On-Chip Organoid Formation to Study CXCR4/CXCL-12 Chemokine Microenvironment Responses for Renal Cancer Drug Testing
Abstract
1. Introduction
2. Materials and Methods
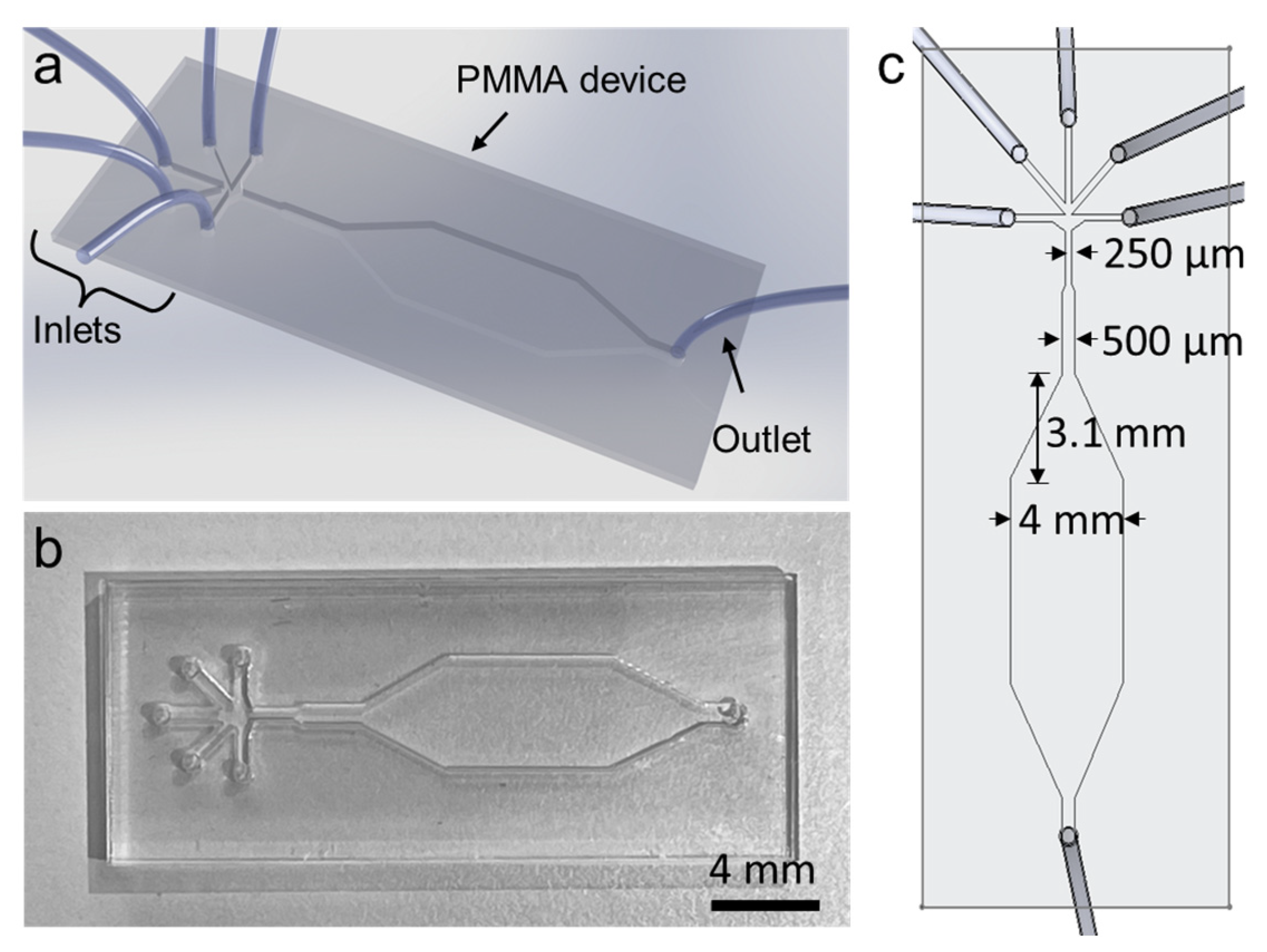
2.1. Microfluidic Device Fabrication
2.2. Cell Culture Conditions
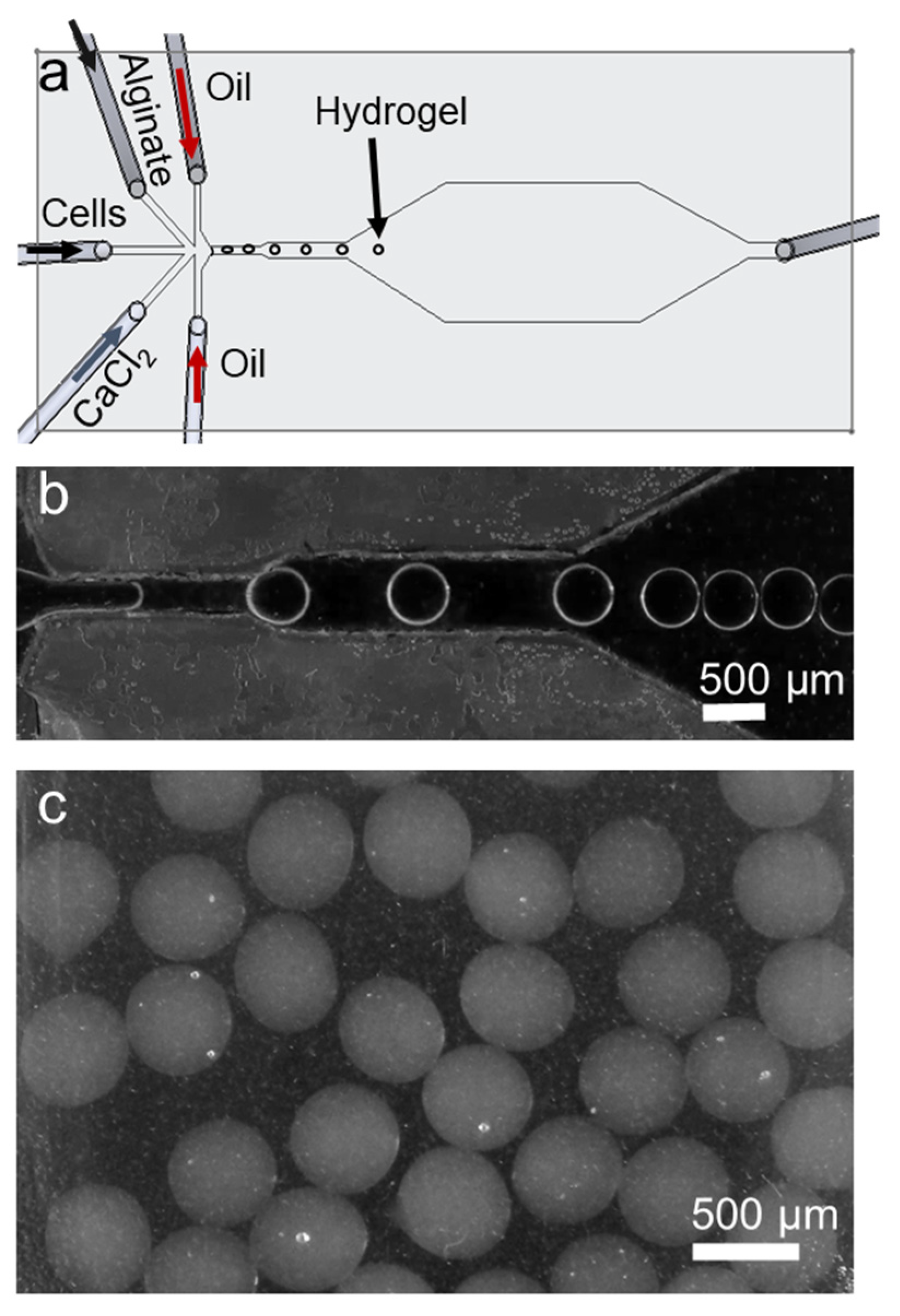
2.3. Organoid Culture with Alginate Beads
2.4. Cell Viability with MTT Assay
2.5. Calcein Dye Staining
2.6. HIF-1α, CXCR4 and CXCL-12 Protein Levels Measurement
2.7. CXCR4 and CXCL-12 Gene Expression Measurement
2.8. Statistical Analysis
3. Results
3.1. Microfluidic Device and Working Principle
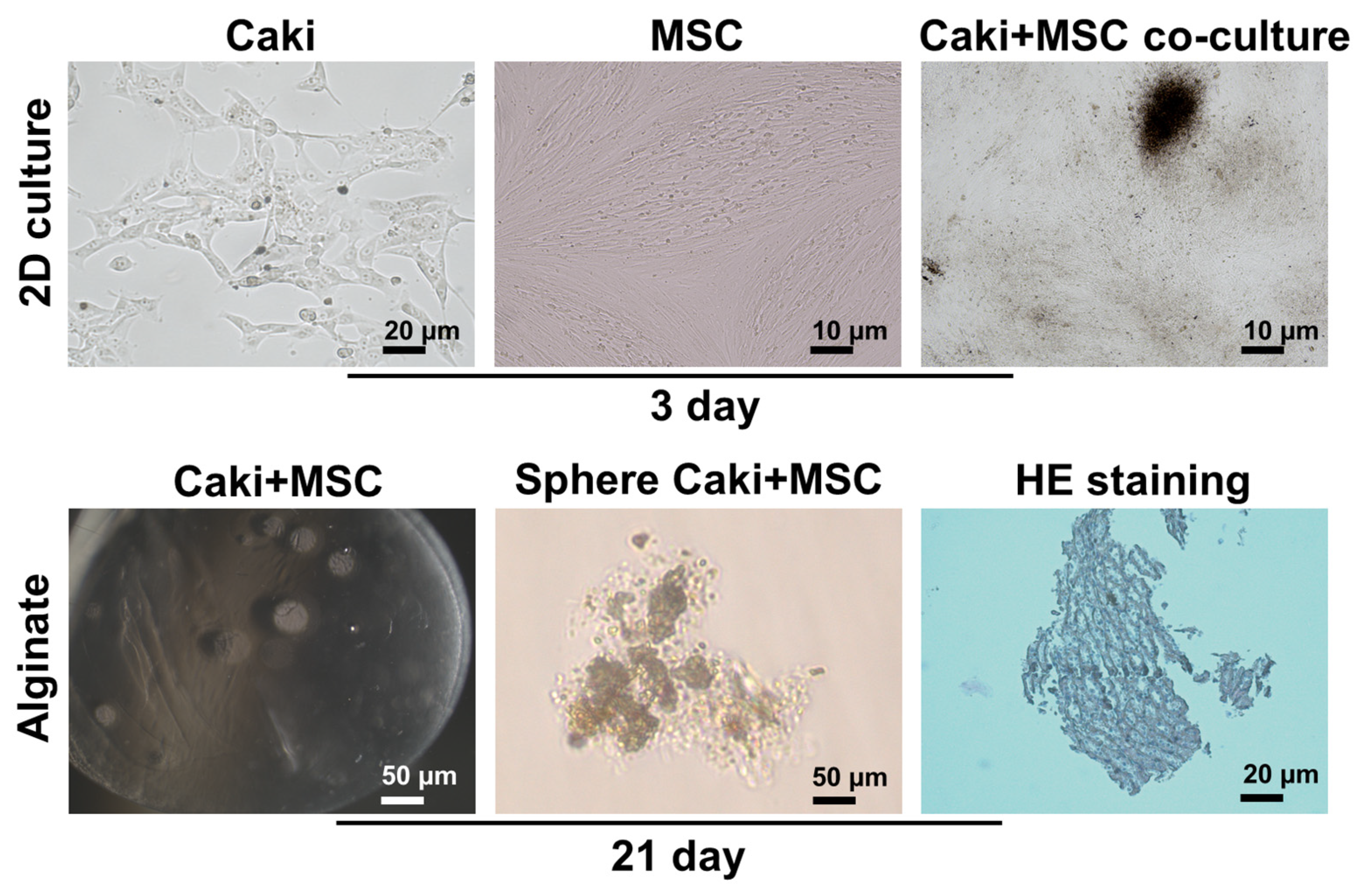
3.2. Characterization of Organoids
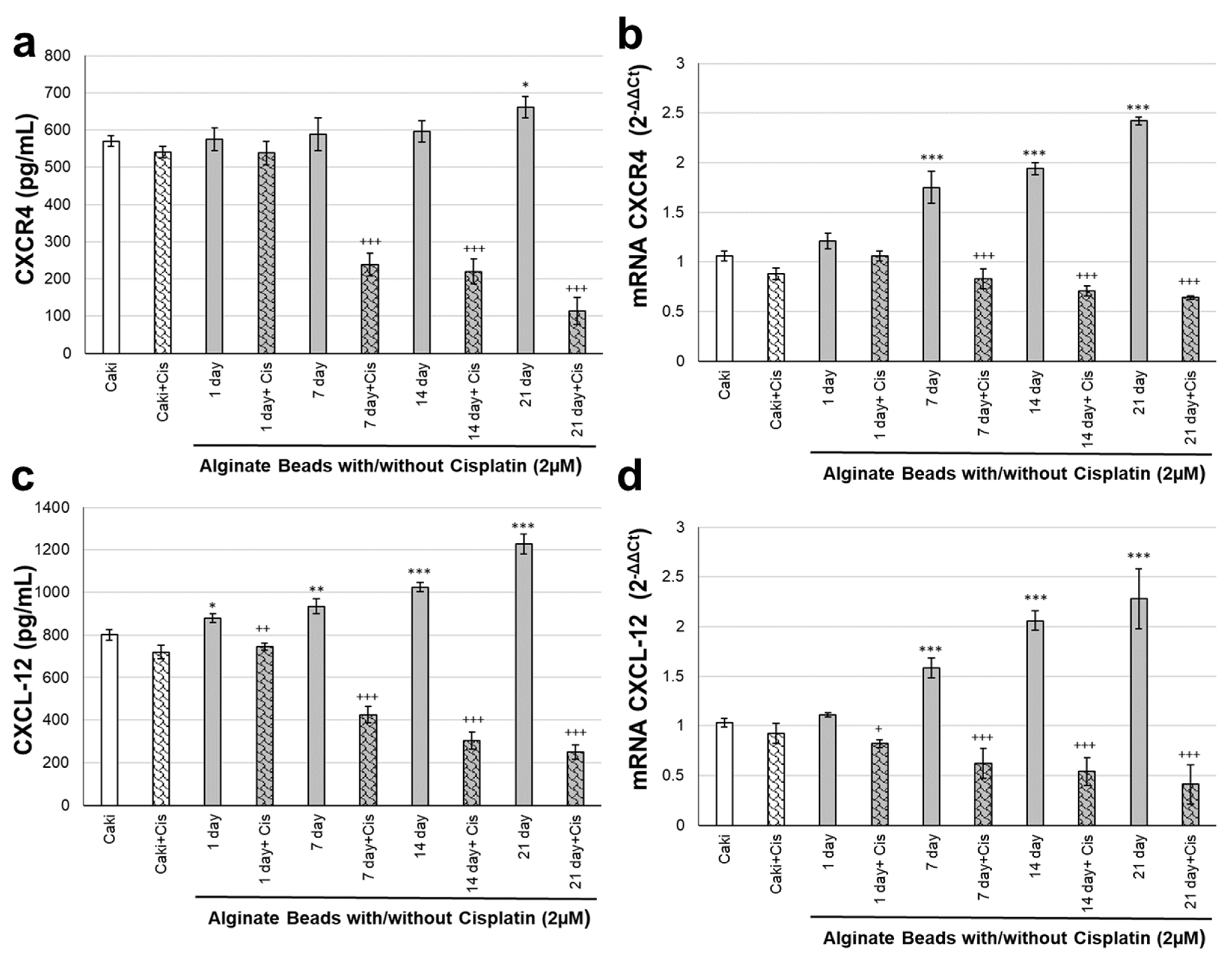
3.3. CXCR4 and CXCL-12 Changes on Renal Organoids
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drost, J.; Clevers, H. Translational applications of adult stem cell-derived organoids. Development 2017, 144, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.J.; Van Es, J.H.; Van Den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef]
- Sachs, N.; Clevers, H. Organoid cultures for the analysis of cancer phenotypes. Curr. Opin. Genet. Dev. 2014, 24, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Fendler, A.; Bauer, D.; Busch, J.; Jung, K.; Wulf-Goldenberg, A.; Kunz, S.; Song, K.; Myszczyszyn, A.; Elezkurtaj, S.; Erguen, B.; et al. Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat. Commun. 2020, 11, 929. [Google Scholar] [CrossRef] [PubMed]
