDA-ESI(-)-MSn/HRMS. J. Agric. Food Chem. 2014, 62, 2935–2945. [Google Scholar] [CrossRef]
- Qu, C.; Fu, F.; Lu, K.; Zhang, K.; Wang, R.; Xu, X.; Wang, M.; Lu, J.; Wan, H.; Tang, Z.; et al. Differential accumulation of phenolic compounds and expression of related genes in black- and yellow-seeded Brassica napus. J. Exp. Bot. 2013, 64, 2885–2898. [Google Scholar] [CrossRef]
- Li, A.; Jiang, J.; Zhang, Y.; Snowdon, R.J.; Liang, G.; Wang, Y. Molecular and cytological characterization of introgression lines in yellow seed derived from somatic hybrids between Brassica napus and Sinapis alba. Mol. Breed. 2012, 29, 209–219. [Google Scholar] [CrossRef]
- Jiang, J.; Shao, Y.; Li, A.; Lu, C.; Zhang, Y.; Wang, Y. Phenolic composition analysis and gene expression in developing seeds of yellow- and black-seeded Brassica napus. J. Integr. Plant Biol. 2013, 55, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Meng, G.; Chen, S.; Chen, Y.; Jiang, J.; Wang, Y.P. Correlation analysis of phenolic contents and antioxidation in yellow- and black-seeded Brassica napus. Molecules 2018, 23, 1815. [Google Scholar] [CrossRef] [PubMed]
- Yin, N.W.; Wang, S.X.; Jia, L.D.; Zhu, M.C.; Yang, J.; Zhou, B.J.; Yin, J.M.; Lu, K.; Wang, R.; Li, J.N.; et al. Identification and characterization of major constituents in different-colored rapeseed petals by UPLC-HESI-MS/MS. J. Agric. Food Chem. 2019, 67, 11053–11065. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Yang, L.; Han, P.; Gu, C.; Li, Y.; Liao, X.; Qin, L. Metabolic profiles reveal changes in the leaves and roots of rapeseed (Brassica napus L.) seedlings under nitrogen deficiency. Int. J. Mol. Sci. 2022, 23, 5784. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.; Lu, X.; Yin, N.; Ma, L.; Lu, J.; Liu, X.; Li, J.; Lu, J.; Lei, B.; Wang, R.; et al. Silencing of BnTT1 family genes affects seed flavonoid biosynthesis and alters seed fatty acid composition in Brassica napus. Plant Sci. 2017, 254, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Lu, K.; Qu, C.; Liang, Y.; Wang, R.; Chai, Y.; Li, J. Gene silencing of BnTT10 family genes causes retarded pigmentation and lignin reduction in the seed coat of Brassica napus. PLoS ONE 2013, 8, e61247. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Chen, D.; Pan, Q.; Li, F.; Zhao, Z.; Ge, X.; Li, Z. Production of red-flowered oilseed rape via the ectopic expression of Orychophragmus violaceus OvPAP2. Plant Biotechnol. J. 2018, 16, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Hua, S.; Ma, T.; Ma, X.; Chen, Y.; Wu, L.; Zhao, L.; Yi, B.; Ma, C.; Tu, J.; et al. Genetic and multi-omics analysis reveal BnaA07.PAP2In-184-317 as the key gene conferring anthocyanin-based color in Brassica napus flowers. J. Exp. Bot. 2022, 73, 6630–6645. [Google Scholar] [CrossRef]
- Li, H.; Yu, K.; Amoo, O.; Yu, Y.; Guo, M.; Deng, S.; Li, M.; Hu, L.; Wang, J.; Fan, C.; et al. Site-directed mutagenesis of the carotenoid isomerase gene BnaCRTISO alters the color of petals and leaves in Brassica napus L. Front. Plant Sci. 2022, 13, 801456. [Google Scholar] [CrossRef]
- Li, H.; Zhu, L.; Yuan, G.; Heng, S.; Yi, B.; Ma, C.; Shen, J.; Tu, J.; Fu, T.; Wen, J. Fine mapping and candidate gene analysis of an anthocyanin-rich gene, BnaA.PL1, conferring purple leaves in Brassica napus L. Mol. Genet. Genomics 2016, 291, 1523–1534. [Google Scholar] [CrossRef]
