Monitoring and Regulating Intracellular GPX4 mRNA Using Gold Nanoflare Probes and Enhancing Erastin-Induced Ferroptosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Instrumentation
2.3. Experimental Sections
2.3.1. Preparation of Gold Nanoparticles and Gold Nanoflare Probes
2.3.2. Incubation of AuNF Probes with Target-DNA or Cell Lysis Solution
2.3.3. Cell Culture, Confocal Imaging, and Flow Cytometric Analysis
2.3.4. qRT-PCR Analysis
2.3.5. Immunofluorescence Labeling
2.3.6. Determining the Content of Glutathione Peroxidases
2.3.7. Inhibiting the Proliferation of Cancer Cells through Erastin or the Synergistic Effect of Erastin and AuNF Probes
2.3.8. Determining the Content Change of Intracellular ROS Caused by Erastin and AuNF Probes
2.3.9. Statistical Analysis
3. Results and Discussion
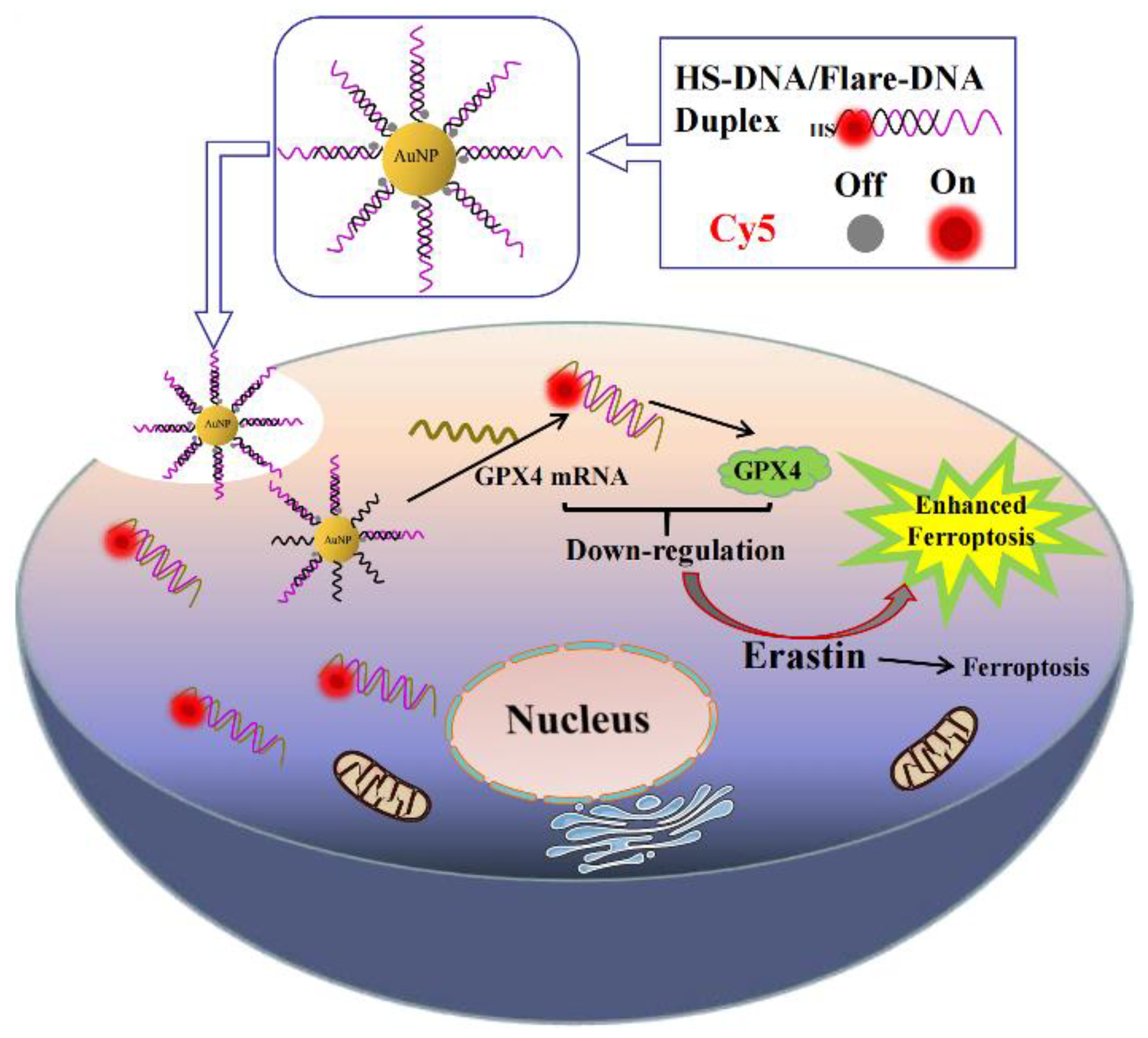
3.1. Design of AuNF Probes and Mechanism of Enhancing Erastin-Induced Ferroptosis
3.2. Characterization of AuNPs and AuNF Probes
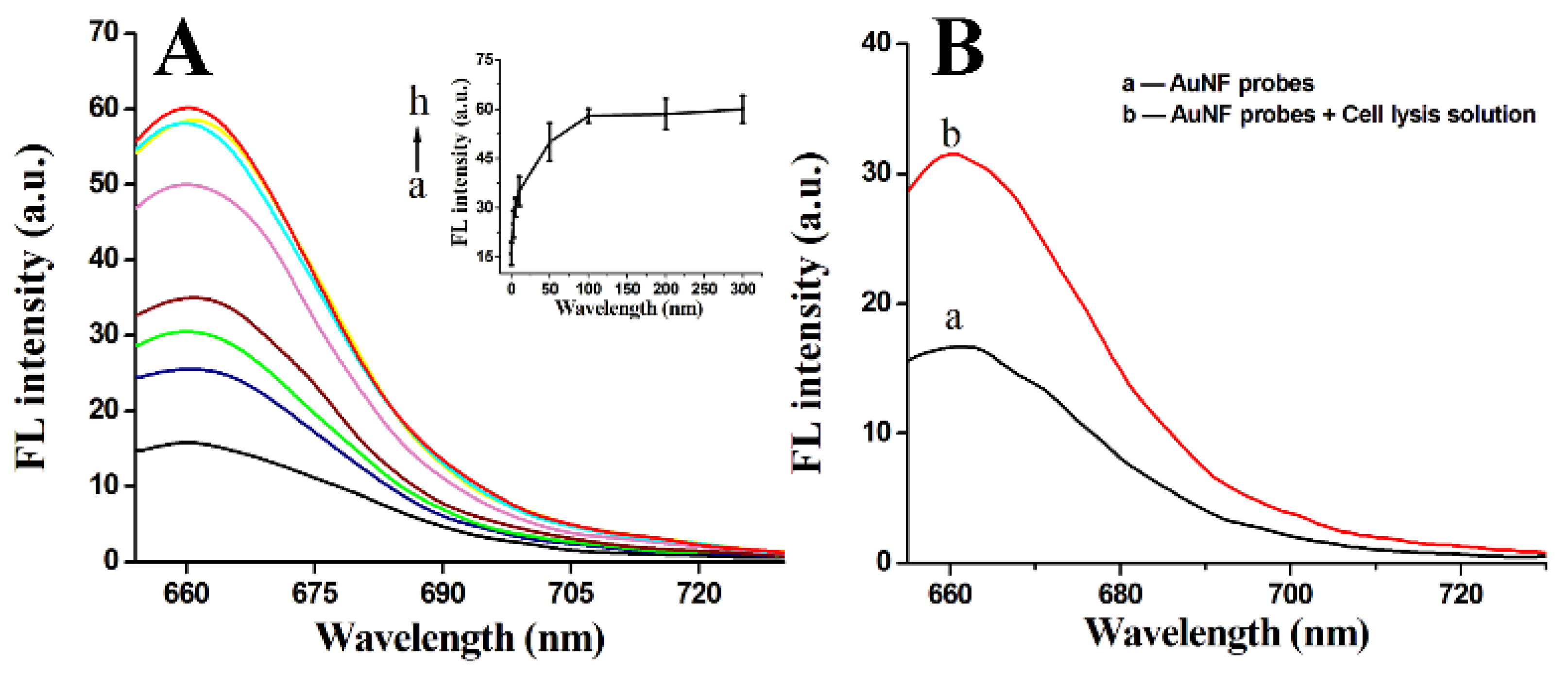
3.3. Fluorescence Response of AuNF Probes to Target-DNA and Cell Lysis Solution