- Calandrini, C.; Schutgens, F.; Oka, R.; Margaritis, T.; Candelli, T.; Mathijsen, L.; Ammerlaan, C.; van Ineveld, R.L.; Derakhshan, S.; de Haan, S.; et al. An organoid biobank for childhood kidney cancers that captures disease and tissue heterogeneity. Nat. Commun. 2020, 11, 1310. [Google Scholar] [CrossRef] [PubMed]
- Eggener, S.E.; Yossepowitch, O.; Pettus, J.A.; Snyder, M.E.; Motzer, R.J.; Russo, P. Renal Cell Carcinoma Recurrence After Nephrectomy for Localized Disease: Predicting Survival From Time of Recurrence. J. Clin. Oncol. 2006, 24, 3101–3106. [Google Scholar] [CrossRef]
- Jonasch, E.; Gao, J.; Rathmell, W.K. Renal cell carcinoma. BMJ 2014, 349, g4797. [Google Scholar] [CrossRef]
- Hoefflin, R.; Harlander, S.; Schäfer, S.; Metzger, P.; Kuo, F.; Schönenberger, D.; Adlesic, M.; Peighambari, A.; Seidel, P.; Chen, C.; et al. HIF-1α and HIF-2α differently regulate tumour development and inflammation of clear cell renal cell carcinoma in mice. Nat. Commun. 2020, 11, 4111. [Google Scholar] [CrossRef]
- Liu, M.; Cardilla, A.; Ngeow, J.; Gong, X.; Xia, Y. Studying Kidney Diseases Using Organoid Models. Front. Cell Dev. Biol. 2022, 10, 845401. [Google Scholar] [CrossRef]
- Wood, L. Sunitinib malate for the treatment of renal cell carcinoma. Expert Opin. Pharmacother. 2012, 13, 1323–1336. [Google Scholar] [CrossRef] [PubMed]
- Grassi, L.; Alfonsi, R.; Francescangeli, F.; Signore, M.; De Angelis, M.L.; Addario, A.; Costantini, M.; Flex, E.; Ciolfi, A.; Pizzi, S.; et al. Organoids as a new model for improving regenerative medicine and cancer personalized therapy in renal diseases. Cell Death Dis. 2019, 10, 201. [Google Scholar] [CrossRef]
- Kim, D.; Dressler, G.R. Nephrogenic Factors Promote Differentiation of Mouse Embryonic Stem Cells into Renal Epithelia. J. Am. Soc. Nephrol. 2005, 16, 3527–3534. [Google Scholar] [CrossRef]
