Metallic Nanoparticle-Based Optical Cell Chip for Nondestructive Monitoring of Intra/Extracellular Signals
Abstract
1. Introduction
2. Fluorescence-Based Analytical Platform on Cell Chip Using MNPs
2.1. Metallic Nanoparticle-Based Fluorescence Sensing Strategies for Intracellular Signal Detection
| Metallic Nanomaterials | Mechanism | Target | Function | Ref. | |
|---|---|---|---|---|---|
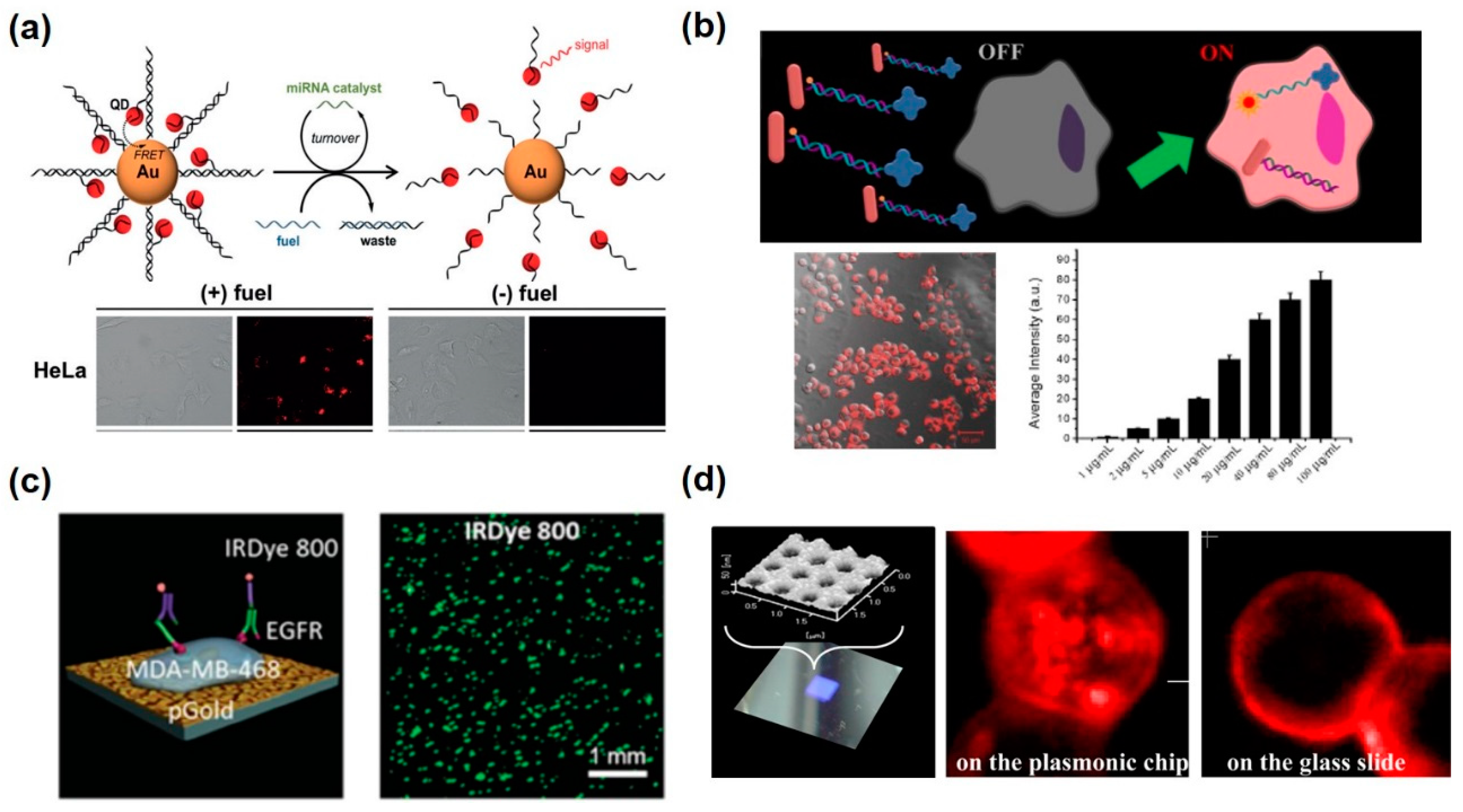
| Solution-based metallic nanoparticles | Au nanoparticle, Quantum dot | Fluorescence resonance energy transfer (FRET), Dequenching-Quenching | miR-21 | Differentiation between cancer cells and normal cells miRNA detection with spatiotemporal control in living cells | [65,66] |
| Au nanocross, Au nanorod | Fluorescence resonance energy transfer and surface-enhanced fluorescence (FRET-SEF) | miR-34a | Measurement of miRNA-34a from the HepG2 and H9C2 cells stimulated by AFB1 and TGF-β1 | [67] | |
| Au nanoparticle | FRET, hairpin-locked-DNAzyme system | miR-141 | Amplified detection of miR-141 by 25pM in living cells | [68] | |
| Au nanoparticle | FRET, aptazyme | Adenosine triphosphate (ATP) | Entering living cells and recognize intracellular target detection | [69] | |
| Nano-platform onto the cell chip | Nanostructured plasmonic gold (pGold) chip (Au nanorods) | Fluorescence enhancement on a plasmonic substrate | Tubulin, HER2, EGFR | Multiple cellular proteins of single cells of various cell types can be detected through a microarray of cells | [70] |
| Au nanoarray chip (Nano-sized holes) | Fluorescence enhancement on a plasmonic substrate | EpCAM | Fluorescence enhancement of APC-EpCAM in the cell membrane in contact with the plasmonic chip | [71] | |
2.2. Fluorescence Detection Platform with Metallic Nanostructures on Cell Chip for Extacellular Signal Detection
3. LSPR-Based Analytical Platform on Cell Chip Using MNPs
3.1. Metallic Nanoparticle-Based LSPR Sensing Strategies onto the Cell for Intracellular Signal Detection
3.2. LSPR-Based Analytical Platform with Metallic Nanostructures on Cell Chip for Extracellular Signal Detection
| Metallic Nanomaterials | Mechanism | Target | Function | Ref. | |
|---|---|---|---|---|---|
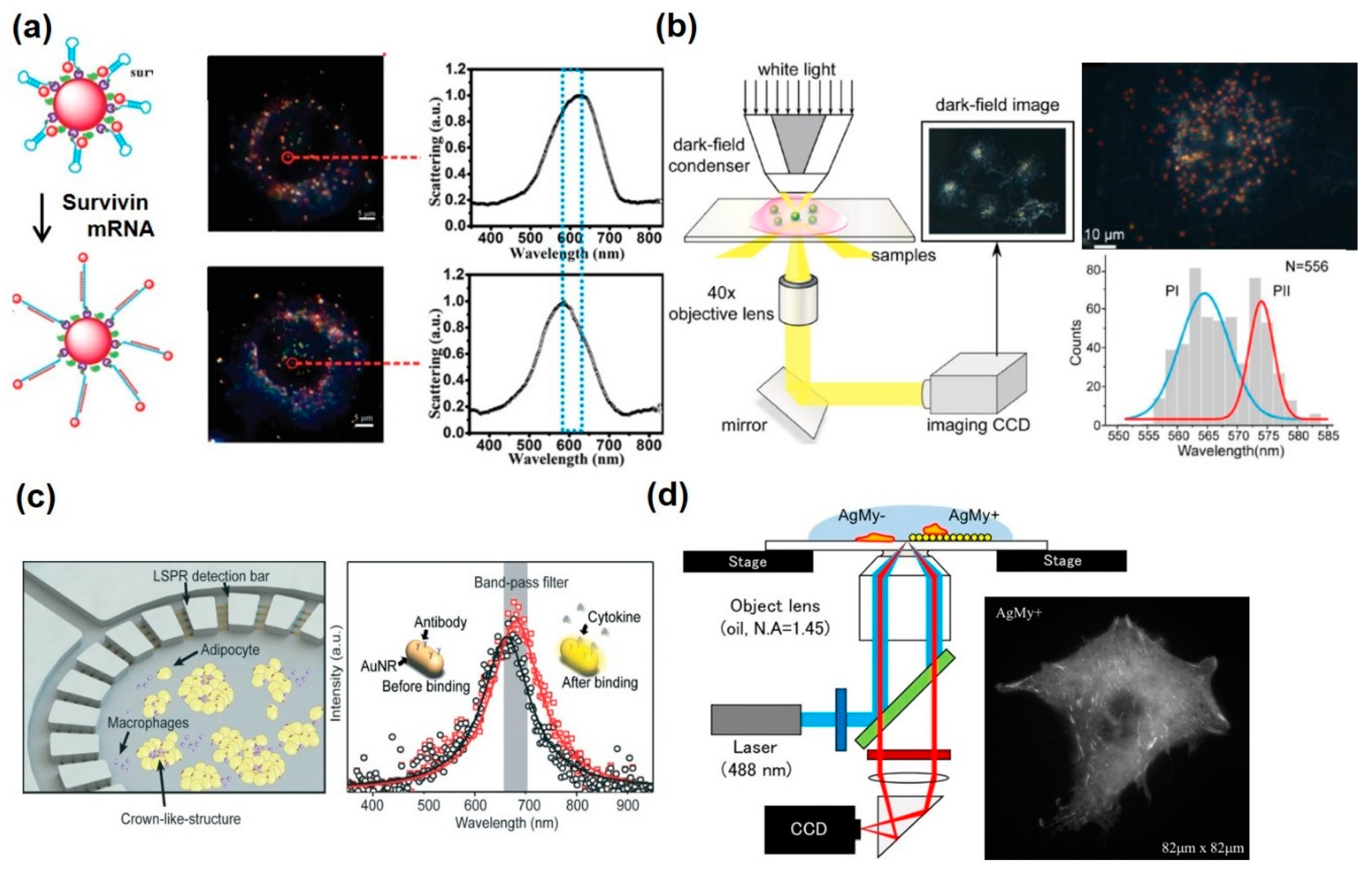
| Solution-based metallic nanoparticles | Crown nanoparticle plasmon rulers (Au nanoparticle) | Scattering spectra by aggregation–dissociation | Caspase-3 | Continuously monitoring of caspase-3 activity in live cells for over 2 h | [84] |
| Plasmonic nanohalo (Large and small Au nanoparticle) | Scattering spectra by aggregation–dissociation | Survivin mRNA | Detecting and imaging survivin mRNA by dark-field analysis in living cells | [85] | |
| Au nanoparticle | Scattering and absorption spectra of Au nanoparticle, bias-modified fuzzy C-means algorithm | HeLa cells, NADH | Fast and high-throughput analysis of cell imaging and presence and location of important biological molecules | [86] | |
| Nano-platform onto the cell chip | Circular sensing array (Au nanorod) | Resonance peak shift by LSPR | IL-6, TNF-α, IL-10, IL-4 | Multiplexed measurements of cytokines from adipocyte and macrophage | [87] |
| 3D multilayered Au nanosquare | Resonance peak shift by LSPR | A549 cells, DNA | 5 × 103 cells mL−1 in 2 μL and c 10−14–10−7 M DNA could be detected | [96] | |
| U-shaped fiber optic with Au nanoparticles | Increase of absorption value | N-Glycan | Label-free and in situ cytosensing of N-glycan expression on the cell surface | [97] | |
| Silver nanoparticle sheet | Confinement and enhancement of the fluorescence by LSPR | RBL-2H3 cell, NIH-3T3 cell | Obtaining high-quality nanointerfacial cell images | [88] | |
4. SERS-Based Analytical Platform on Cell Chip Using MNPs
4.1. Metallic Nanoparticle-Based SERS Sensing Strategies for Intracellular Signal Detection
4.2. SERS-Based Analytical Platform with Metallic Nanostructures on Cell Chip for Extracellular Signal Detection
5. Future Perspective and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hansen, A.; Eder, A.; Bönstrup, M.; Flato, M.; Mewe, M.; Schaaf, S.; Aksehirlioglu, B.; Schwörer, A.; Uebeler, J.; Eschenhagen, T. Development of a drug screening platform based on engineered heart tissue. Circ. Res. 2010, 107, 35. [Google Scholar] [CrossRef] [PubMed]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T. Comparison of 2d-and 3d-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef] [PubMed]
