Combined Delivery of miR-15/16 through Humanized Ferritin Nanocages for the Treatment of Chronic Lymphocytic Leukemia
Abstract
1. Introduction
2. Materials and Methods
2.1. Production and Purification of HumAfFt
2.2. Functionalization of HumAfFt with Polyamines-Based Thiol-Reactive Linker
2.3. Incorporation of miRNAs into PA-HumAfFt
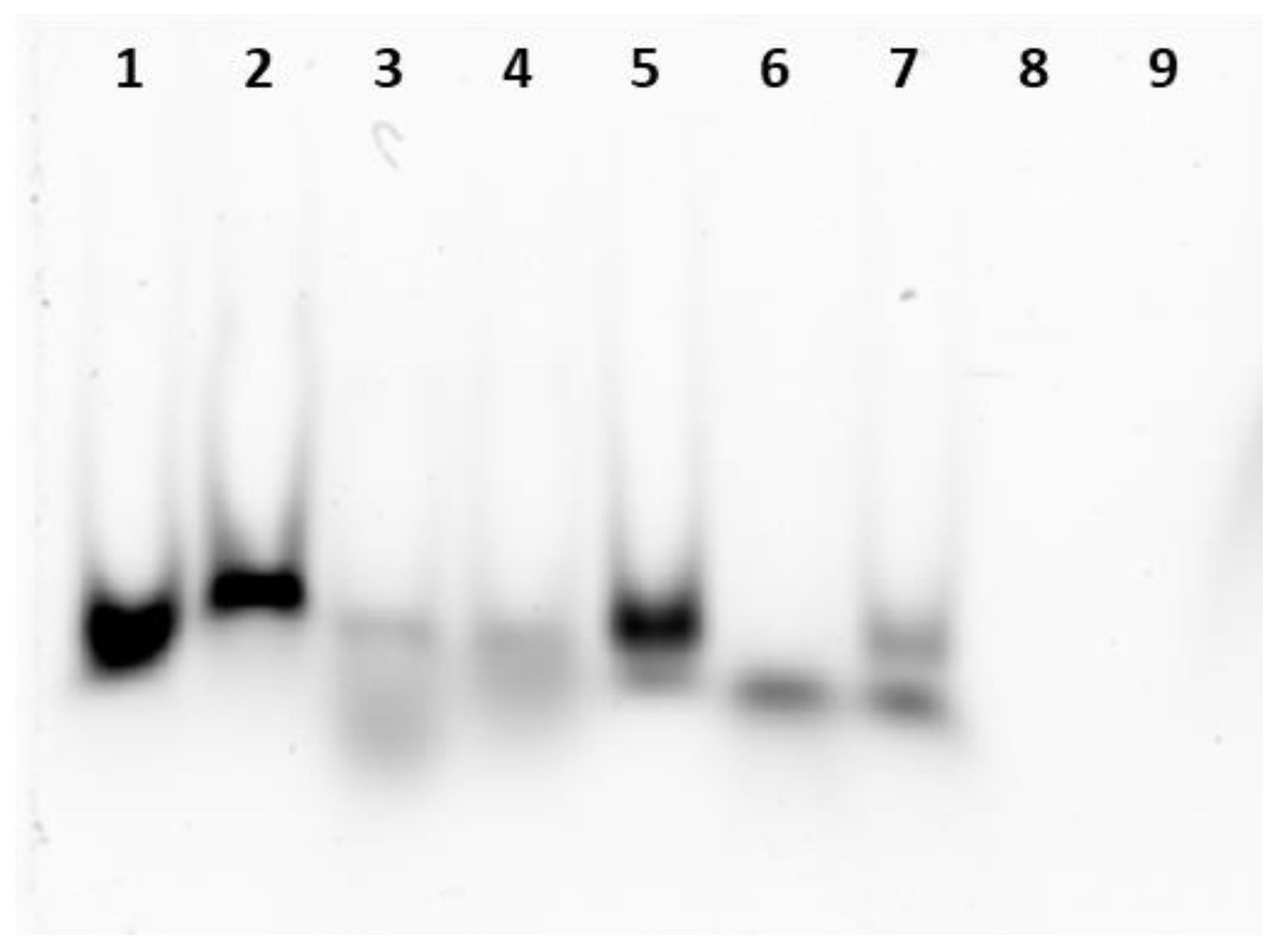
2.4. Native Electrophoresis Gel Analysis and RNase Digestion
2.5. Protein Extraction and Western Blotting Analysis
2.6. Cell Cultures and Transfection
2.7. Viability Assay
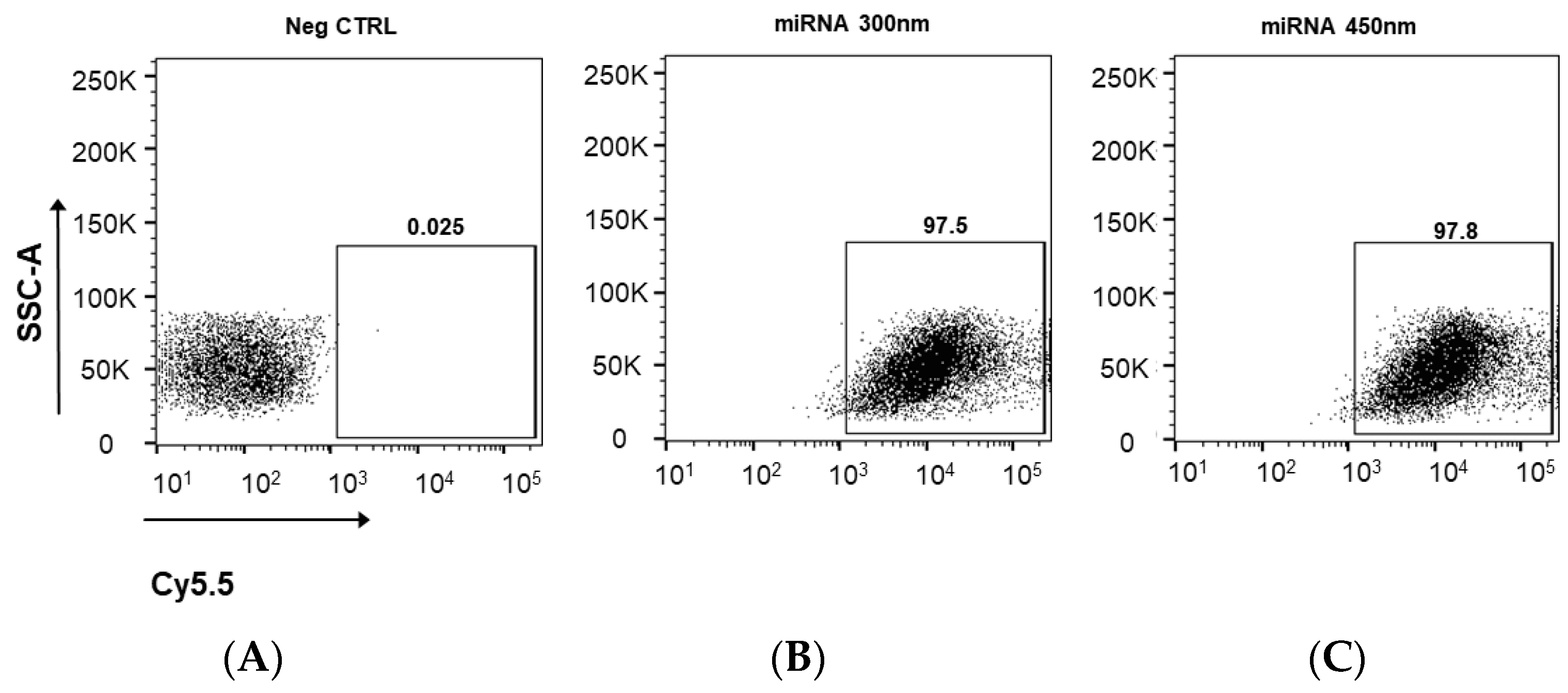
2.8. Cytometric Analysis
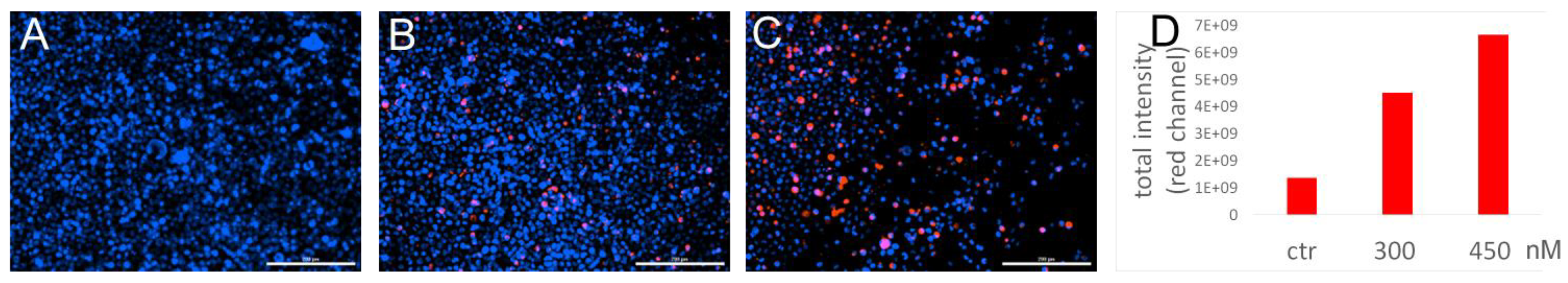
2.9. Immunofluorescence
2.10. Statistical Analysis
3. Results
3.1. miRNA Encapsulation into Ferritin Nanocages
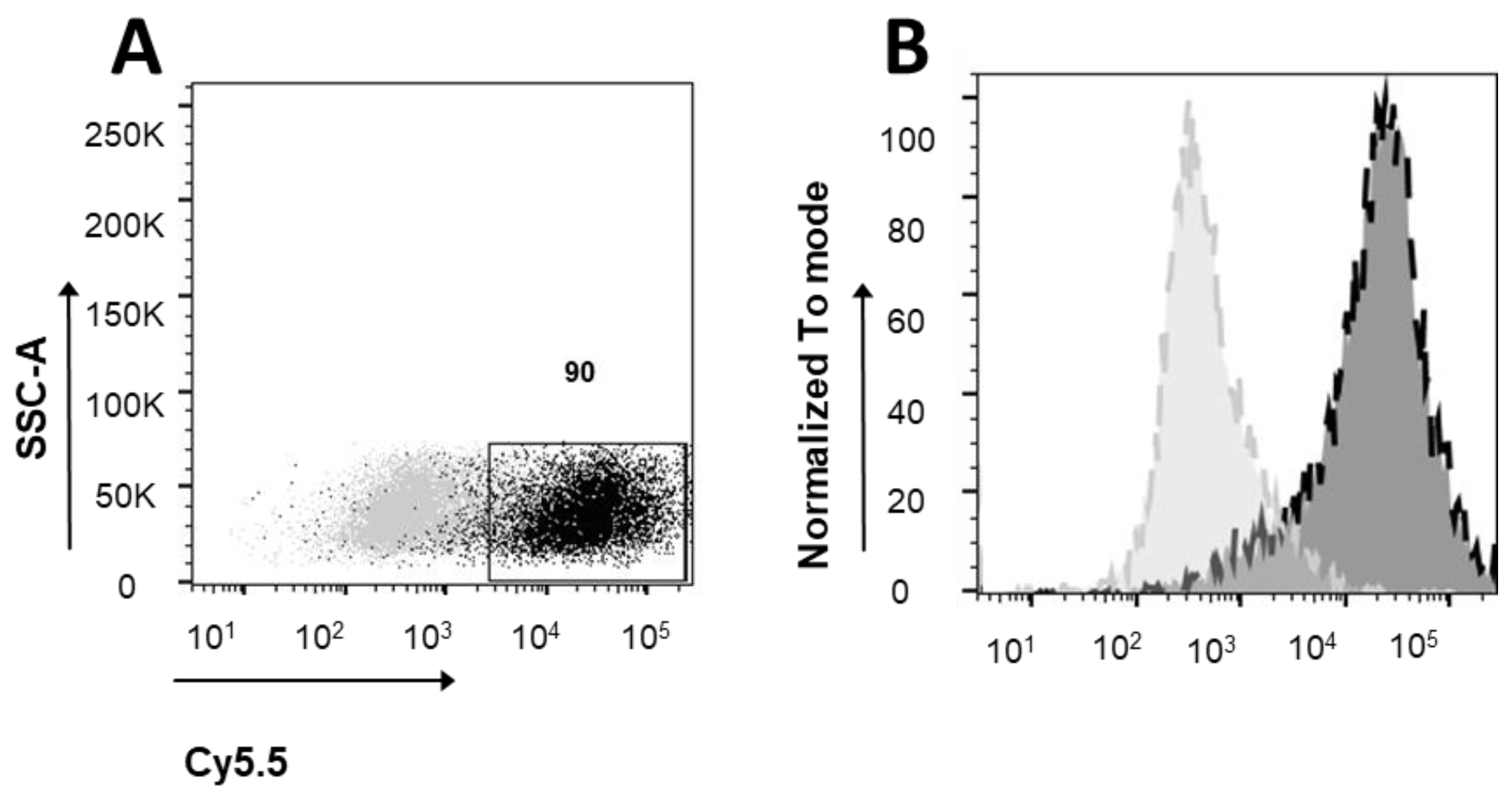
3.2. Ferritin Receptor Expression in MEG01 Cell Line
3.3. Cellular Uptake of Cy5.5 Labeled miRNA-PA-HumAfFt Nanoparticles
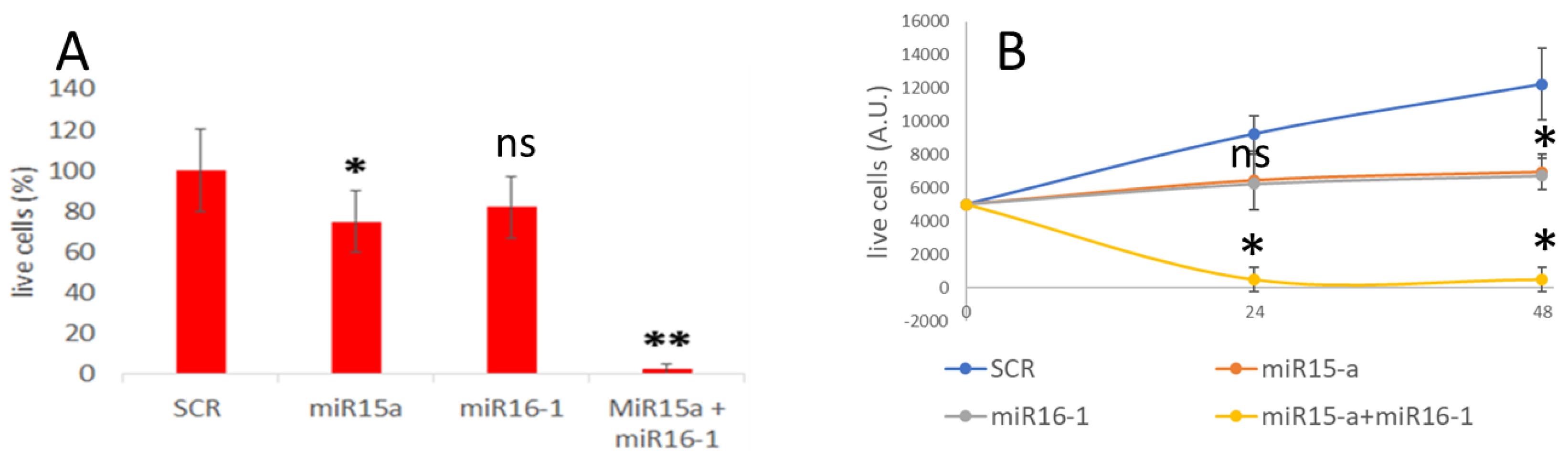
3.4. MiRNA Delivery and Effect on Apoptosis
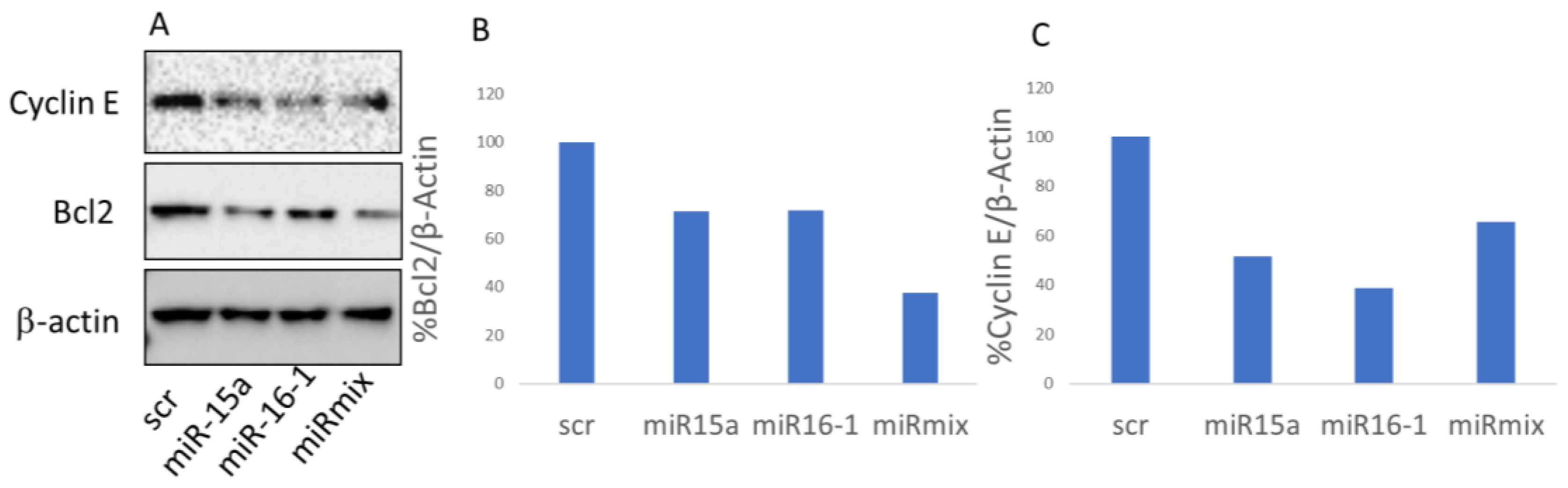
3.5. BCL-2 Expression Is Down-Regulated via miR-Pa-HumAfFt Treatment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Konno, M.; Koseki, J.; Asai, A.; Yamagata, A.; Shimamura, T.; Motooka, D.; Okuzaki, D.; Kawamoto, K.; Mizushima, T.; Eguchi, H.; et al. Distinct methylation levels of mature microRNAs in gastrointestinal cancers. Nat. Commun. 2019, 10, 3888. [Google Scholar] [CrossRef]