- Taguchi, A.; Nishinakamura, R. Higher-Order Kidney Organogenesis from Pluripotent Stem Cells. Cell Stem Cell 2017, 21, 730–746. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. Applications of patient-derived tumor xenograft models and tumor organoids. J. Hematol. Oncol. 2020, 13, 4. [Google Scholar] [CrossRef] [PubMed]
- Bolck, H.A.; Corrò, C.; Kahraman, A.; von Teichman, A.; Toussaint, N.C.; Kuipers, J.; Chiovaro, F.; Koelzer, V.H.; Pauli, C.; Moritz, W.; et al. Tracing Clonal Dynamics Reveals that Two- and Three-dimensional Patient-derived Cell Models Capture Tumor Heterogeneity of Clear Cell Renal Cell Carcinoma. Eur. Urol. Focus 2021, 7, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xu, H.; Yu, L.; Wang, J.; Meng, Q.; Mei, H.; Cai, Z.; Chen, W.; Huang, W. Patient-derived renal cell carcinoma organoids for personalized cancer therapy. Clin. Transl. Med. 2022, 12, e970. [Google Scholar] [CrossRef]
- Guo, F.; Li, P.; French, J.B.; Mao, Z.; Zhao, H.; Li, S.; Nama, N.; Fick, J.R.; Benkovic, S.J.; Huang, T.J. Controlling cell–cell interactions using surface acoustic waves. Proc. Natl. Acad. Sci. USA 2015, 112, 43–48. [Google Scholar] [CrossRef]
- Wu, Y.; Ao, Z.; Chen, B.; Muhsen, M.; Bondesson, M.; Lu, X.; Guo, F. Acoustic assembly of cell spheroids in disposable capillaries. Nanotechnology 2018, 29, 504006. [Google Scholar] [CrossRef]
- Regmi, S.; Poudel, C.; Adhikari, R.; Luo, K.Q. Applications of Microfluidics and Organ-on-a-Chip in Cancer Research. Biosensors 2022, 12, 459. [Google Scholar] [CrossRef]