- Chen, D.; Liu, Y.; Yin, S.; Qiu, J.; Jin, Q.; King, G.J.; Wang, J.; Ge, X.; Li, Z. Alternatively spliced BnaPAP2.A7 isoforms play opposing roles in anthocyanin biosynthesis of Brassica napus L. Front. Plant Sci. 2020, 11, 983. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Guo, Y.; Wang, J.; Li, D.; Liu, K.; Qi, S.; Jin, C.; Duan, S.; Gong, J.; Li, Z.; et al. Brassica napus GLABRA3-1 promotes anthocyanin biosynthesis and trichome formation in true leaves when expressed in Arabidopsis thaliana. Plant Biol. 2018, 20, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gao, M.J.; Pan, H.Y.; Cui, D.J.; Gruber, M.Y. Purple canola: Arabidopsis PAP1 increases antioxidants and phenolics in Brassica napus leaves. J. Agric. Food Chem. 2010, 58, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Chatterjee, M.; Burman, N.; Khurana, J.P. Cryptochrome 1 regulates growth and development in Brassica through alteration in the expression of genes involved in light, phytohormone and stress signalling. Plant Cell Environ. 2014, 37, 961–977. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Mishra, S.; Burman, N.; Chatterjee, M.; Singh, S.; Pradhan, A.K.; Khurana, P.; Khurana, J.P. Characterization of Cry2 genes (CRY2a and CRY2b) of B. napus and comparative analysis of BnCRY1 and BnCRY2a in regulating seedling photomorphogenesis. Plant Mol. Biol. 2022, 110, 161–186. [Google Scholar] [CrossRef]
- Kim, J.; Lee, W.J.; Vu, T.T.; Jeong, C.Y.; Hong, S.W.; Lee, H. High accumulation of anthocyanins via the ectopic expression of AtDFR confers significant salt stress tolerance in Brassica napus L. Plant Cell Rep. 2017, 36, 1215–1224. [Google Scholar] [CrossRef]
- Vu, T.T.; Jeong, C.Y.; Nguyen, H.N.; Lee, D.; Lee, S.A.; Kim, J.H.; Hong, S.W.; Lee, H. Characterization of Brassica napus flavonol synthase involved in flavonol biosynthesis in Brassica napus L. J. Agric. Food Chem. 2015, 63, 7819–7829. [Google Scholar] [CrossRef]
- Ellerstrom, M.; Reidt, W.; Ivanov, R.; Tiedemann, J.; Melzer, M.; Tewes, A.; Moritz, T.; Mock, H.P.; Sitbon, F.; Rask, L.; et al. Ectopic expression of EFFECTOR OF TRANSCRIPTION perturbs gibberellin-mediated plant developmental processes. Plant Mol. Biol. 2005, 59, 663–681. [Google Scholar] [CrossRef]
- Schilbert, H.M.; Schone, M.; Baier, T.; Busche, M.; Viehover, P.; Weisshaar, B.; Holtgrawe, D. Characterization of the Brassica napus flavonol synthase gene family reveals bifunctional flavonol synthases. Front. Plant Sci. 2021, 12, 733762. [Google Scholar] [CrossRef]
- Jia, L.; Wu, Q.; Ye, N.; Liu, R.; Shi, L.; Xu, W.; Zhi, H.; Rahman, A.N.; Xia, Y.; Zhang, J. Proanthocyanidins inhibit seed germination by maintaining a high level of abscisic acid in Arabidopsis thaliana. J. Integr. Plant Biol. 2012, 54, 663–673. [Google Scholar] [CrossRef]