3.4. Evaluation the Number of HS-DNA/Flare-DNA Duplexes Bound on AuNF Probes
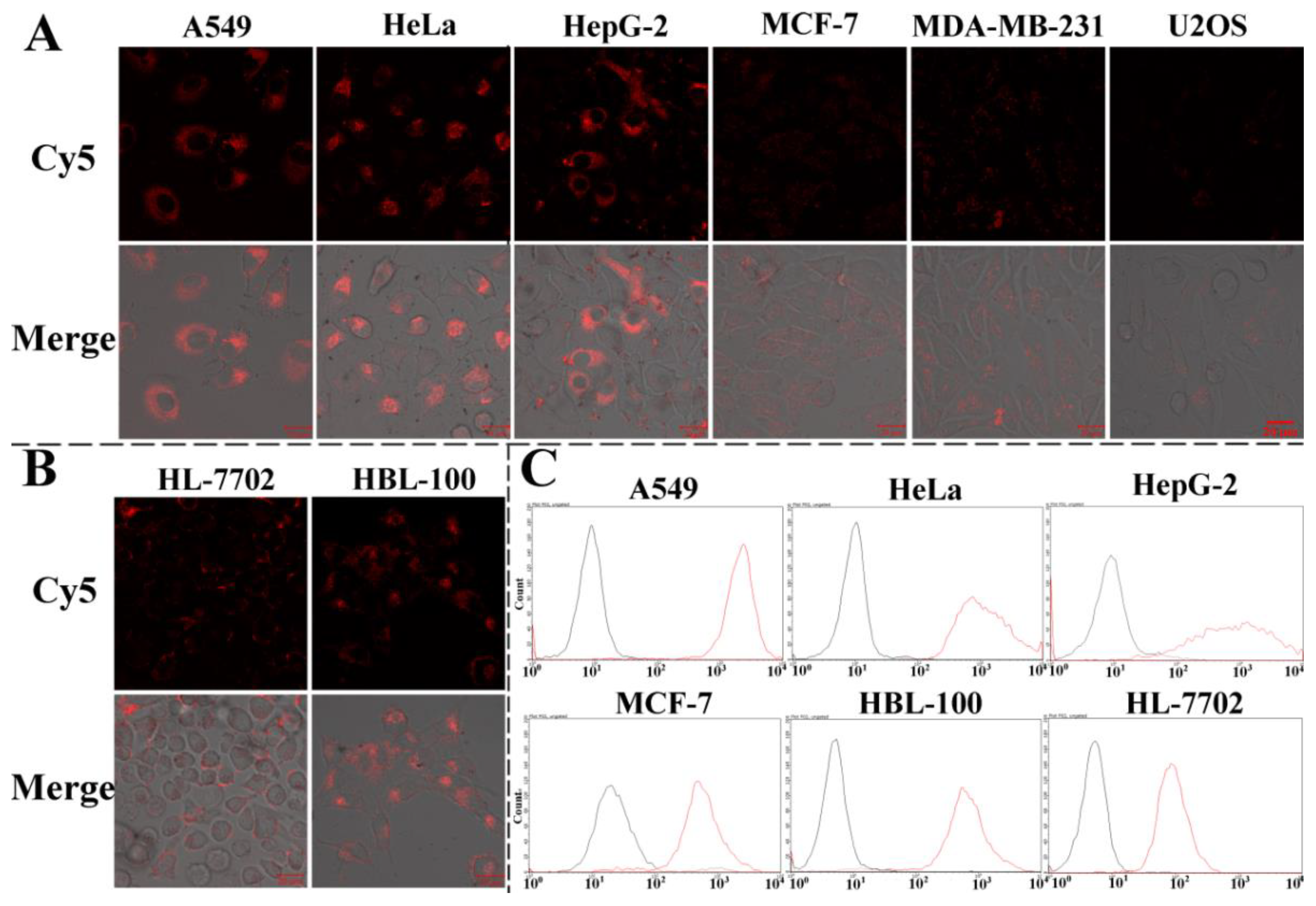
3.5. Fluorescence Imaging of Intracellular GPX4 mRNA by CLSM
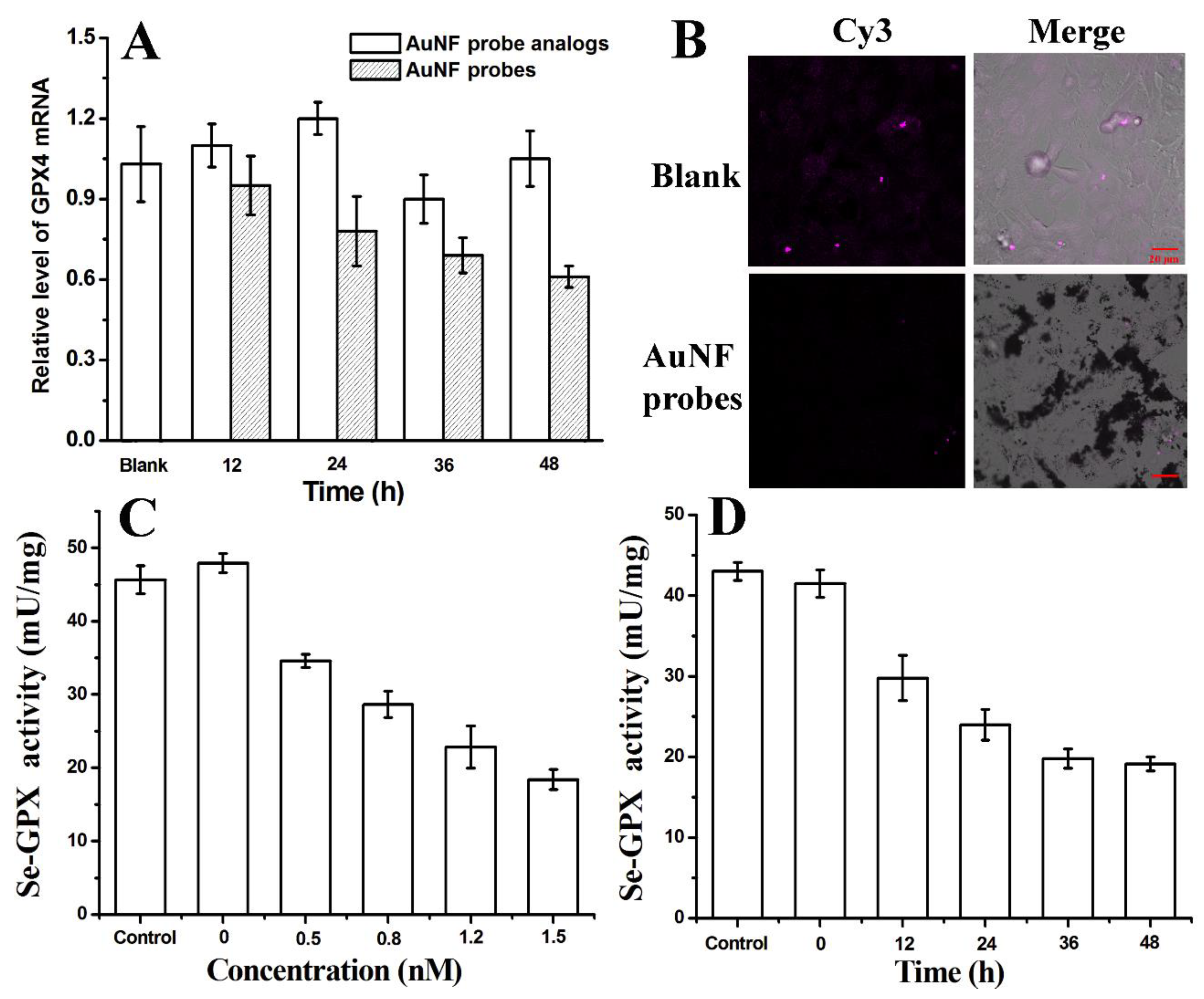
3.6. Expression Analysis of Intracellular GPX4 mRNA, GPX4 and Total GPX
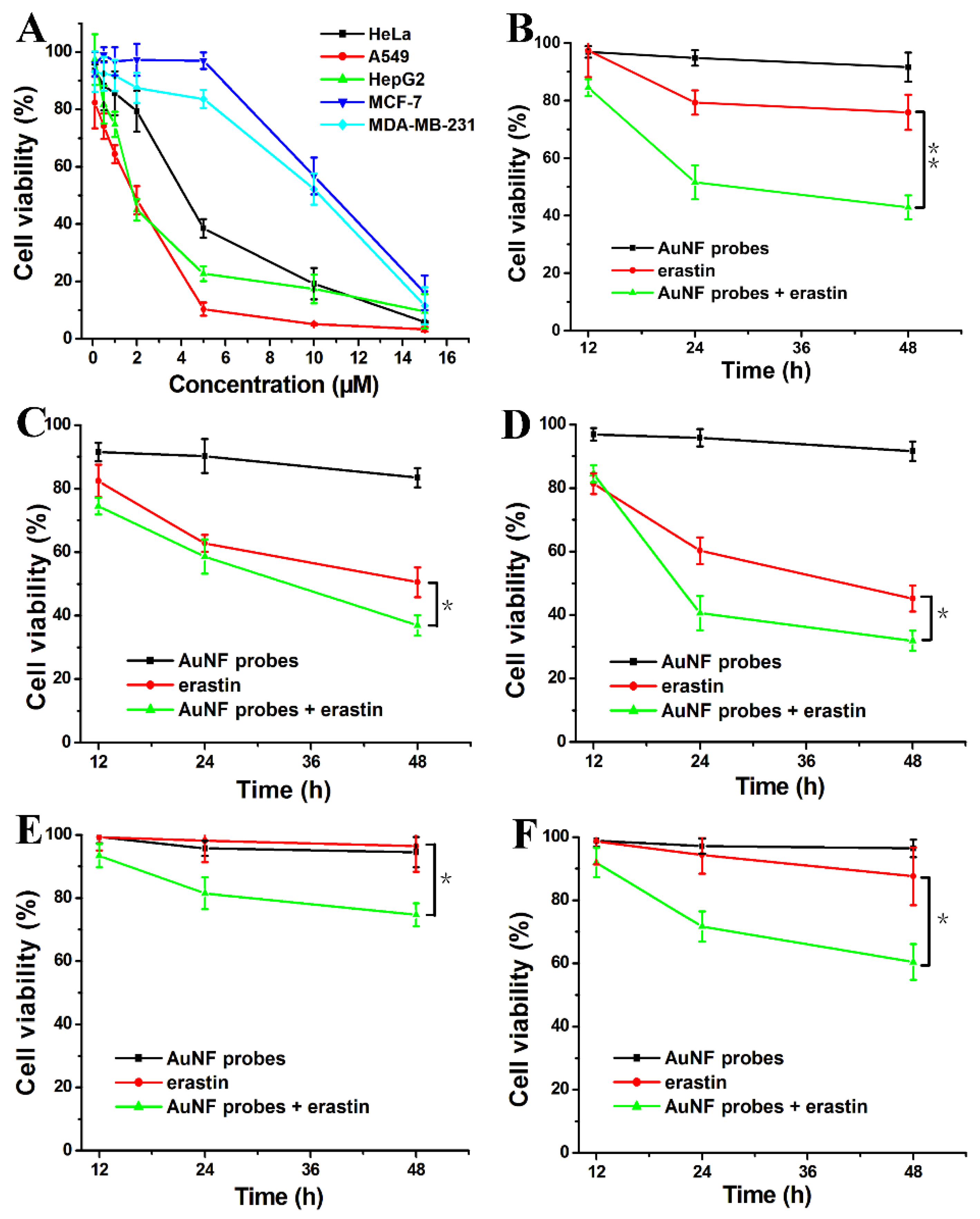
3.7. Enhancing Erastin-Induced Ferroptosis Using the Synergistic Effect of AuNF Probes
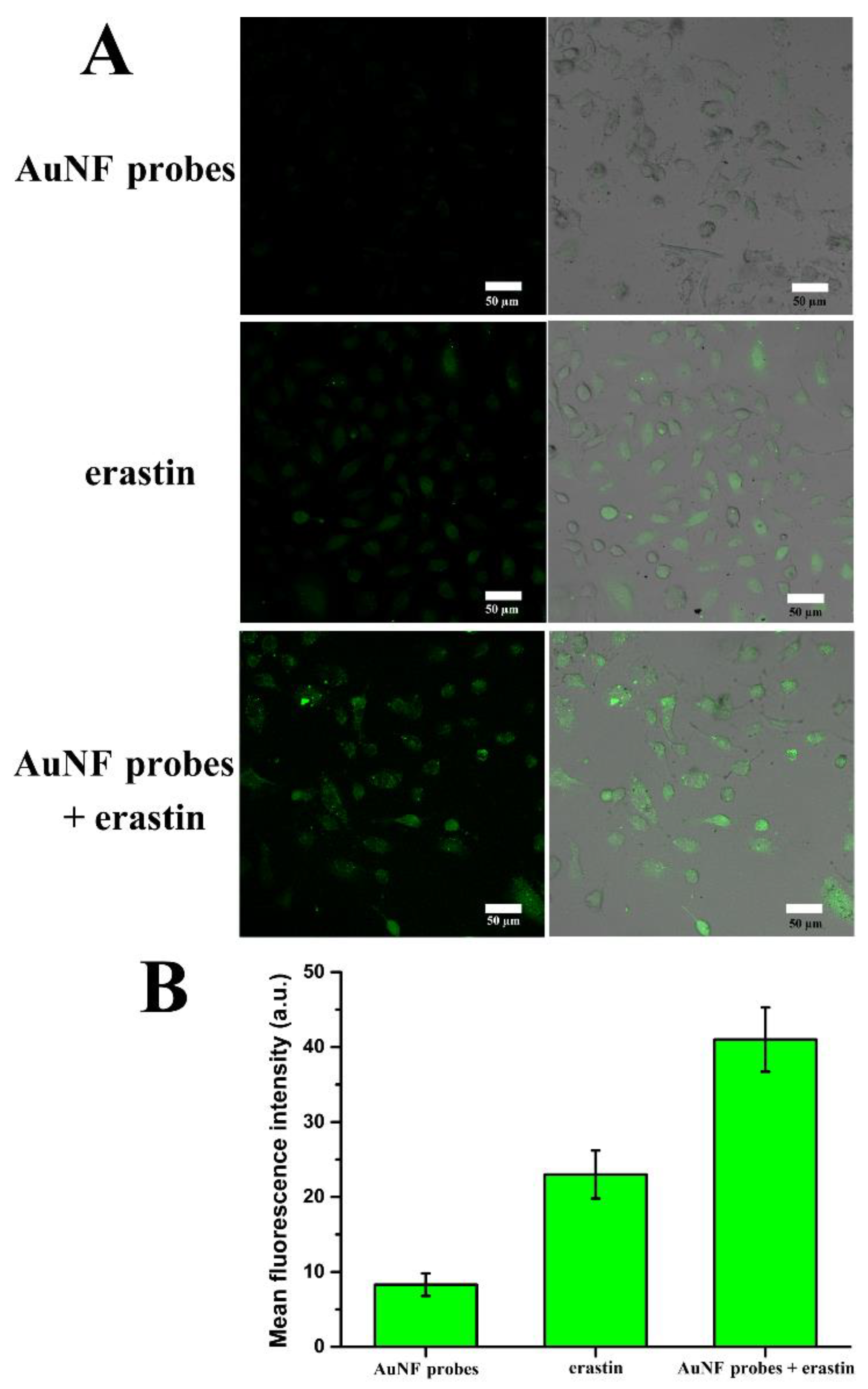
3.8. ROS Generation Caused by AuNF Probes and Erastin
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Den Bulk, J.; Verdegaal, E.M.; De Miranda, N.F. Cancer immunotherapy: Broadening the scope of targetable tumours. Open. Biol. 2018, 8, 180037–180044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodall, G.J.; Wickramasinghe, V.O. RNA in cancer. Nat. Rev. Cancer. 2021, 21, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.D.; Kathryn, M.L.; Michael, R.L.; Rachid, S.; Eleina, M.Z.; Caroline, E.G.; Darpan, N.P.; Andras, J.B.; Alexandra, M.C.; Wan, S.Y.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar]