- Chan, J.J.; Tay, Y. Noncoding RNA: RNA regulatory networks in cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Placzek, W.J. Post-transcriptional regulation of anti-apoptotic BCL2 family members. Int. J. Mol. Sci. 2018, 19, 308. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Paone, A.; Galli, R.; Fabbri, M. MicroRNAs as new characters in the plot between epigenetics and prostate cancer. Front. Genet. 2011, 2, 62. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Khalid, K.; Padda, J.; Syam, M.; Moosa, A.; Kakani, V.; Sanka, S.; Zubair, U.; Padda, S.; Cooper, A.C.; Jean-Charles, G. 13q14 Deletion and Its Effect on Prognosis of Chronic Lymphocytic Leukemia. Cureus 2021, 13, e16839. [Google Scholar] [CrossRef]
- Lovat, F.; Nigita, G.; Distefano, R.; Nakamura, T.; Gasparini, P.; Tomasello, L.; Fadda, P.; Ibrahimova, N.; Catricalà, S.; Palamarchuk, A.; et al. Combined loss of function of two different loci of miR-15/16 drives the pathogenesis of acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2020, 117, 12332–12340. [Google Scholar] [CrossRef]
- Hampel, P.J.; Parikh, S.A. Chronic lymphocytic leukemia treatment algorithm 2022. Blood Cancer J. 2022, 12, 161, Erratum in Blood Cancer J. 2022, 12, 172. [Google Scholar] [CrossRef]
- Hallek, M. Chronic lymphocytic leukemia: 2020 update on diagnosis, risk stratification and treatment. Am. J. Hematol. 2019, 94, 1266–1287. [Google Scholar] [CrossRef]
- Hallek, M.; Al-Sawaf, O. Chronic lymphocytic leukemia: 2022 update on diagnostic and therapeutic procedures. Am. J. Hematol. 2021, 96, 1679–1705. [Google Scholar] [CrossRef]