- Bhinder, G.; Sharma, S.; Kaur, H.; Akhatar, J.; Mittal, M.; Sandhu, S. Genomic regions associated with seed meal quality traits in Brassica napus germplasm. Front. Plant Sci. 2022, 13, 882766. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.Y. Molecular mechanism of manipulating seed coat coloration in oilseed Brassica species. J. Appl. Genet. 2013, 54, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.B.; Li, J.N.; Zhang, X.K.; Wang, R.; Xie, L.L.; Chai, Y.R. Cloning and molecular characterization of a functional flavonoid 3′-hydroxylase gene from Brassica napus. J. Plant Physiol. 2007, 164, 350–363. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.L.; Li, J.N.; Lu, J.; Tang, Z.L.; Pu, D.C.; Chai, Y.R. Molecular cloning of Brassica napus TRANSPARENT TESTA 2 gene family encoding potential MYB regulatory proteins of proanthocyanidin biosynthesis. Mol. Biol. Rep. 2007, 34, 105–120. [Google Scholar] [CrossRef] [PubMed]
- Auger, B.; Baron, C.; Lucas, M.O.; Vautrin, S.; Berges, H.; Chalhoub, B.; Fautrel, A.; Renard, M.; Nesi, N. Brassica orthologs from BANYULS belong to a small multigene family, which is involved in procyanidin accumulation in the seed. Planta 2009, 230, 1167–1183. [Google Scholar] [CrossRef][Green Version]
- Chai, Y.R.; Lei, B.; Huang, H.L.; Li, J.N.; Yin, J.M.; Tang, Z.L.; Wang, R.; Chen, L. TRANSPARENT TESTA 12 genes from Brassica napus and parental species: Cloning, evolution, and differential involvement in yellow seed trait. Mol. Genet. Genomics 2009, 281, 109–123. [Google Scholar] [CrossRef]
- Ni, Y.; Jiang, H.; Li, J.; Chai, Y. Molecular cloning, characterization and expression of two rapeseed (Brassica napus L.) cDNAs orthologous to Arabidopsis thaliana phenylalanine ammonialyase 1. Euphytica 2007, 159, 1–16. [Google Scholar] [CrossRef]
- Lu, J.; Li, J.; Lei, B.; Wang, S.; Chai, Y. Molecular cloning and characterization of two Brassica napus TTG1 genes reveal genus-specific nucleotide preference, extreme protein-level conservation and fast divergence of organ-specificity. Genes Genomics 2009, 31, 129–142. [Google Scholar] [CrossRef]
- Liu, X.; Lu, Y.; Yan, M.; Sun, D.; Hu, X.; Liu, S.; Chen, S.; Guan, C.; Liu, Z. Genome-wide identification, localization, and expression analysis of proanthocyanidin-associated genes in Brassica. Front Plant Sci. 2016, 7, 1831. [Google Scholar] [CrossRef]
- Hong, M.; Hu, K.; Tian, T.; Li, X.; Chen, L.; Zhang, Y.; Yi, B.; Wen, J.; Ma, C.; Shen, J.; et al. Transcriptomic analysis of seed coats in yellow-seeded Brassica napus reveals novel genes that influence proanthocyanidin biosynthesis. Front. Plant Sci. 2017, 8, 1674. [Google Scholar] [CrossRef]
- Jiang, J.; Zhu, S.; Yuan, Y.; Wang, Y.; Zeng, L.; Batley, J.; Wang, Y.P. Transcriptomic comparison between developing seeds of yellow- and black-seeded Brassica napus reveals that genes influence seed quality. BMC Plant Biol. 2019, 19, 203. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Ma, J.; Xu, F.; Xu, W.; Jiang, H.; Zhang, H.; Qu, C.; Wei, L.; Li, J. Differences in alternative splicing between yellow and black-seeded rapeseed. Plants 2020, 9, 977. [Google Scholar] [CrossRef] [PubMed]
- Badani, A.G.; Snowdon, R.J.; Wittkop, B.; Lipsa, F.D.; Baetzel, R.; Horn, R.; De Haro, A.; Font, R.; Luhs, W.; Friedt, W. Colocalization of a partially dominant gene for yellow seed colour with a major QTL influencing acid detergent fibre (ADF) content in different crosses of oilseed rape (Brassica napus). Genome 2006, 49, 1499–1509. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X.; Chen, W.; Yi, B.; Wen, J.; Shen, J.X.; Ma, C.Z.; Chen, B.Y.; Tu, J.X.; Fu, T.D. Identification of two major QTL for yellow seed color in two crosses of resynthesized Brassica napus line No. 2127-17. Mol. Breed. 2011, 28, 335–342. [Google Scholar] [CrossRef]