- Fan, S.; Yang, Q.; Song, Q.; Hong, M.; Liu, X.; Chen, H.; Wang, J.; Li, C.; Cheng, S. Multi-pathway inducing ferroptosis by MnO2-based nanodrugs for targeted cancer therapy. Chem. Commun. 2022, 58, 6486–6489. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qi, H.; Liu, Y.; Duan, C.; Liu, X.; Xia, T.; Chen, D.; Piao, H.; Liu, H. The double-edged roles of ROS in cancer prevention and therapy. Theranostics 2021, 11, 4839–4857. [Google Scholar] [CrossRef]
- Li, Y.; Yan, H.; Xu, X.; Liu, H.; Wu, C.; Zhao, L. Erastin/sorafenib induces cis-platin-resistant non-small cell lung cancer cell ferroptosis through inhibition of the Nrf2/xCT pathway. Oncol. Lett. 2020, 19, 323–333. [Google Scholar]
- Yang, W.S.; Kim, K.J.; Gaschler, M.M.; Patel, M.; Shchepinov, S.M.; Stockwell, B.R. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. USA 2016, 113, 4966–4975. [Google Scholar] [CrossRef] [Green Version]
- Reed, J.C.; Pellecchia, M. Ironing out cell death mechanisms. Cell 2012, 149, 963–965. [Google Scholar] [CrossRef] [Green Version]