- Homan, K.A.; Gupta, N.; Kroll, K.T.; Kolesky, D.B.; Skylar-Scott, M.; Miyoshi, T.; Mau, D.; Valerius, M.T.; Ferrante, T.; Bonventre, J.V.; et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods 2019, 16, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Man, Y.; Maji, D.; An, R.; Ahuja, S.P.; Little, J.A.; Mohseni, P.; Suster, M.A.; Gurkan, U.A. Assessment of Red Blood Cell-Mediated Microvascular Occlusion in Sickle Cell Disease By a Novel Electrical Impedance-Based Microfluidic Device. Blood 2020, 136, 10. [Google Scholar] [CrossRef]
- Man, Y.; An, R.; Monchamp, K.; Sekyonda, Z.; Kucukal, E.; Federici, C.; Wulftange, W.J.; Goreke, U.; Bode, A.; Sheehan, V.A.; et al. OcclusionChip: A functional microcapillary occlusion assay complementary to ektacytometry for detection of small-fraction red blood cells with abnormal deformability. Front. Physiol. 2022, 13, 954106. [Google Scholar] [CrossRef] [PubMed]
- Wulftange, W.J.; Kucukal, E.; Man, Y.; An, R.; Monchamp, K.; Sevrain, C.D.; Dashora, H.R.; Owusu-Ansah, A.T.; Bode, A.; Ilich, A.; et al. Antithrombin-III mitigates thrombin-mediated endothelial cell contraction and sickle red blood cell adhesion in microscale flow. Br. J. Haematol. 2022, 198, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Man, Y.; Kucukal, E.; An, R.; Bode, A.; Little, J.A.; Gurkan, U.A. Standardized microfluidic assessment of red blood cell–mediated microcapillary occlusion: Association with clinical phenotype and hydroxyurea responsiveness in sickle cell disease. Microcirculation 2021, 28, e12662. [Google Scholar] [CrossRef]
- Borók, A.; Laboda, K.; Bonyár, A. PDMS Bonding Technologies for Microfluidic Applications: A Review. Biosensors 2021, 11, 292. [Google Scholar] [CrossRef]