- Du, G.; Fang, Q.; den Toonder, J.M. Microfluidics for cell-based high throughput screening platforms—a review. Anal. Chim. Acta 2016, 903, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Streit, J.; Kleinlogel, S. Dynamic all-optical drug screening on cardiac voltage-gated ion channels. Sci. Rep. 2018, 8, 1153. [Google Scholar] [CrossRef] [PubMed]
- Popova, A.A.; Schillo, S.M.; Demir, K.; Ueda, E.; Nesterov-Mueller, A.; Levkin, P.A. Droplet-array (da) sandwich chip: A versatile platform for high-throughput cell screening based on superhydrophobic–superhydrophilic micropatterning. Adv. Mater. 2015, 27, 5217–5222. [Google Scholar] [CrossRef]
- El-Said, W.A.; Yoon, J.; Choi, J.-W. Nanostructured surfaces for analysis of anticancer drug and cell diagnosis based on electrochemical and sers tools. Nano Converg. 2018, 5, 1–19. [Google Scholar] [CrossRef]
- Arandian, A.; Bagheri, Z.; Ehtesabi, H.; Najafi Nobar, S.; Aminoroaya, N.; Samimi, A.; Latifi, H. Optical imaging approaches to monitor static and dynamic cell-on-chip platforms: A tutorial review. Small 2019, 15, e1900737. [Google Scholar] [CrossRef]
- Schubert, M.; Steude, A.; Liehm, P.; Kronenberg, N.M.; Karl, M.; Campbell, E.C.; Powis, S.J.; Gather, M.C. Lasing within live cells containing intracellular optical microresonators for barcode-type cell tagging and tracking. Nano Lett. 2015, 15, 5647–5652. [Google Scholar] [CrossRef]
- Mousavi Shaegh, S.A.; De Ferrari, F.; Zhang, Y.S.; Nabavinia, M.; Binth Mohammad, N.; Ryan, J.; Pourmand, A.; Laukaitis, E.; Banan Sadeghian, R.; Nadhman, A. A microfluidic optical platform for real-time monitoring of ph and oxygen in microfluidic bioreactors and organ-on-chip devices. Biomicrofluidics 2016, 10, 044111. [Google Scholar] [CrossRef]
- Chen, Y.X.; Huang, K.J.; Niu, K.X. Recent advances in signal amplification strategy based on oligonucleotide and nanomaterials for microrna detection-a review. Biosens. Bioelectron. 2018, 99, 612–624. [Google Scholar] [CrossRef]
- Hasanzadeh, M.; Shadjou, N. Advanced nanomaterials for use in electrochemical and optical immunoassays of carcinoembryonic antigen. A review. Microchim. Acta 2017, 184, 389–414. [Google Scholar] [CrossRef]
- Tripathy, N.; Kim, D.-H. Metal oxide modified zno nanomaterials for biosensor applications. Nano Converg. 2018, 5, 27. [Google Scholar] [CrossRef] [PubMed]
- Nunna, B.B.; Mandal, D.; Lee, J.U.; Singh, H.; Zhuang, S.; Misra, D.; Bhuyian, M.N.U.; Lee, E.S. Detection of cancer antigens (ca-125) using gold nano particles on interdigitated electrode-based microfluidic biosensor. Nano Converg. 2019, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Chen, X.; Ren, T.; Zhang, P.; Yang, D. Carbon nanotube based biosensors. Sens. Actuators B: Chem. 2015, 207, 690–715. [Google Scholar] [CrossRef]
- Aldewachi, H.; Chalati, T.; Woodroofe, M.N.; Bricklebank, N.; Sharrack, B.; Gardiner, P. Gold nanoparticle-based colorimetric biosensors. Nanoscale 2017, 10, 18–33. [Google Scholar] [CrossRef]
- Haun, J.B.; Yoon, T.J.; Lee, H.; Weissleder, R. Magnetic nanoparticle biosensors. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnology 2010, 2, 291–304. [Google Scholar] [CrossRef]
- Justino, C.I.; Gomes, A.R.; Freitas, A.C.; Duarte, A.C.; Rocha-Santos, T.A. Graphene based sensors and biosensors. TrAC Trends Anal. Chem. 2017, 91, 53–66. [Google Scholar] [CrossRef]
- Jiang, Z.; Le, N.D.; Gupta, A.; Rotello, V.M. Cell surface-based sensing with metallic nanoparticles. Chem. Soc. Rev. 2015, 44, 4264–4274. [Google Scholar] [CrossRef]
- Rosarin, F.S.; Mirunalini, S. Nobel metallic nanoparticles with novel biomedical properties. J. Bioanal. Biomed. 2011, 3, 85–91. [Google Scholar] [CrossRef]
- Song, B.; Zhou, T.; Liu, J.; Shao, L. Involvement of programmed cell death in neurotoxicity of metallic nanoparticles: Recent advances and future perspectives. Nanoscale Res. Lett. 2016, 11, 1–12. [Google Scholar] [CrossRef]
- Scanlon, M.D.; Peljo, P.; Méndez, M.A.; Smirnov, E.; Girault, H.H. Charging and discharging at the nanoscale: Fermi level equilibration of metallic nanoparticles. Chem. Sci. 2015, 6, 2705–2720. [Google Scholar] [CrossRef] [PubMed]
- Bigot, J.Y.; Halte, V.; Merle, J.C.; Daunois, A. Electron dynamics in metallic nanoparticles. Chem. Phys. 2000, 251, 181–203. [Google Scholar] [CrossRef]
- Scholl, J.A.; Koh, A.L.; Dionne, J.A. Quantum plasmon resonances of individual metallic nanoparticles. Nature 2012, 483, 421. [Google Scholar] [CrossRef] [PubMed]
- Kango, S.; Kalia, S.; Celli, A.; Njuguna, J.; Habibi, Y.; Kumar, R. Surface modification of inorganic nanoparticles for development of organic–inorganic nanocomposites—A review. Prog. Polym. Sci. 2013, 38, 1232–1261. [Google Scholar] [CrossRef]