- Liu, L.; Stein, A.; Wittkop, B.; Sarvari, P.; Li, J.; Yan, X.; Dreyer, F.; Frauen, M.; Friedt, W.; Snowdon, R.J. A knockout mutation in the lignin biosynthesis gene CCR1 explains a major QTL for acid detergent lignin content in Brassica napus seeds. Theor. Appl. Genet. 2012, 124, 1573–1586. [Google Scholar] [CrossRef]
- Stein, A.; Wittkop, B.; Liu, L.Z.; Obermeier, C.; Friedt, W.; Snowdon, R.J. Dissection of a major QTL for seed colour and fibre content in Brassica napus reveals colocalization with candidate genes for phenylpropanoid biosynthesis and flavonoid deposition. Plant Breed. 2013, 132, 382–389. [Google Scholar] [CrossRef]
- Liu, L.; Qu, C.; Wittkop, B.; Yi, B.; Xiao, Y.; He, Y.; Snowdon, R.J.; Li, J. A high-density SNP map for accurate mapping of seed fibre QTL in Brassica napus L. PLoS ONE 2013, 8, e83052. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.; Zhao, H.; Fu, F.; Zhang, K.; Yuan, J.; Liu, L.; Wang, R.; Xu, X.; Lu, K.; Li, J.N. Molecular mapping and QTL for expression profiles of flavonoid genes in Brassica napus. Front. Plant Sci. 2016, 7, 1691. [Google Scholar] [CrossRef]
- Wang, J.; Xian, X.; Xu, X.; Qu, C.; Lu, K.; Li, J.; Liu, L. Genome-wide association mapping of seed coat color in Brassica napus. J. Agric. Food Chem. 2017, 65, 5229–5237. [Google Scholar] [CrossRef]
- Gacek, K.; Bayer, P.E.; Anderson, R.; Severn-Ellis, A.A.; Wolko, J.; Lopatynska, A.; Matuszczak, M.; Bocianowski, J.; Edwards, D.; Batley, J. QTL genetic mapping study for traits affecting meal quality in winter oilseed rape (Brassica napus L.). Genes 2021, 12, 1235. [Google Scholar] [CrossRef]
- Chao, H.; Guo, L.; Zhao, W.; Li, H.; Li, M. A major yellow-seed QTL on chromosome A09 significantly increases the oil content and reduces the fiber content of seed in Brassica napus. Theor. Appl. Genet. 2022, 135, 1293–1305. [Google Scholar] [CrossRef]
- Tohge, T.; de Souza, L.P.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028. [Google Scholar] [CrossRef] [PubMed]
- Iwashina, T. Contribution to flower colors of flavonoids including anthocyanins: A review. Nat. Prod. Commun. 2015, 10, 529–544. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Zhang, Z.; Zhang, L. Anthocyanin accumulation, antioxidant ability and stability, and a transcriptional analysis of anthocyanin biosynthesis in purple heading chinese cabbage (Brassica rapa L. ssp. pekinensis). J. Agric. Food Chem. 2016, 64, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, Z.; Zhu, M.; Zhu, Z.; Wang, Z.; Tian, S.; Chen, G. Anthocyanin accumulation and molecular analysis of correlated genes in purple kohlrabi (Brassica oleracea var. gongylodes L.). J. Agric. Food Chem. 2015, 63, 4160–4169. [Google Scholar] [CrossRef]
- Heng, S.; Cheng, Q.; Zhang, T.; Liu, X.; Huang, H.; Yao, P.; Liu, Z.; Wan, Z.; Fu, T. Fine-mapping of the BjPur gene for purple leaf color in Brassica juncea. Theor. Appl. Genet. 2020, 133, 2989–3000. [Google Scholar] [CrossRef]
- Zhang, K.; Yang, D.; Hu, Y.; Njogu, M.K.; Qian, J.; Jia, L.; Yan, C.; Li, Z.; Wang, X.; Wang, L. Integrated analysis of transcriptome and metabolome reveals new insights into the formation of purple leaf veins and leaf edge cracks in Brassica juncea. Plants 2022, 11, 2229. [Google Scholar] [CrossRef]