- Kikushige, Y. Pathogenesis of chronic lymphocytic leukemia and the development of novel therapeutic strategies. J. Clin. Exp. Hematop. 2020, 60, 146–158. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Croce, C.M. Role of miR-15/16 in CLL. Cell Death Differ. 2015, 22, 6–11. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Balatti, V.; Croce, C.M. BCL2 and miR-15/16: From gene discovery to treatment. Cell Death Differ. 2018, 25, 21–26. [Google Scholar] [CrossRef]
- Ghaffari, M.; Kalantar, S.M.; Hemati, M.; Firoozabadi, A.D.; Asri, A.; Shams, A.; Ghalekohneh, S.J.; Haghiralsadat, F. Co-delivery of miRNA-15a and miRNA-16–1 using cationic PEGylated niosomes downregulates Bcl-2 and induces apoptosis in prostate cancer cells. Biotechnol Lett. 2021, 43, 981–994. [Google Scholar] [CrossRef]
- Katsaraki, K.; Karousi, P.; Artemaki, P.I.; Scorilas, A.; Pappa, V.; Kontos, C.K.; Papageorgiou, S.G. Micrornas: Tiny regulators of gene expression with pivotal roles in normal b-cell development and b-cell chronic lymphocytic leukemia. Cancers 2021, 13, 593. [Google Scholar] [CrossRef]
- Jain, C.K.; Srivastava, P.; Pandey, A.K.; Singh, N.; Kumar, R.S. miRNA therapeutics in precision oncology: A natural premium to nurture. Explor. Target Anti-Tumor Ther. 2022, 3, 511–532. [Google Scholar] [CrossRef] [PubMed]
- Raue, R.; Frank, A.C.; Syed, S.N.; Brüne, B. Therapeutic targeting of micrornas in the tumor microenvironment. Int. J. Mol. Sci. 2021, 22, 2210. [Google Scholar] [CrossRef] [PubMed]