- Chen, Y.; Xianyu, Y.; Jiang, X. Surface modification of gold nanoparticles with small molecules for biochemical analysis. Acc. Chem. Res. 2017, 50, 310–319. [Google Scholar] [CrossRef]
- Salatin, S.; Maleki Dizaj, S.; Yari Khosroushahi, A. Effect of the surface modification, size, and shape on cellular uptake of nanoparticles. Cell Biol. Int. 2015, 39, 881–890. [Google Scholar] [CrossRef]
- Klar, T.; Perner, M.; Grosse, S.; Von Plessen, G.; Spirkl, W.; Feldmann, J. Surface-plasmon resonances in single metallic nanoparticles. Phys. Rev. Lett. 1998, 80, 4249. [Google Scholar] [CrossRef]
- García, M.A. Surface plasmons in metallic nanoparticles: Fundamentals and applications. J. Phys. D: Appl. Phys. 2011, 44, 283001. [Google Scholar] [CrossRef]
- Trügler, A.; Hohenester, U. Strong coupling between a metallic nanoparticle and a single molecule. Phys. Rev. B 2008, 77, 115403. [Google Scholar] [CrossRef]
- Kailasa, S.K.; Koduru, J.R.; Desai, M.L.; Park, T.J.; Singhal, R.K.; Basu, H. Recent progress on surface chemistry of plasmonic metal nanoparticles for colorimetric assay of drugs in pharmaceutical and biological samples. TrAC Trends Anal. Chem. 2018, 105, 106–120. [Google Scholar] [CrossRef]
- Sepúlveda, B.; Angelomé, P.C.; Lechuga, L.M.; Liz-Marzán, L.M. Lspr-based nanobiosensors. Nano Today 2009, 4, 244–251. [Google Scholar] [CrossRef]
- Kerker, M. Electromagnetic model for surface-enhanced raman scattering (sers) on metal colloids. Acc. Chem. Res. 1984, 17, 271–277. [Google Scholar] [CrossRef]
- Quilis, N.G.; Lequeux, M.; Venugopalan, P.; Khan, I.; Knoll, W.; Boujday, S.; de La Chapelle, M.L.; Dostalek, J. Tunable laser interference lithography preparation of plasmonic nanoparticle arrays tailored for sers. Nanoscale 2018, 10, 10268–10276. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.J.; Hong, Z.-Y.; Chang, M.-H.; Liu, C.-C.; Cheng, H.-F.; Loh, X.J.; Chen, C.-H.; Liao, C.-D.; Kong, K.V. Metal carbonyl-gold nanoparticle conjugates for highly sensitive sers detection of organophosphorus pesticides. Biosens. Bioelectron. 2017, 96, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Jeong, W.-J.; Bu, J.; Kubiatowicz, L.J.; Chen, S.S.; Kim, Y.; Hong, S. Peptide–nanoparticle conjugates: A next generation of diagnostic and therapeutic platforms? Nano Converg. 2018, 5, 38. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; Kook, Y.-M.; Lee, K.; Koh, W.-G. Metal enhanced fluorescence (mef) for biosensors: General approaches and a review of recent developments. Biosens. Bioelectron. 2018, 111, 102–116. [Google Scholar] [CrossRef] [PubMed]
- Liaw, J.-W.; Wu, H.-Y.; Huang, C.-C.; Kuo, M.-K. Metal-enhanced fluorescence of silver island associated with silver nanoparticle. Nanoscale Res. Lett. 2016, 11, 26. [Google Scholar] [CrossRef]
- Lee, J.-H.; Choi, J.-H.; Chueng, S.-T.D.; Pongkulapa, T.; Yang, L.; Cho, H.-Y.; Choi, J.-W.; Lee, K.-B. Nondestructive characterization of stem cell neurogenesis by a magneto-plasmonic nanomaterial-based exosomal mirna detection. ACS Nano 2019, 13, 8793–8803. [Google Scholar] [CrossRef]
- Palermo, G.; Pagnotto, D.; Ricciardi, L.; Pezzi, L.; La Deda, M.; De Luca, A. Thermoplasmonic effects in gain-assisted nanoparticle solutions. J. Phys. Chem. C 2017, 121, 24185–24191. [Google Scholar] [CrossRef]
- Ricciardi, L.; Sancey, L.; Palermo, G.; Termine, R.; De Luca, A.; Szerb, E.I.; Aiello, I.; Ghedini, M.; Strangi, G.; La Deda, M. Plasmon-mediated cancer phototherapy: The combined effect of thermal and photodynamic processes. Nanoscale 2017, 9, 19279–19289. [Google Scholar] [CrossRef]
- Rehberg, M.; Nekolla, K.; Sellner, S.; Praetner, M.; Mildner, K.; Zeuschner, D.; Krombach, F. Intercellular transport of nanomaterials is mediated by membrane nanotubes in vivo. Small 2016, 12, 1882–1890. [Google Scholar] [CrossRef] [PubMed]
- Arcuino, G.; Lin, J.H.; Takano, T.; Liu, C.; Jiang, L.; Gao, Q.; Kang, J.; Nedergaard, M. Intercellular calcium signaling mediated by point-source burst release of atp. Proc. Natl. Acad. Sci. USA 2002, 99, 9840–9845. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Rosser, E.W.; Matsunaga, T.; Pacheco, A.; Akaike, T.; Xian, M. The development of fluorescent probes for visualizing intracellular hydrogen polysulfides. Angew. Chem. Int. Ed. Engl. 2015, 54, 13961–13965. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Song, Y.; Zhu, C.; Song, J.; Du, D.; Su, X.; Lin, Y. Graphene quantum dot–mno2 nanosheet based optical sensing platform: A sensitive fluorescence “turn off–on” nanosensor for glutathione detection and intracellular imaging. ACS Appl. Mater. Interfaces 2016, 8, 21990–21996. [Google Scholar] [CrossRef] [PubMed]