- Goswami, G.; Nath, U.K.; Park, J.I.; Hossain, M.R.; Biswas, M.K.; Kim, H.T.; Kim, H.R.; Nou, I.S. Transcriptional regulation of anthocyanin biosynthesis in a high-anthocyanin resynthesized Brassica napus cultivar. J. Biol. Res. 2018, 25, 19. [Google Scholar] [CrossRef]
- He, D.; Zhang, D.; Li, T.; Liu, L.; Zhou, D.; Kang, L.; Wu, J.; Liu, Z.; Yan, M. Whole-genome identification and comparative expression analysis of anthocyanin biosynthetic genes in Brassica napus. Front. Genet. 2021, 12, 764835. [Google Scholar] [CrossRef]
- Yu, E.; Fan, C.; Yang, Q.; Li, X.; Wan, B.; Dong, Y.; Wang, X.; Zhou, Y. Identification of heat responsive genes in Brassica napus siliques at the seed-filling stage through transcriptional profiling. PLoS ONE 2014, 9, e101914. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, Z.; Anjum, S.; Skalicky, M.; Waraich, E.A.; Muhammad Sabir Tariq, R.; Ayub, M.A.; Hossain, A.; Hassan, M.M.; Brestic, M.; Sohidul Islam, M.; et al. Selenium alleviates the adverse effect of drought in oilseed crops camelina (Camelina sativa L.) and canola (Brassica napus L.). Molecules 2021, 26, 1699. [Google Scholar] [CrossRef] [PubMed]
- Mi, W.; Liu, Z.; Jin, J.; Dong, X.; Xu, C.; Zou, Y.; Xu, M.; Zheng, G.; Cao, X.; Fang, X.; et al. Comparative proteomics analysis reveals the molecular mechanism of enhanced cold tolerance through ROS scavenging in winter rapeseed (Brassica napus L.). PLoS ONE 2021, 16, e0243292. [Google Scholar] [CrossRef] [PubMed]
- Sami, A.; Shah, F.A.; Abdullah, M.; Zhou, X.; Yan, Y.; Zhu, Z.; Zhou, K. Melatonin mitigates cadmium and aluminium toxicity through modulation of antioxidant potential in Brassica napus L. Plant Biol. 2020, 22, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Miao, Y.; Ayyaz, A.; Hannan, F.; Huang, Q.; Ulhassan, Z.; Zhou, Y.; Islam, F.; Hong, Z.; Farooq, M.A.; et al. Purple stem Brassica napus exhibits higher photosynthetic efficiency, antioxidant potential and anthocyanin biosynthesis related genes expression against drought stress. Front. Plant Sci. 2022, 13, 936696. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, K.E.; Lampi, M.A.; Greenberg, B.M. The effects of far-red light on plant growth and flavonoid accumulation in Brassica napus in the presence of ultraviolet B radiation. Photochem. Photobiol. 2008, 84, 1445–1454. [Google Scholar] [CrossRef]
- Luo, Y.; Teng, S.; Yin, H.; Zhang, S.; Tuo, X.; Tran, L.P. Transcriptome analysis reveals roles of anthocyanin- and jasmonic acid-biosynthetic pathways in rapeseed in response to high light stress. Int. J. Mol. Sci. 2021, 22, 13027. [Google Scholar] [CrossRef]
- Groenbaek, M.; Tybirk, E.; Neugart, S.; Sundekilde, U.K.; Schreiner, M.; Kristensen, H.L. Flavonoid glycosides and hydroxycinnamic acid derivatives in baby leaf rapeseed from white and yellow flowering cultivars with repeated harvest in a 2-years field study. Front. Plant Sci. 2019, 10, 355. [Google Scholar] [CrossRef]
- Brazaityte, A.; Miliauskiene, J.; Vastakaite-Kairiene, V.; Sutuliene, R.; Lauzike, K.; Duchovskis, P.; Malek, S. Effect of different ratios of blue and red LED light on Brassicaceae microgreens under a controlled environment. Plants 2021, 10, 801. [Google Scholar] [CrossRef]