- Bellini, M.; Mazzucchelli, S.; Galbiati, E.; Sommaruga, S.; Fiandra, L.; Truffi, M.; Rizzuto, M.A.; Colombo, M.; Tortora, P.; Corsi, F.; et al. Protein nanocages for self-triggered nuclear delivery of DNA-targeted chemotherapeutics in Cancer Cells. J. Control Release 2014, 196, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Fang, C.J.; Ryan, J.C.; Niemi, E.C.; Lebrón, J.A.; Björkman, P.J.; Arase, H.; Torti, F.M.; Torti, S.V.; Nakamura, M.C.; et al. Binding and uptake of H-ferritin are mediated by human transferrin receptor-1. Proc. Natl. Acad. Sci. USA 2010, 107, 3505. [Google Scholar] [CrossRef]
- Shesh, B.P.; Connor, J.R. A novel view of ferritin in cancer. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188917. [Google Scholar] [CrossRef]
- Johnson, E.; Cascio, D.; Sawaya, M.R.; Gingery, M.; Schröder, I. Crystal structures of a tetrahedral open pore ferritin from the hyperthermophilic Archaeon Archaeoglobus fulgidus. Structure 2005, 13, 637–648. [Google Scholar] [CrossRef]
- De Turris, V.; Cardoso Trabuco, M.; Peruzzi, G.; Testi, C.; Vallone, B.; Montemiglio, L.C.; Georges, A.D.; Calisti, L.; Benni, I. Humanized archaeal ferritin as a tool for cell targeted delivery. Nanoscale 2017, 9, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Montemiglio, L.C.; Testi, C.; Ceci, P.; Falvo, E.; Pitea, M.; Savino, C.; Arcovito, A.; Peruzzi, G.; Baiocco, P.; Mancia, F.; et al. Cryo-EM structure of the human ferritin–transferrin receptor 1 complex. Nat. Commun. 2019, 10, 1121. [Google Scholar] [CrossRef] [PubMed]
- Embaby, A.M.; Schoffelen, S.; Kofoed, C.; Meldal, M.; Diness, F. Rational Tuning of Fluorobenzene Probes for Cysteine-Selective Protein Modification. Angew. Chemie Int. Ed. 2018, 57, 8022–8026. [Google Scholar] [CrossRef] [PubMed]