- Buchwalow, I.B.; Minin, E.A.; Boecker, W. A multicolor fluorescence immunostaining technique for simultaneous antigen targeting. Acta Histochem. 2005, 107, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Aw, S.S.; Tang, M.X.; Teo, Y.N.; Cohen, S.M. A conformation-induced fluorescence method for microrna detection. Nucleic Acids Res. 2016, 44, e92. [Google Scholar] [CrossRef]
- Greenwald, E.C.; Mehta, S.; Zhang, J. Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem. Rev. 2018, 118, 11707–11794. [Google Scholar] [CrossRef]
- Ma, F.; Li, Y.; Tang, B.; Zhang, C.Y. Fluorescent biosensors based on single-molecule counting. Acc. Chem. Res. 2016, 49, 1722–1730. [Google Scholar] [CrossRef]
- Newman, R.H.; Fosbrink, M.D.; Zhang, J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem. Rev. 2011, 111, 3614–3666. [Google Scholar] [CrossRef]
- Lee, S.; Sun, Y.; Cao, Y.; Kang, S.H. Plasmonic nanostructure-based bioimaging and detection techniques at the single-cell level. TrAC Trends Anal. Chem. 2019. [Google Scholar] [CrossRef]
- Li, Q.; Chen, P.; Fan, Y.; Wang, X.; Xu, K.; Li, L.; Tang, B. Multicolor fluorescence detection-based microfluidic device for single-cell metabolomics: Simultaneous quantitation of multiple small molecules in primary liver cells. Anal. Chem. 2016, 88, 8610–8616. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Yang, R.; Wang, H.L.; Fang, S.Y.; Liu, Y.P.; Long, F.; Zhu, A.N. Development of dual-color total internal reflection fluorescence biosensor for simultaneous quantitation of two small molecules and their affinity constants with antibodies. Biosens. Bioelectron. 2019, 126, 824–830. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.K.; Huang, X.H.; El-Sayed, I.H.; El-Sayed, M.A. Noble metals on the nanoscale: Optical and photothermal properties and some applications in imaging, sensing, biology, and medicine. Acc. Chem. Res. 2008, 41, 1578–1586. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Jang, D.-J. Metal-enhanced fluorescence of gold nanoclusters adsorbed onto ag@ sio 2 core–shell nanoparticles. J. Mater. Chem. C 2017, 5, 6037–6046. [Google Scholar] [CrossRef]
- Xu, J.; Yu, H.; Hu, Y.; Chen, M.; Shao, S. A gold nanoparticle-based fluorescence sensor for high sensitive and selective detection of thiols in living cells. Biosens. Bioelectron. 2016, 75, 1–7. [Google Scholar] [CrossRef]
- Hu, J.; Wu, M.; Jiang, L.; Zhong, Z.; Zhou, Z.; Rujiralai, T.; Ma, J. Combining gold nanoparticle antennas with single-molecule fluorescence resonance energy transfer (smfret) to study DNA hairpin dynamics. Nanoscale 2018, 10, 6611–6619. [Google Scholar] [CrossRef]
- Takalkar, S.; Baryeh, K.; Liu, G. Fluorescent carbon nanoparticle-based lateral flow biosensor for ultrasensitive detection of DNA. Biosens. Bioelectron. 2017, 98, 147–154. [Google Scholar] [CrossRef]
- Kyriazi, M.E.; Giust, D.; El-Sagheer, A.H.; Lackie, P.M.; Muskens, O.L.; Brown, T.; Kanaras, A.G. Multiplexed mrna sensing and combinatorial-targeted drug delivery using DNA-gold nanoparticle dimers. ACS Nano 2018, 12, 3333–3340. [Google Scholar] [CrossRef]
- Yang, Y.; Zhong, S.; Wang, K.; Huang, J. Gold nanoparticle based fluorescent oligonucleotide probes for imaging and therapy in living systems. Analyst 2019, 144, 1052–1072. [Google Scholar] [CrossRef]
- Yuan, Z.; Du, Y.; Tseng, Y.T.; Peng, M.; Cai, N.; He, Y.; Chang, H.T.; Yeung, E.S. Fluorescent gold nanodots based sensor array for proteins discrimination. Anal. Chem. 2015, 87, 4253–4259. [Google Scholar] [CrossRef]
- Choi, J.H.; Kim, H.S.; Choi, J.W.; Hong, J.W.; Kim, Y.K.; Oh, B.K. A novel au-nanoparticle biosensor for the rapid and simple detection of psa using a sequence-specific peptide cleavage reaction. Biosens. Bioelectron. 2013, 49, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Hong, M.Y.; Lee, D.; Nam, S.H.; Yoon, H.C.; Kim, H.S. Inhibition assay of biomolecules based on fluorescence resonance energy transfer (fret) between quantum dots and gold nanoparticles. J. Am. Chem. Soc. 2005, 127, 3270–3271. [Google Scholar] [CrossRef]