- Seibt, T.M.; Proneth, B.; Conrad, M. Role of GPX4 in ferroptosis and its pharmacological implication. Free. Radic. Biol. Med. 2019, 133, 144–152. [Google Scholar] [CrossRef]
- Krümmel, B.; Plötz, T.; Jörns, A.; Lenzen, S.; Mehmeti, I. The central role of glutathione peroxidase 4 in the regulation of ferroptosis and its implications for pro-inflammatory cytokine-mediated beta-cell death. Biochim. Biophys. Acta-Mol. Basis Dis. 2021, 1867, 166114–166136. [Google Scholar] [CrossRef]
- Yang, W.S.; Stockwell, B.R. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem. Biol. 2008, 15, 234–245. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamj, A.F.; Clish, C.B.; et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef]
- Gaschler, M.M.; Andia, A.A.; Liu, H.; Csuka, J.M.; Hurlocker, B.; Vaiana, C.A.; Heindel, D.W.; Zuckerman, D.S.; Bos, P.H.; Reznik, E.; et al. FINO2 initiates ferroptosis through GPX4 inactivation and iron oxidation. Nat. Chem. Biol. 2018, 14, 507–515. [Google Scholar] [CrossRef]
- Briley, W.E.; Bondy, M.H.; Randeria, P.S.; Dupper, J.; Mirkin, C.A. Quantification and real-time tracking of RNA in live cells using Sticky-flares. Proc. Natl. Acad. Sci. USA 2015, 112, 9591–9595. [Google Scholar] [CrossRef] [Green Version]
- Grabar, K.C.; Freeman, R.G.; Hommer, M.B.; Natan, M.J. Preparation and Characterization of Au Colloid Monolayers. Anal. Chem. 1995, 67, 735–743. [Google Scholar] [CrossRef]
- Hong, M.; Sun, H.; Yang, Q.; Cheng, S.; Yu, S.; Fan, S.; Li, C.; Cui, C.; Tan, W. A microRNA-21-responsive doxorubicin-releasing sticky-flare for synergistic anticancer with silencing of microRNA and chemotherapy. Sci. China-Chem. 2021, 64, 1009–1019. [Google Scholar] [CrossRef]
- Stryer, L.; Haugland, R.P. Energy transfer: A spectroscopic ruler. Proc. Natl. Acad. Sci. USA 1967, 58, 719–726. [Google Scholar] [CrossRef] [Green Version]
- Hangauer, M.J.; Viswanathan, V.S.; Ryan, M.J.; Bole, D.; Eato, J.K.; Matov, A.; Galeas, J.; Dhruv, H.D.; Berans, M.E.; Schreiber, S.L.; et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 2017, 551, 247–250. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Fu, X.; Jia, J.; Wikerholmen, T.; Xi, K.; Kong, Y.; Wang, J.; Chen, H.; Ma, Y.; Li, Z.; et al. Glioblastoma Therapy Using Codelivery of Cisplatin and Glutathione Peroxidase Targeting siRNA from Iron Oxide Nanoparticles. ACS. Appl. Mater. Interfaces 2020, 12, 43408–43421. [Google Scholar] [CrossRef]
- Crooke, S.T.; Witztum, J.L.; Bennett, C.F.; Baker, B.F. RNA-targeted therapeutics. Cell. Metab. 2018, 27, 714–739. [Google Scholar] [CrossRef] [Green Version]
- Zou, Y.; Palte, M.J.; Deik, A.A.; Li, H.; Eaton, J.K.; Wang, W.; Tseng, Y.; Deasy, R.; Kost-Alimova, M.; Dančík, V.; et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat. Commun. 2019, 10, 1617–1622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, C.; Zhang, X.; Yang, M.; Dong, C. Recent progress in ferroptosis inducers for cancer therapy. Adv. Mater. 2019, 31, e1904197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tan, H.; Daniels, J.D.; Zandkarimi, F.; Liu, H.; Brown, L.M.; Uchida, K.; O’Connor, O.A.; Stockwell, B.R. Imidazole ketone erastin induces ferroptosis and slows tumor growth in a mouse lymphoma model. Cell Chem. Biol. 