- Pediconi, N.; Ghirga, F.; Del Plato, C.; Peruzzi, G.; Athanassopoulos, C.M.; Mori, M.; Crestoni, M.E.; Corinti, D.; Ugozzoli, F.; Massera, C.; et al. Design and Synthesis of Piperazine-Based Compounds Conjugated to Humanized Ferritin as Delivery System of siRNA in Cancer Cells. Bioconjug. Chem. 2021, 32, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Doll, T.A.P.F.; Raman, S.; Dey, R.; Burkhard, P. Nanoscale assemblies and their biomedical applications. J. R. Soc. Interface 2013, 10, 20120740. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Zhang, J.; Zhai, J.; Hong, J.; Yuan, C.; Liang, M. Ferritin: A Multifunctional Nanoplatform for Biological Detection, Imaging Diagnosis, and Drug Delivery. Acc. Chem. Res. 2021, 54, 3313–3325. [Google Scholar] [CrossRef] [PubMed]
- Palombarini, F.; Masciarelli, S.; Incocciati, A.; Liccardo, F.; Di Fabio, E.; Iazzetti, A.; Fabrizi, G.; Fazi, F.; Macone, A.; Bonamore, A.; et al. Self-assembling ferritin-dendrimer nanoparticles for targeted delivery of nucleic acids to myeloid leukemia cells. J. Nanobiotechnol. 2021, 19, 172. [Google Scholar] [CrossRef]
- Ofir, M.; Hacohen, D.; Ginsberg, D. miR-15 and miR-16 are direct transcriptional targets of E2F1 that limit E2F-induced proliferation by targeting cyclin E. Mol. Cancer Res. 2011, 9, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, F.; Zhang, T.; Yu, M.; Sun, Y. Recent advances in anti-multidrug resistance for nano-drug delivery system. Drug Deliv. 2022, 29, 1684–1697. [Google Scholar] [CrossRef] [PubMed]