- Medintz, I.L.; Clapp, A.R.; Mattoussi, H.; Goldman, E.R.; Fisher, B.; Mauro, J.M. Self-assembled nanoscale biosensors based on quantum dot fret donors. Nat. Mater. 2003, 2, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Rabie, H.; Zhang, Y.; Pasquale, N.; Lagos, M.J.; Batson, P.E.; Lee, K.B. Nir biosensing of neurotransmitters in stem cell-derived neural interface using advanced core–shell upconversion nanoparticles. Adv. Mater. 2019, 31, 1806991. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wei, M.; Li, Y.; Liu, A.; Wei, W.; Zhang, Y.; Liu, S. Application of spectral crosstalk correction for improving multiplexed microrna detection using a single excitation wavelength. Anal. Chem. 2017, 89, 3430–3436. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.E.; Koo, H.; Sun, I.C.; Ryu, J.H.; Kim, K.; Kwon, I.C. Multifunctional nanoparticles for multimodal imaging and theragnosis. Chem. Soc. Rev. 2012, 41, 2656–2672. [Google Scholar] [CrossRef]
- He, X.; Zeng, T.; Li, Z.; Wang, G.; Ma, N. Catalytic molecular imaging of microrna in living cells by DNA-programmed nanoparticle disassembly. Angew. Chem. Int. Ed. Engl. 2016, 55, 3073–3076. [Google Scholar] [CrossRef]
- Shen, Y.; Li, Z.; Wang, G.; Ma, N. Photocaged nanoparticle sensor for sensitive microrna imaging in living cancer cells with temporal control. ACS Sens. 2018, 3, 494–503. [Google Scholar] [CrossRef]
- Sun, J.; Pi, F.; Ji, J.; Lei, H.; Gao, Z.; Zhang, Y.; Habimana, J.D.D.; Li, Z.; Sun, X. Ultrasensitive “fret-sef” probe for sensing and imaging micrornas in living cells based on gold nanoconjugates. Anal. Chem. 2018, 90, 3099–3108. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, J.; Yang, X.; He, X.; Quan, K.; Xie, N.; Ou, M.; Wang, K. Gold nanoparticle based hairpin-locked-dnazyme probe for amplified mirna imaging in living cells. Anal. Chem. 2017, 89, 5850–5856. [Google Scholar] [CrossRef]
- Yang, Y.; Huang, J.; Yang, X.; Quan, K.; Wang, H.; Ying, L.; Xie, N.; Ou, M.; Wang, K. Aptazyme-gold nanoparticle sensor for amplified molecular probing in living cells. Anal. Chem. 2016, 88, 5981–5987. [Google Scholar] [CrossRef] [PubMed]
- Koh, B.; Li, X.; Zhang, B.; Yuan, B.; Lin, Y.; Antaris, A.L.; Wan, H.; Gong, M.; Yang, J.; Zhang, X.; et al. Visible to near-infrared fluorescence enhanced cellular imaging on plasmonic gold chips. Small 2016, 12, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Tawa, K.; Yamamura, S.; Sasakawa, C.; Shibata, I.; Kataoka, M. Sensitive detection of cell surface membrane proteins in living breast cancer cells using multicolor fluorescence microscopy with a plasmonic chip. ACS Appl. Mater. Interfaces 2016, 8, 29893–29898. [Google Scholar] [CrossRef] [PubMed]
- Prodan, E.; Radloff, C.; Halas, N.J.; Nordlander, P. A hybridization model for the plasmon response of complex nanostructures. Science 2003, 302, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Yong, K.T.; Roy, I.; Ding, H.; He, S.; Prasad, P.N. Metallic nanostructures as localized plasmon resonance enhanced scattering probes for multiplex dark-field targeted imaging of cancer cells. J. Phys. Chem. C 2009, 113, 2676–2684. [Google Scholar] [CrossRef]
- Raphael, M.P.; Christodoulides, J.A.; Mulvaney, S.P.; Miller, M.M.; Long, J.P.; Byers, J.M. A new methodology for quantitative lspr biosensing and imaging. Anal. Chem. 2012, 84, 1367–1373. [Google Scholar] [CrossRef]
- Chung, T.; Lee, S.Y.; Song, E.Y.; Chun, H.; Lee, B. Plasmonic nanostructures for nano-scale bio-sensing. Sensors 2011, 11, 10907–10929. [Google Scholar] [CrossRef]
- Gao, P.F.; Gao, M.X.; Zou, H.Y.; Li, R.S.; Zhou, J.; Ma, J.; Wang, Q.; Liu, F.; Li, N.; Li, Y.F.; et al. Plasmon-induced light concentration enhanced imaging visibility as observed by a composite-field microscopy imaging system. Chem. Sci. 2016, 7, 5477–5483. [Google Scholar] [CrossRef]
- Zhang, S.; Huang, Q.; Zhang, L.; Zhang, H.; Han, Y.; Sun, Q.; Cheng, Z.; Qin, H.; Dou, S.; Li, Z. Vacancy engineering of cu 2− x se nanoparticles with tunable lspr and magnetism for dual-modal imaging guided photothermal therapy of cancer. Nanoscale 2018, 10, 3130–3143. [Google Scholar] [CrossRef]
- Cheng, X.D.; Cao, X.; Xiong, B.; He, Y.; Yeung, E.S. Background-free three-dimensional selective imaging of anisotropic plasmonic nanoparticles. Nano Res. 2017, 10, 1423–1433. [Google Scholar] [CrossRef]
- Du, J.; Shao, Q.; Yin, S.; Jiang, L.; Ma, J.; Chen, X. Colorimetric chemodosimeter based on diazonium-gold-nanoparticle complexes for sulfite ion detection in solution. Small 2012, 8, 3412–3416. [Google Scholar] [CrossRef]
- Liu, J.W.; Lu, Y. A colorimetric lead biosensor using dnazyme-directed assembly of gold nanoparticles. J. Am. Chem. Soc. 2003, 125, 6642–6643. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Xue, X.; Li, T.; Zeng, H.; Liu, X. Ultrasensitive and selective colorimetric DNA detection by nicking endonuclease assisted nanoparticle amplification. Angew. Chem. Int. Ed. Engl. 2009, 48, 6849–6852. [Google Scholar] [CrossRef] [PubMed]