- Ao, Z.; Cai, H.; Wu, Z.; Song, S.; Karahan, H.; Kim, B.; Lu, H.-C.; Kim, J.; Mackie, K.; Guo, F. Tubular human brain organoids to model microglia-mediated neuroinflammation. Lab Chip 2021, 21, 2751–2762. [Google Scholar] [CrossRef]
- Cai, H.; Ao, Z.; Wu, Z.; Song, S.; Mackie, K.; Guo, F. Intelligent acoustofluidics enabled mini-bioreactors for human brain organoids. Lab Chip 2021, 21, 2194–2205. [Google Scholar] [CrossRef]
- Ao, Z.; Cai, H.; Wu, Z.; Ott, J.; Wang, H.; Mackie, K.; Guo, F. Controllable fusion of human brain organoids using acoustofluidics. Lab Chip 2021, 21, 688–699. [Google Scholar] [CrossRef]
- Fang, G.; Lu, H.; Rodriguez de la Fuente, L.; Law, A.M.K.; Lin, G.; Jin, D.; Gallego-Ortega, D. Mammary Tumor Organoid Culture in Non-Adhesive Alginate for Luminal Mechanics and High-Throughput Drug Screening. Adv. Sci. 2021, 8, 2102418. [Google Scholar] [CrossRef]
- Abas, B.I.; Demirbolat, G.M.; Cevik, O. Wharton jelly-derived mesenchymal stem cell exosomes induce apoptosis and suppress EMT signaling in cervical cancer cells as an effective drug carrier system of paclitaxel. PLoS ONE 2022, 17, e0274607. [Google Scholar] [CrossRef] [PubMed]
- Abas, B.I.; Cevik, E.; Kocabiyik, B.; Cenik, M.; Cevik, O.; Gumus, E. Alginate encapsulation induce colony formation with umbilical cord-derived mesenchymal stem cells. Exp. Biomed. Res. 2021, 4, 113–121. [Google Scholar] [CrossRef]
- Akkoyun, F.; Ozcelik, A. A Simple Approach for Controlling an Open-Source Syringe Pump. Eur. Mech. Sci. 2020, 4, 166–170. [Google Scholar] [CrossRef]