- Lee, J.H.; Shibata, S.; Goto, E. Time-course of changes in photosynthesis and secondary metabolites in canola (Brassica napus) under different UV-B irradiation levels in a plant factory with artificial light. Front. Plant Sci. 2021, 12, 786555. [Google Scholar] [CrossRef]
- Chatterjee, M.; Sharma, P.; Khurana, J.P. Cryptochrome 1 from Brassica napus is up-regulated by blue light and controls hypocotyl/stem growth and anthocyanin accumulation. Plant Physiol. 2006, 141, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.W.; Malchev, I.T.; Rajcan, I.; Kott, L.S. Identification of putative quantitative trait loci associated with a flavonoid related to resistance to cabbage seedpod weevil (Ceutorhynchus obstrictus) in canola derived from an intergeneric cross, Sinapis alba × Brassica napus. Theor. Appl. Genet. 2014, 127, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ye, C.; Zhao, R.; Li, X.; Liu, W.Z.; Wu, F.; Yan, J.; Jiang, Y.Q.; Yang, B. Mitogen-activated protein kinase kinase kinase (MAPKKK) 4 from rapeseed (Brassica napus L.) is a novel member inducing ROS accumulation and cell death. Biochem. Biophys. Res. Commun. 2015, 467, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.T.; Lee, B.R.; Park, S.H.; La, V.H.; Jung, W.J.; Bae, D.W.; Kim, T.H. Hormonal regulations in soluble and cell-wall bound phenolic accumulation in two cultivars of Brassica napus contrasting susceptibility to Xanthomonas campestris pv. campestris. Plant Sci. 2019, 285, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Ding, L.N.; Li, M.; Cao, W.; Wang, Y.K.; Wang, W.J.; Yu, Y.-K.; Wang, Z.; Zhu, K.M.; Tan, X.L. Characterization of a rapeseed anthocyanin-more mutant with enhanced resistance to Sclerotinia sclerotiorum. J. Plant Growth Regul. 2019, 39, 703–716. [Google Scholar] [CrossRef]
- Ding, L.N.; Liu, R.; Li, T.; Li, M.; Liu, X.Y.; Wang, W.J.; Yu, Y.K.; Cao, J.; Tan, X.L. Physiological and comparative transcriptome analyses reveal the mechanisms underlying waterlogging tolerance in a rapeseed anthocyanin-more mutant. Biotechnol. Biofuels Bioprod. 2022, 15, 55. [Google Scholar] [CrossRef]
| Gene | Origin | Function | Phenotype | Reference |
|---|---|---|---|---|
| BnTT8 | Brassica napus | BnTT8 mutation suppressed the phenylpropanoid and flavonoid biosynthetic gene expression, and inhibited proanthocyanidin accumulation in seed coat of rapeseed. | Seed color | [20] |
| BnTT1 | Brassica napus | Silencing of BnTT1 reduced flavonoid accumulation and fatty acid biosynthesis through altering gene expression in flavonoid and fatty acid biosynthesis. | Seed color | [46] |
| BnTT10 | Brassica napus | Silencing of BnTT10 increased soluable proanthocyanidins, decreased extractable lignin, and retarded pigmentation in seed coat of B. napus. | Seed color | [47] |
| BnTT2 | Brassica napus | Mutation of BnTT2 reduced flavonoids and improved fatty acid composition in seed of B. napus. | Seed color | [19] |
| OvPAP2 | Orychophragmus violaceus | Ectopic expression of OvPAP2 led to red anthers and petals in B. napus. | Petal color | [48] |
| BnaA03.ANS | Brassica napus | RNA interference of BnaA03.ANS repressed anthocyanin accumulation in red petal rapeseed. | Petal color | [16] |