- Jun, Y.W.; Sheikholeslami, S.; Hostetter, D.R.; Tajon, C.; Craik, C.S.; Alivisatos, A.P. Continuous imaging of plasmon rulers in live cells reveals early-stage caspase-3 activation at the single-molecule level. Proc. Natl. Acad. Sci. USA 2009, 106, 17735–17740. [Google Scholar] [CrossRef] [PubMed]
- Qian, G.S.; Kang, B.; Zhang, Z.L.; Li, X.L.; Zhao, W.; Xu, J.J.; Chen, H.Y. Plasmonic nanohalo optical probes for highly sensitive imaging of survivin mrna in living cells. Chem. Commun. 2016, 52, 11052–11055. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Jing, C.; Ying, Y.L.; He, P.; Long, Y.T. In situ high throughput scattering light analysis of single plasmonic nanoparticles in living cells. Theranostics 2015, 5, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; He, J.; Verano, M.; Brimmo, A.T.; Glia, A.; Qasaimeh, M.A.; Chen, P.; Aleman, J.O.; Chen, W. An integrated adipose-tissue-on-chip nanoplasmonic biosensing platform for investigating obesity-associated inflammation. Lab Chip 2018, 18, 3550–3560. [Google Scholar] [CrossRef] [PubMed]
- Usukura, E.; Yanase, Y.; Ishijima, A.; Kuboki, T.; Kidoaki, S.; Okamoto, K.; Tamada, K. Lspr-mediated high axial-resolution fluorescence imaging on a silver nanoparticle sheet. PLoS ONE 2017, 12, e0189708. [Google Scholar] [CrossRef]
- Li, L.; Wang, W.; Ding, M.; Luo, G.; Liang, Q. Single-cell-arrayed agarose chip for in situ analysis of cytotoxicity and genotoxicity of DNA cross-linking agents. Anal. Chem. 2016, 88, 6734–6742. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Wang, Z.Y.; Wu, L.; Zong, S.F.; Yun, B.F.; Cui, Y.P. Combining multiplex sers nanovectors and multivariate analysis for in situ profiling of circulating tumor cell phenotype using a microfluidic chip. Small 2018, 14, 1704433. [Google Scholar] [CrossRef]
- Wu, X.; Schneider, N.; Platen, A.; Mitra, I.; Blazek, M.; Zengerle, R.; Schule, R.; Meier, M. In situ characterization of the mtorc1 during adipogenesis of human adult stem cells on chip. Proc. Natl. Acad. Sci. USA 2016, 113, E4143–E4150. [Google Scholar] [CrossRef] [PubMed]
- Bian, S.; Zhou, Y.; Hu, Y.; Cheng, J.; Chen, X.; Xu, Y.; Liu, P. High-throughput in situ cell electroporation microsystem for parallel delivery of single guide rnas into mammalian cells. Sci. Rep. 2017, 7, 42512. [Google Scholar] [CrossRef] [PubMed]
- Heath, J.R.; Ribas, A.; Mischel, P.S. Single-cell analysis tools for drug discovery and development. Nat. Rev. Drug. Discov. 2016, 15, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; George, J.; Li, B.; Amini, N.; Paluh, J.; Wang, J. Clickable multifunctional dumbbell particles for in situ multiplex single-cell cytokine detection. ACS Appl. Mater. Interfaces 2017, 9, 32482–32488. [Google Scholar] [CrossRef]
- Sriram, M.; Markhali, B.P.; Nicovich, P.R.; Bennett, D.T.; Reece, P.J.; Hibbert, D.B.; Tilley, R.D.; Gaus, K.; Vivekchand, S.R.C.; Gooding, J.J. A rapid readout for many single plasmonic nanoparticles using dark-field microscopy and digital color analysis. Biosens. Bioelectron. 2018, 117, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.Y.; Li, H.L.; Yang, M.S.; Pang, S.W. Label-free detection of live cancer cells and DNA hybridization using 3d multilayered plasmonic biosensor. Nanotechnology 2018, 29, 365503. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.W.; Wang, Y.; Xu, Y.; Wang, X.; Huang, Z.J.; Chen, J.M.; Li, Y.X.; Duan, Y.X. Ultrasensitive u-shaped fiber optic lspr cytosensing for label-free and in situ evaluation of cell surface n-glycan expression. Sens. Actuators B-Chem. 2019, 284, 582–588. [Google Scholar] [CrossRef]
- Long, D.A.; Long, D. Raman Spectroscopy; McGraw-Hill New York: New York, NY, USA, 1977; Volume 276. [Google Scholar]
- Tu, A.T.; Tu, A. Raman Spectroscopy in Biology: Principles and Applications; Wiley: New York, NY, USA, 1982. [Google Scholar]
- Stiles, P.L.; Dieringer, J.A.; Shah, N.C.; Van Duyne, R.P. Surface-enhanced raman spectroscopy. Annu. Rev. Anal. Chem. 2008, 1, 601–626. [Google Scholar] [CrossRef]
- Moskovits, M. Surface-enhanced raman spectroscopy: A brief retrospective. J. Raman Spectrosc. 2005, 36, 485–496. [Google Scholar] [CrossRef]
- Schlücker, S. Surface-enhanced raman spectroscopy: Concepts and chemical applications. Angew. Chem. Int. Ed. 2014, 53, 4756–4795. [Google Scholar] [CrossRef]
- Ding, S.Y.; Yi, J.; Li, J.F.; Ren, B.; Wu, D.Y.; Panneerselvam, R.; Tian, Z.Q. Nanostructure-based plasmon-enhanced raman spectroscopy for surface analysis of materials. Nat. Rev. Mater. 2016, 1, 16021. [Google Scholar] [CrossRef]