- Waclawiczek, A.; Leppä, A.M.; Renders, S.; Stumpf, K.; Reyneri, C.; Betz, B.; Janssen, M.; Shahswar, R.; Donato, E.; Karpova, D.; et al. Combinatorial BCL2 Family Expression in Acute Myeloid Leukemia Stem Cells Predicts Clinical Response to Azacitidine/Venetoclax. Cancer Discov. 2023, 13, 1408–1427. [Google Scholar] [CrossRef] [PubMed]
- Lew, T.E.; Seymour, J.F. Clinical experiences with venetoclax and other pro-apoptotic agents in lymphoid malignancies: Lessons from monotherapy and chemotherapy combination. J. Hematol. Oncol. 2022, 15, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Brackman, D.; Eckert, D.; Menon, R.; Salem, A.H.; Potluri, J.; Smith, B.D.; Wei, A.H.; Hayslip, J.; Miles, D.; Mensing, S.; et al. Venetoclax exposure-efficacy and exposure-safety relationships in patients with treatment-naïve acute myeloid leukemia who are ineligible for intensive chemotherapy. Hematol. Oncol. 2022, 40, 269–279. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Kondo, Y.; Ishiyama, K.; Alatrash, G.; Lu, S.; Cox, K.; Qiao, N.; Clise-Dwyer, K.; John, L.S.; Sukhumalchandra, P.; et al. Two unique HLA-A*0201 restricted peptides derived from cyclin E as immunotherapeutic targets in leukemia. Leukemia 2020, 34, 1626–1636. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Y.; Lu, J.; Xiao, L.; Chen, H.; Li, Q.; Li, Y.-Y.; Xu, P.; Ruan, C.; Zhou, H.; et al. A conserved ZFX/WNT3 axis modulates the growth and imatinib response of chronic myeloid leukemia stem/progenitor cells. Cell Mol. Biol. Lett. 2023, 28, 83. [Google Scholar] [CrossRef]