2019, 26, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gai, C.; Ding, D.; Wang, F.; Li, W. Targeted p53 on small-molecules induced ferroptosis in cancers. Front. Oncol. 2018, 8, 507–511. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Qiao, L.; Bian, Y.; Sun, X. GDF15 knockdown promotes erastin-induced ferroptosis by decreasing SLC7A11 expression. Biochem. Biophys. Res. Commun. 2020, 526, 293–299. [Google Scholar] [CrossRef]

| Names | Sequences |
|---|---|
| HS-DNA | 5′-TCA GCG TAT CAA AAA AAA AA-SH-3′ |
| HS-DNA’ | 5′-GAT CTA AGC TAA AAA AAA AA-SH-3′ |
| Flare-DNA | 5′-GAT ACG CTG AGT GTG GTT T-Cy5-3′ |
| Flare-DNA’ | 5′-AGC TTA GAT CGC TAT GTA C-Cy5-3′ |
| Flare-DNA-N | 5′-GAT ACG CTG AGT GTG GTT T-3′ |
| Target-DNA | 5′-AAA CCA CAC TCA GCG TAT C-3′ |
| Random-DNA | 5′-GGC CGA TTG TGA ACA TGG A -3′ |
| GPX4-F-primer | 5′-AGA GAT CAA AGA GTT CGC CGC-3′ |
| GPX4-R-primer | 5′-TCT TCA TCC ACT TCC ACA GCG-3′ |
| GAPDH-F-primer | 5′-CTC AGA CAC CAT GGG GAA GGT GA-3′ |