- Fuller, M.A.; Köper, I. Biomedical applications of polyelectrolyte coated spherical gold nanoparticles. Nano Converg. 2019, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Zong, S.; Wang, Z.; Yang, J.; Cui, Y. Intracellular ph sensing using p-aminothiophenol functionalized gold nanorods with low cytotoxicity. Anal. Chem. 2011, 83, 4178–4183. [Google Scholar] [CrossRef] [PubMed]
- Huefner, A.; Kuan, W.-L.; Barker, R.A.; Mahajan, S. Intracellular sers nanoprobes for distinction of different neuronal cell types. Nano Lett. 2013, 13, 2463–2470. [Google Scholar] [CrossRef]
- El-Said, W.A.; Cho, H.Y.; Yea, C.H.; Choi, J.W. Synthesis of metal nanoparticles inside living human cells based on the intracellular formation process. Adv. Mater. 2014, 26, 910–918. [Google Scholar] [CrossRef]
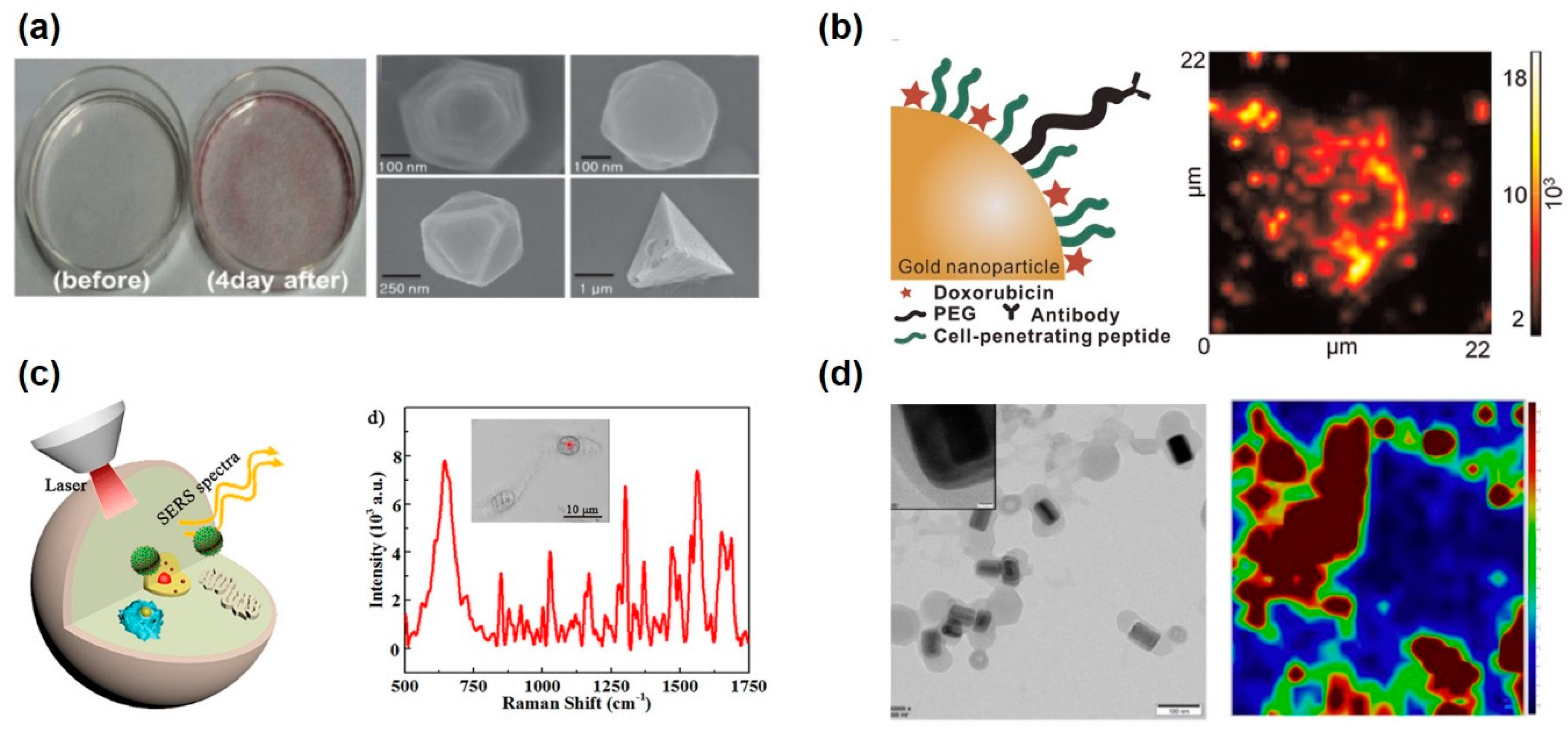
- Hossain, M.K.; Cho, H.-Y.; Kim, K.-J.; Choi, J.-W. In situ monitoring of doxorubicin release from biohybrid nanoparticles modified with antibody and cell-penetrating peptides in breast cancer cells using surface-enhanced raman spectroscopy. Biosens. Bioelectron. 2015, 71, 300–305. [Google Scholar] [CrossRef]
- Zeng, F.; Xu, D.; Zhan, C.; Liang, C.; Zhao, W.; Zhang, J.; Feng, H.; Ma, X. Surfactant-free synthesis of graphene oxide coated silver nanoparticles for sers biosensing and intracellular drug delivery. ACS Appl. Nano Mater. 2018, 1, 2748–2753. [Google Scholar] [CrossRef]
- Hada, A.-M.; Potara, M.; Suarasan, S.; Vulpoi, A.; Nagy-Simon, T.; Licarete, E.; Astilean, S. Fabrication of gold–silver core–shell nanoparticles for performing as ultrabright sers-nanotags inside human ovarian cancer cells. Nanotechnology 2019, 30, 315701. [Google Scholar] [CrossRef]
- Jiang, P.; Hu, Y.; Li, G. Biocompatible au@ ag nanorod@ zif-8 core-shell nanoparticles for surface-enhanced raman scattering imaging and drug delivery. Talanta 2019, 200, 212–217. [Google Scholar] [CrossRef]
- Kim, T.-H.; Lee, K.-B.; Choi, J.-W. 3d graphene oxide-encapsulated gold nanoparticles to detect neural stem cell differentiation. Biomaterials 2013, 34, 8660–8670. [Google Scholar] [CrossRef]
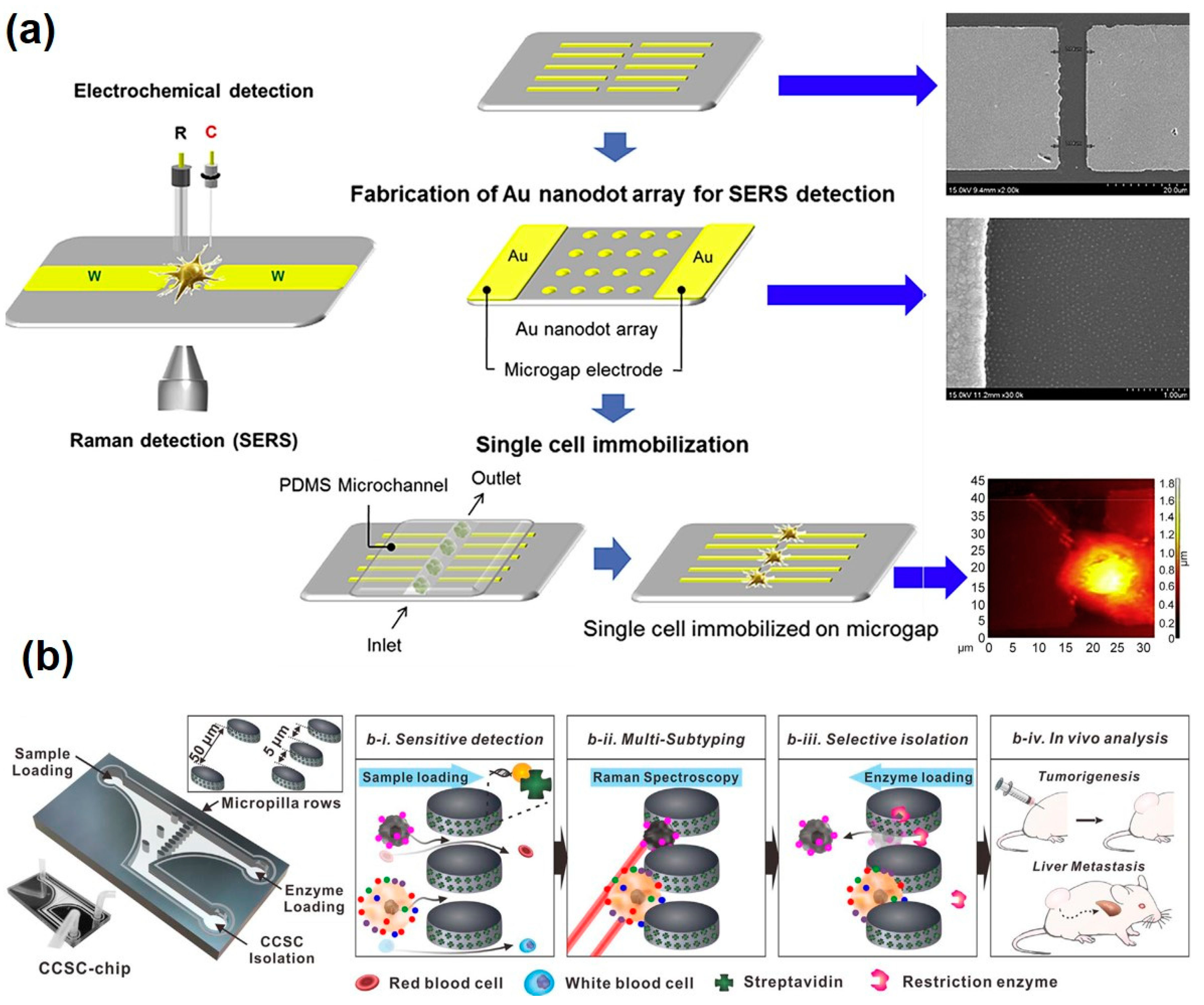
- El-Said, W.A.; Kim, T.H.; Chung, Y.H.; Choi, J.W. Fabrication of new single cell chip to monitor intracellular and extracellular redox state based on spectroelectrochemical method. Biomaterials 2015, 40, 80–87. [Google Scholar] [CrossRef]
- Hossain, M.; Cho, H.-Y.; Kim, K.-J.; Choi, J.-W. Silver nanostar patterned substrate for label-free characterization of breast cancer cells based on surface-enhanced raman spectroscopy. Sci. Adv. Mater. 2014, 6, 2491–2495. [Google Scholar] [CrossRef]
- Hossain, M.; Cho, H.-Y.; Choi, J.-W. Gold nanosphere-deposited substrate for distinguishing of breast cancer subtypes using surface-enhanced raman spectroscopy. J. Nanosci. Nanotechnol. 2016, 16, 6299–6303. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Hossain, M.K.; Lee, J.H.; Han, J.; Lee, H.J.; Kim, K.J.; Kim, J.H.; Lee, K.B.; Choi, J.W. Selective isolation and noninvasive analysis of circulating cancer stem cells through raman imaging. Biosens. Bioelectron. 2018, 102, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Seok, J.; Warren, H.S.; Cuenca, A.G.; Mindrinos, M.N.; Baker, H.V.; Xu, W.; Richards, D.R.; McDonald-Smith, G.P.; Gao, H.; Hennessy, L.; et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA 2013, 110, 3507–3512. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Bhise, N.S.; Ribas, J.; Manoharan, V.; Zhang, Y.S.; Polini, A.; Massa, S.; Dokmeci, M.R.; Khademhosseini, A. Organ-on-a-chip platforms for studying drug delivery systems. J. Control. Release 2014, 190, 82–93. [Google Scholar] [CrossRef]
- Choi, J.H.; Lee, J.; Shin, W.; Choi, J.W.; Kim, H.J. Priming nanoparticle-guided diagnostics and therapeutics towards human organs-on-chips microphysiological system. Nano Converg. 2016, 3, 24. [Google Scholar] [CrossRef]
- Bhise, N.S.; Manoharan, V.; Massa, S.; Tamayol, A.; Ghaderi, M.; Miscuglio, M.; Lang, Q.; Shrike Zhang, Y.; Shin, S.R.; Calzone, G.; et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 2016, 8, 014101. [Google Scholar] [CrossRef]