- Chen, L.; Chen, W.; Mysliwski, M.; Serio, J.; Ropa, J.; A Abulwerdi, F.; Chan, R.J.; Patel, J.P.; Tallman, M.S.; Paietta, E.; et al. Mutated Ptpn11 alters leukemic stem cell frequency and reduces the sensitivity of acute myeloid leukemia cells to Mcl1 inhibition. Leukemia 2015, 29, 1290–1300. [Google Scholar] [CrossRef]
- Ahn, I.E.; Brown, J.R. Targeting Bruton’s Tyrosine Kinase in CLL. Front. Immunol. 2021, 12, 687458. [Google Scholar] [CrossRef]
- Patel, V.; Balakrishnan, K.; Bibikova, E.; Ayres, M.; Keating, M.J.; Wierda, W.G.; Gandhi, V. Comparison of acalabrutinib, a selective Bruton tyrosine kinase inhibitor, with ibrutinib in chronic lymphocytic leukemia cells. Clin. Cancer Res. 2017, 23, 3734–3743. [Google Scholar] [CrossRef]
- Moia, R.; Boggione, P.; Mahmoud, A.M.; Kodipad, A.A.; Adhinaveni, R.; Sagiraju, S.; Patriarca, A.; Gaidano, G. Targeting p53 in chronic lymphocytic leukemia. Expert. Opin. Ther. Targets 2020, 24, 1239–1250. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Rai, K.R. Chronic lymphocytic leukemia (CLL) treatment: So many choices, such great options. Cancer 2019, 125, 1432–1440. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Gong, H.; Zhou, J.; Zhang, Q.; Gao, W. Targeted gene silencing in vivo by platelet membrane—Coated metal-organic framework nanoparticles. Sci. Adv. 2020, 6, eaaz6108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Nakauchi, Y.; Köhnke, T.; Stafford, M.; Bottomly, D.; Thomas, R.; Wilmot, B.; McWeeney, S.K.; Majeti, R.; Tyner, J.W. Integrated analysis of patient samples identifies biomarkers for venetoclax efficacy and combination strategies in acute myeloid leukemia. Nat. Cancer 2020, 1, 826–839. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liberati, F.R.; Di Russo, S.; Barolo, L.; Peruzzi, G.; Farina, M.V.; Spizzichino, S.; Di Fonzo, F.; Quaglio, D.; Pisano, L.; Botta, B.; et al. Combined Delivery of miR-15/16 through Humanized Ferritin Nanocages for the Treatment of Chronic Lymphocytic Leukemia. Pharmaceutics 2024, 16, 402. https://doi.org/10.3390/pharmaceutics16030402
Liberati FR, Di Russo S, Barolo L, Peruzzi G, Farina MV, Spizzichino S, Di Fonzo F, Quaglio D, Pisano L, Botta B, et al. Combined Delivery of miR-15/16 through Humanized Ferritin Nanocages for the Treatment of Chronic Lymphocytic Leukemia. Pharmaceutics. 2024; 16(3):402. https://doi.org/10.3390/pharmaceutics16030402
Chicago/Turabian StyleLiberati, Francesca Romana, Sara Di Russo, Lorenzo Barolo, Giovanna Peruzzi, Maria Vittoria Farina, Sharon Spizzichino, Federica Di Fonzo, Deborah Quaglio, Luca Pisano, Bruno Botta, and et al. 2024. "Combined Delivery of miR-15/16 through Humanized Ferritin Nanocages for the Treatment of Chronic Lymphocytic Leukemia" Pharmaceutics 16, no. 3: 402. https://doi.org/10.3390/pharmaceutics16030402
APA StyleLiberati, F. R., Di Russo, S., Barolo, L., Peruzzi, G., Farina, M. V., Spizzichino, S., Di Fonzo, F., Quaglio, D., Pisano, L., Botta, B., Giorgi, A., Boffi, A., Cutruzzolà, F., Paone, A., & Baiocco, P. (2024). Combined Delivery of miR-15/16 through Humanized Ferritin Nanocages for the Treatment of Chronic Lymphocytic Leukemia. Pharmaceutics, 16(3), 402. https://doi.org/10.3390/pharmaceutics16030402