- Galateanu, B.; Dimonie, D.; Vasile, E.; Nae, S.; Cimpean, A.; Costache, M. Layer-shaped alginate hydrogels enhance the biological performance of human adipose-derived stem cells. BMC Biotechnol. 2012, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Duzagac, F.; Saorin, G.; Memeo, L.; Canzonieri, V.; Rizzolio, F. Microfluidic Organoids-on-a-Chip: Quantum Leap in Cancer Research. Cancers 2021, 13, 737. [Google Scholar] [CrossRef] [PubMed]
- Schuster, B.; Junkin, M.; Kashaf, S.S.; Romero-Calvo, I.; Kirby, K.; Matthews, J.; Weber, C.R.; Rzhetsky, A.; White, K.P.; Tay, S. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat. Commun. 2020, 11, 5271. [Google Scholar] [CrossRef] [PubMed]
- Zlotnik, A. Chemokines and cancer. Int. J. Cancer 2006, 119, 2026–2029. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wei, Y.; Liu, J.; Zhang, H. Chemotactic Responses of Neural Stem Cells to SDF-1α Correlate Closely with Their Differentiation Status. J. Mol. Neurosci. 2014, 54, 219–233. [Google Scholar] [CrossRef]
- Righetti, A.; Giulietti, M.; Šabanović, B.; Occhipinti, G.; Principato, G.; Piva, F. CXCL12 and Its Isoforms: Different Roles in Pancreatic Cancer? J. Oncol. 2019, 2019, 9681698. [Google Scholar] [CrossRef]
- Tirone, M.; Tran, N.L.; Ceriotti, C.; Gorzanelli, A.; Canepari, M.; Bottinelli, R.; Raucci, A.; Di Maggio, S.; Santiago, C.; Mellado, M.; et al. High mobility group box 1 orchestrates tissue regeneration via CXCR4. J. Exp. Med. 2018, 215, 303–318. [Google Scholar] [CrossRef]