- Agarwal, A.; Goss, J.A.; Cho, A.; McCain, M.L.; Parker, K.K. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 2013, 13, 3599–3608. [Google Scholar] [CrossRef]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, B.K.; Jeong, G.S.; Hyun, J.K.; Lee, C.J.; Lee, S.H. Three-dimensional brain-on-a-chip with an interstitial level of flow and its application as an in vitro model of alzheimer’s disease. Lab Chip 2015, 15, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; LesherPerez, S.C.; Kim, B.C.C.; Yamanishi, C.; Labuz, J.M.; Leung, B.; Takayama, S. Pharmacokinetic profile that reduces nephrotoxicity of gentamicin in a perfused kidney-on-a-chip. Biofabrication 2016, 8, 015021. [Google Scholar] [CrossRef] [PubMed]
- Henry, O.Y.F.; Villenave, R.; Cronce, M.J.; Leineweber, W.D.; Benz, M.A.; Ingber, D.E. Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (teer) measurements of human epithelial barrier function. Lab Chip 2017, 17, 2264–2271. [Google Scholar] [CrossRef]
| Metallic Nanomaterials | Mechanism | Target | Function | Ref. | |
|---|---|---|---|---|---|
| Solution-based metallic nanoparticles | Au nanorod | Change of SERS spectra by aggregation–dissociation | pH | Measurement of intracellular pH from pH 3 to 8 | [105] |
| Au nanoparticle | Scattering spectra by aggregation–dissociation | Cell nucleus | Segregation of undifferentiated from differentiated cells in a human neuronal cell line using PCA analysis | [106] | |
| Au and Ag nanoparticle | SERS spectrum from different shape and size of intracellular-synthesized Au nanoparticles | HEK293T cells | Green synthesis method for production of different metal nanoparticles inside living cells and detection by SERS | [107] | |
| Au nanoparticle | Monitoring of SERS intensity of doxorubicin inside the cancer cell | Doxorubicin | Label-free in situ monitoring of intracellular anti-cancer drug-releasing by Au nanoparticles based on SERS | [108] | |
| Graphene oxide coated silver nanoparticle | Monitoring of SERS intensity of doxorubicin inside the cancer cell | Doxorubicin | SERS effect for monitoring of the loading and releasing of doxorubicin attached to the surface of nanoparticle | [109] | |
| Au-Ag core-shell nanoparticle | Shift of the Raman peaks of 4MBA by the intracellular pH value | pH | High SERS activity of the bimetallic nano-construct with 4MBA for the intracellular pH measurement | [110] | |
| Au-Ag nanorod-ZIF-8 core-shell nanoparticles | SERS effect of the 4-ATP on the surface of nanoparticle | Cancer cells | Targeted SERS imaging of the cancer cells | [111] | |
| Nano-platform onto the cell chip | 3D graphene oxide-encapsulated Au nanoparticle | Change the SERS intensity by the C=C bond of differentiated and undifferentiated cells | Number of C=C bonds | Detection of the differentiation potential of neural stem cells based on SERS | [112] |
| Au nanodot array and microgap electrode | Change of the SERS spectrum by oxidization and reduction of cell | PC12 cell | Measurement of intracellular and extracellular redox state of PC12 cells using SERS and electrochemical techniques | [113] | |
| Ag nanostar patterned ITO substrate | Measurement of biomolecules inside the cells by SERS | SK-BR-3, MCF-7 | Biomolecular detection and characterization of different breast cancer cell lines | [114] | |
| Au nanosphere deposited ITO substrate | Measurement of biomolecules inside the cells by SERS | MDA-MB-231, MCF-7 | Biomolecular detection and characterization of different breast cancer cell lines | [115] | |
| Au nanoparticle | Detection of Raman reporter-labelled Au nanoparticle on the different kind of CCSC | MCF-7, MDA-MB-231, SK-BR-3, Humanbreast CCSC | Nanoparticle-mediated Raman imaging for CCSC characterization which profiles based on the surface marker expression phenotype | [116] | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-N.; Choi, J.-H.; Cho, H.-Y.; Choi, J.-W. Metallic Nanoparticle-Based Optical Cell Chip for Nondestructive Monitoring of Intra/Extracellular Signals. Pharmaceutics 2020, 12, 50. https://doi.org/10.3390/pharmaceutics12010050
Lee S-N, Choi J-H, Cho H-Y, Choi J-W. Metallic Nanoparticle-Based Optical Cell Chip for Nondestructive Monitoring of Intra/Extracellular Signals. Pharmaceutics. 2020; 12(1):50. https://doi.org/10.3390/pharmaceutics12010050
Chicago/Turabian StyleLee, Sang-Nam, Jin-Ha Choi, Hyeon-Yeol Cho, and Jeong-Woo Choi. 2020. "Metallic Nanoparticle-Based Optical Cell Chip for Nondestructive Monitoring of Intra/Extracellular Signals" Pharmaceutics 12, no. 1: 50. https://doi.org/10.3390/pharmaceutics12010050
APA StyleLee, S.-N., Choi, J.-H., Cho, H.-Y., & Choi, J.-W. (2020). Metallic Nanoparticle-Based Optical Cell Chip for Nondestructive Monitoring of Intra/Extracellular Signals. Pharmaceutics, 12(1), 50. https://doi.org/10.3390/pharmaceutics12010050