| BnaA07.PAP2 | Brassica napus | The insertions in −184 and −371 bp were responsible for the transcriptional activation of BnaA07.PAP2 and anthocyanin-related genes, and resulted apricot petal color in rapeseed. | Petal color | [49] |
| BnaA09.ZEP/BnaC09.ZEP | Brassica napus | BnaA09.ZEP and BnaC09.ZEP negatively regulated the orange color in rapeseed petals by affecting the carotenoid and flavonoid content, as well as the expression of carotenoid and flavonoid biosynthetic genes. | Petal color | [15] |
| BnaCRTISO | Brassica napus | BnaCRTISO mutation reduced chalcone content and increased carotene content, thus changing the petal and leaf color of rapeseed. | Petal/leaf color | [50] |
| BnaA.PL1 | Brassica napus | A QTL locus for anthocyanin-rich mutant of rapeseed, including a candidate gene BnAPR2 that encoded adenosine 5’-phosphosulfate reductase. | Leaf color | [51] |
| BnaPAP2.A7 | Brassica napus | Three isoforms of BnaPAP2.A7 identified in rapeseed introgression line were confirmed with different roles in manuplating anthocyanin accumulation in leaves. | Leaf color | [52] |
| BnGL3-1 | Brassica napus | Ectopic expression of BnGL3-1 increased the trichome number and anthocyanin accumulation in true leaves of Arabidopsis gl3-3 mutant. | Leaf color | [53] |
| AtPAP1 | Arabidopsis thaliana | Overexpression of Arabidopsis PAP1 increased flavonoid and sinapic acid accumulation in leaves and stems of rapeseed. | Leaf/stem color | [54] |
| BnCRY1/BnCRY2 | Brassica napus | Overexpression of BnCRY1 and BnCRY2a increased anthocyanin content and regulated seedling photomorphogenesis of B. napus. | Seedling development | [55,56] |
| AtDFR | Arabidopsis thaliana | Overexpression of Arabidopsis AtDFR increased anthocyanin accumulation and improved salt tolerance of B. napus. | Salt tolerance | [57] |
| BnFLS | Brassica napus | Overexpression of BnFLS recovered the flavonol content in Arabidopsis atfls1-ko mutant. | -- | [58] |
| BnET | Brassica napus | Overexpression of BnET promoted anthocyanin accumulation in Arabidopsis. | -- | [59] |
| BnFLS1-1/1-2 | Brassica napus | BnFLS1-1 and BnFLS1-2 restored the flavonoid content in Arabidopsis ans/fls1 and f3h mutants. | -- | [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-Y.; Lu, H.-Q.; Jiang, K.-X.; Wang, Y.-R.; Wang, Y.-P.; Jiang, J.-J. The Flavonoid Biosynthesis and Regulation in Brassica napus: A Review. Int. J. Mol. Sci. 2023, 24, 357. https://doi.org/10.3390/ijms24010357
Chen Y-Y, Lu H-Q, Jiang K-X, Wang Y-R, Wang Y-P, Jiang J-J. The Flavonoid Biosynthesis and Regulation in Brassica napus: A Review. International Journal of Molecular Sciences. 2023; 24(1):357. https://doi.org/10.3390/ijms24010357
Chicago/Turabian StyleChen, Yuan-Yuan, Hai-Qin Lu, Kai-Xuan Jiang, Yi-Ran Wang, You-Ping Wang, and Jin-Jin Jiang. 2023. "The Flavonoid Biosynthesis and Regulation in Brassica napus: A Review" International Journal of Molecular Sciences 24, no. 1: 357. https://doi.org/10.3390/ijms24010357
APA StyleChen, Y.-Y., Lu, H.-Q., Jiang, K.-X., Wang, Y.-R., Wang, Y.-P., & Jiang, J.-J. (2023). The Flavonoid Biosynthesis and Regulation in Brassica napus: A Review. International Journal of Molecular Sciences, 24(1), 357. https://doi.org/10.3390/ijms24010357