- Zhu, C.; Yao, W.-L.; Tan, W.; Zhang, C.-H. SDF-1 and CXCR4 play an important role in adult SVZ lineage cell proliferation and differentiation. Brain Res. 2017, 1657, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Kryczek, I.; Lange, A.; Mottram, P.; Alvarez, X.; Cheng, P.; Hogan, M.; Moons, L.; Wei, S.; Zou, L.; Machelon, V.; et al. CXCL12 and vascular endothelial growth factor synergistically induce neoangiogenesis in human ovarian cancers. Cancer Res. 2005, 65, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Gassenmaier, M.; Chen, D.; Buchner, A.; Henkel, L.; Schiemann, M.; Mack, B.; Schendel, D.J.; Zimmermann, W.; Pohla, H. CXC Chemokine Receptor 4 is Essential for Maintenance of Renal cell Carcinoma-Initiating Cells and Predicts Metastasis. Stem Cells 2013, 31, 1467–1476. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ozcelik, A.; Abas, B.I.; Erdogan, O.; Cevik, E.; Cevik, O. On-Chip Organoid Formation to Study CXCR4/CXCL-12 Chemokine Microenvironment Responses for Renal Cancer Drug Testing. Biosensors 2022, 12, 1177. https://doi.org/10.3390/bios12121177
Ozcelik A, Abas BI, Erdogan O, Cevik E, Cevik O. On-Chip Organoid Formation to Study CXCR4/CXCL-12 Chemokine Microenvironment Responses for Renal Cancer Drug Testing. Biosensors. 2022; 12(12):1177. https://doi.org/10.3390/bios12121177
Chicago/Turabian StyleOzcelik, Adem, Burcin Irem Abas, Omer Erdogan, Evrim Cevik, and Ozge Cevik. 2022. "On-Chip Organoid Formation to Study CXCR4/CXCL-12 Chemokine Microenvironment Responses for Renal Cancer Drug Testing" Biosensors 12, no. 12: 1177. https://doi.org/10.3390/bios12121177
APA StyleOzcelik, A., Abas, B. I., Erdogan, O., Cevik, E., & Cevik, O. (2022). On-Chip Organoid Formation to Study CXCR4/CXCL-12 Chemokine Microenvironment Responses for Renal Cancer Drug Testing. Biosensors, 12(12), 1177. https://doi.org/10.3390/bios12121177