| GAPDH-R-primer | 5′-ATG ATC TTG AGG CTG TTG TCA TA-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Yang, Q.; Sui, Y.; Yue, Q.; Yan, S.; Li, C.; Hong, M. Monitoring and Regulating Intracellular GPX4 mRNA Using Gold Nanoflare Probes and Enhancing Erastin-Induced Ferroptosis. Biosensors 2022, 12, 1178. https://doi.org/10.3390/bios12121178
Liu X, Yang Q, Sui Y, Yue Q, Yan S, Li C, Hong M. Monitoring and Regulating Intracellular GPX4 mRNA Using Gold Nanoflare Probes and Enhancing Erastin-Induced Ferroptosis. Biosensors. 2022; 12(12):1178. https://doi.org/10.3390/bios12121178
Chicago/Turabian StyleLiu, Xiaoyan, Qiangqiang Yang, Yanan Sui, Qiaoli Yue, Shuqing Yan, Chuan Li, and Min Hong. 2022. "Monitoring and Regulating Intracellular GPX4 mRNA Using Gold Nanoflare Probes and Enhancing Erastin-Induced Ferroptosis" Biosensors 12, no. 12: 1178. https://doi.org/10.3390/bios12121178
APA StyleLiu, X., Yang, Q., Sui, Y., Yue, Q., Yan, S., Li, C., & Hong, M. (2022). Monitoring and Regulating Intracellular GPX4 mRNA Using Gold Nanoflare Probes and Enhancing Erastin-Induced Ferroptosis. Biosensors, 12(12), 1178. https://doi.org/10.3390/bios12121178





