Cleavage and Structural Transitions during Maturation of Staphylococcus aureus Bacteriophage 80α and SaPI1 Capsids
Abstract
1. Introduction
2. Materials and Methods
2.1. Allelic Exchange
2.2. Preparation and Titering of Phage Samples
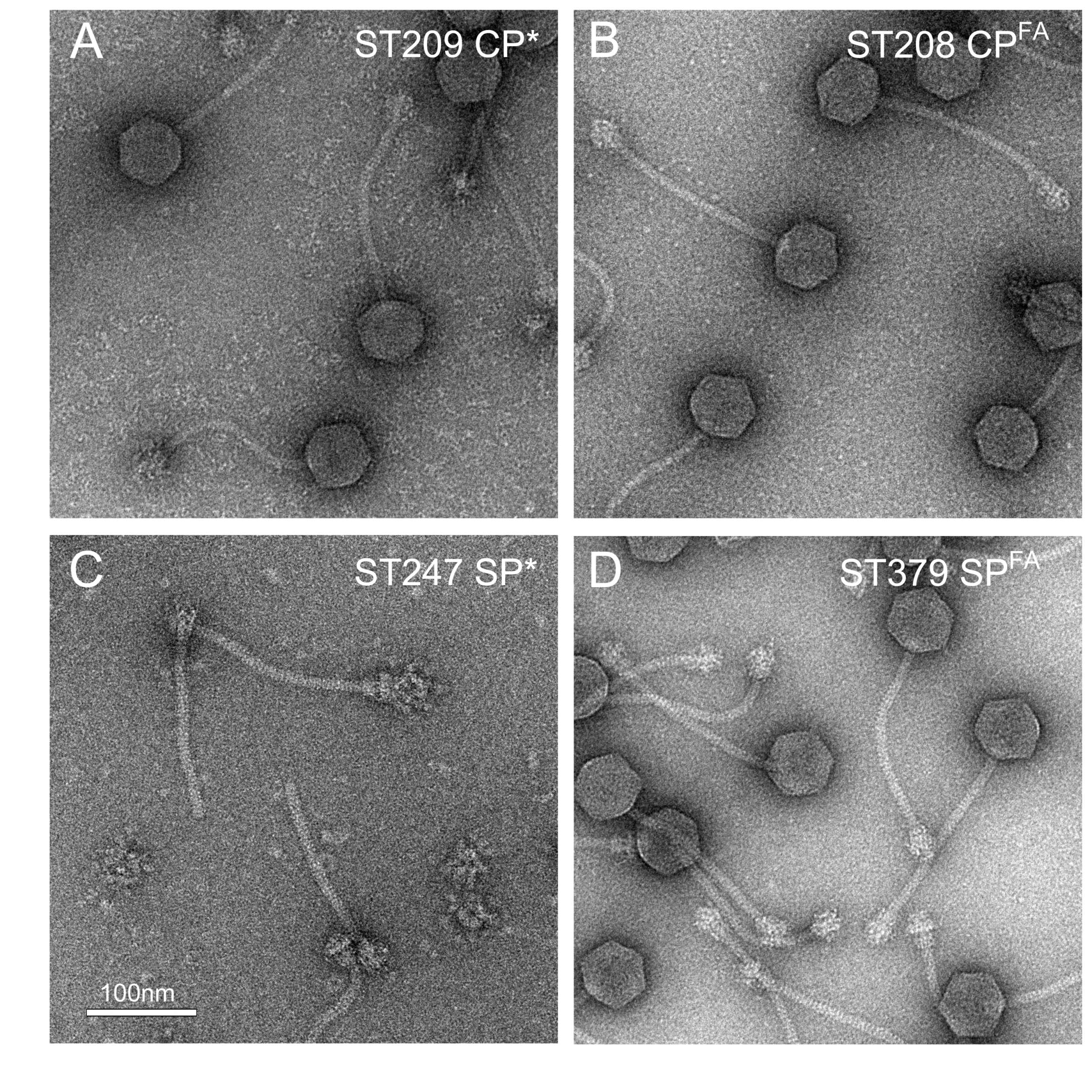
2.3. Phage Stability Assays
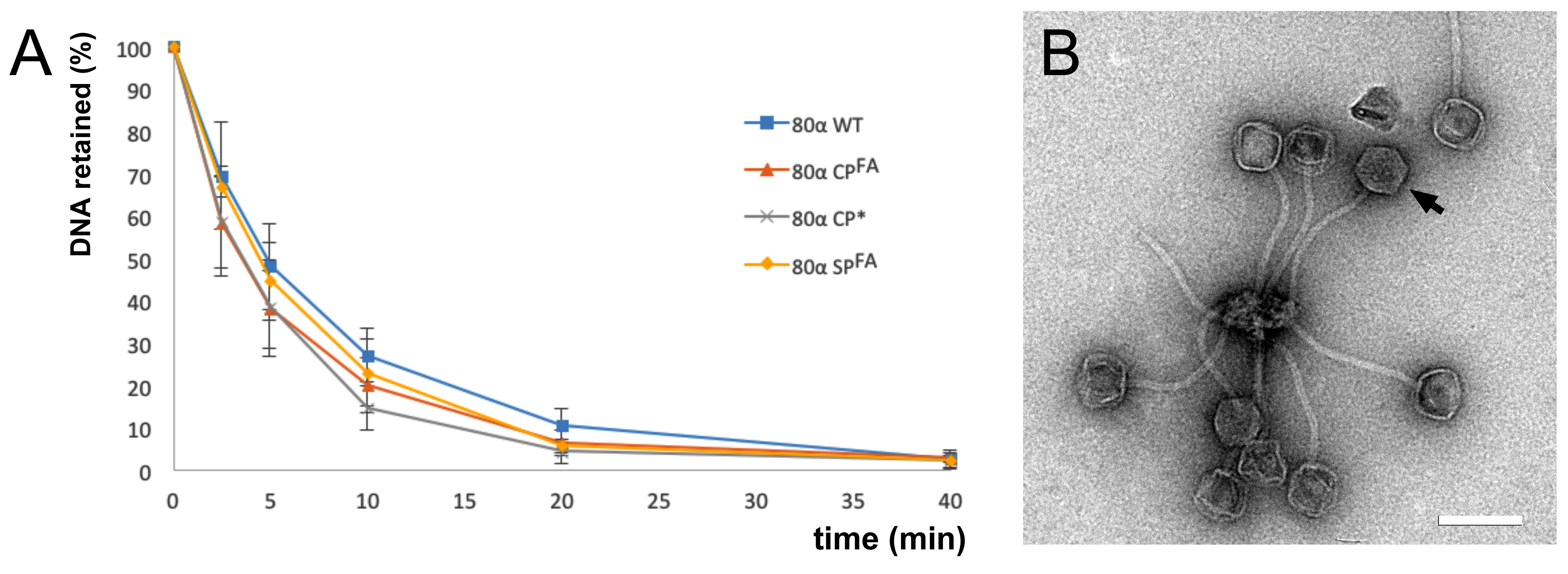
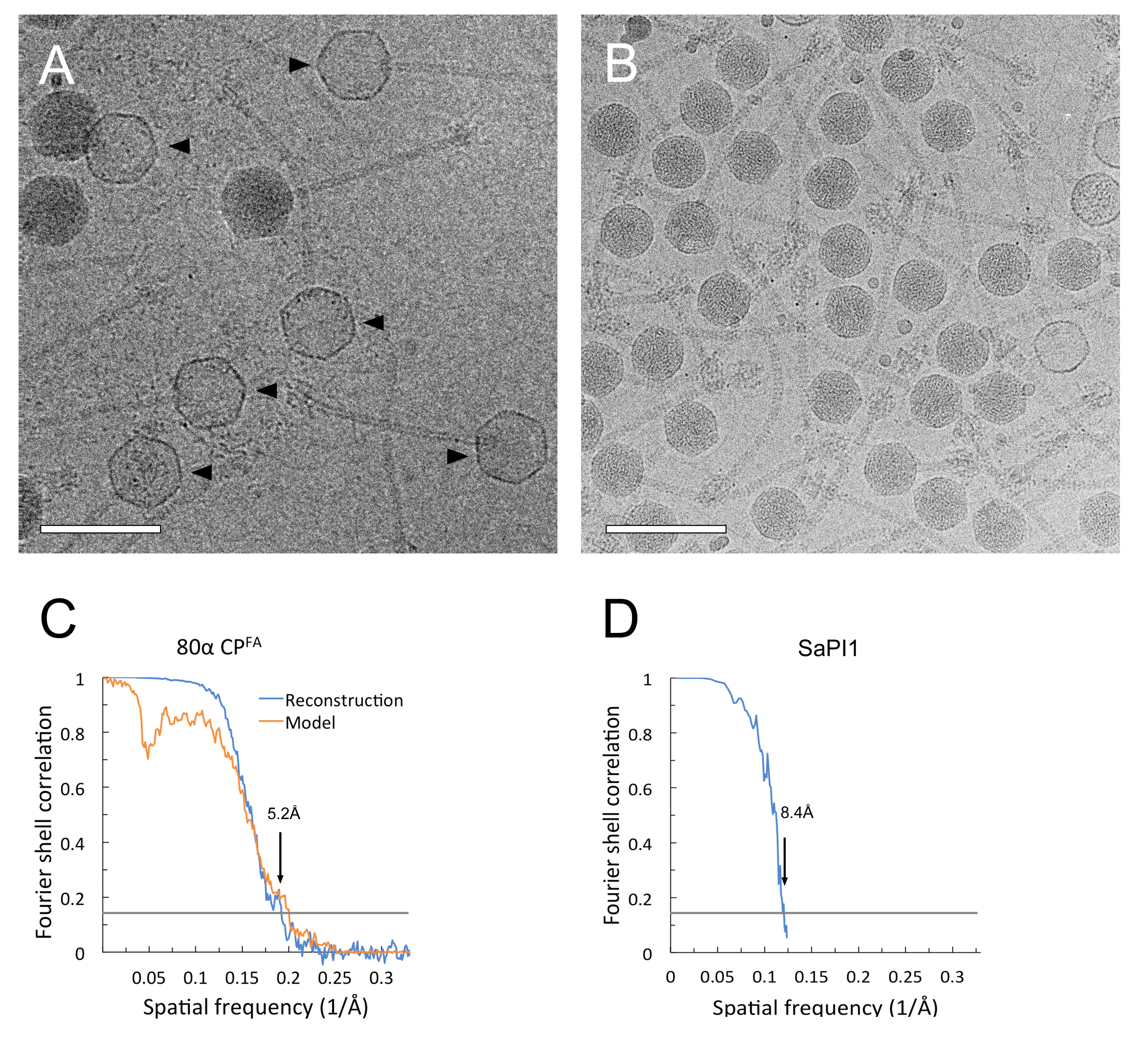
2.4. Electron Microscopy
2.5. Three-Dimensional Reconstruction and Model Building
3. Results
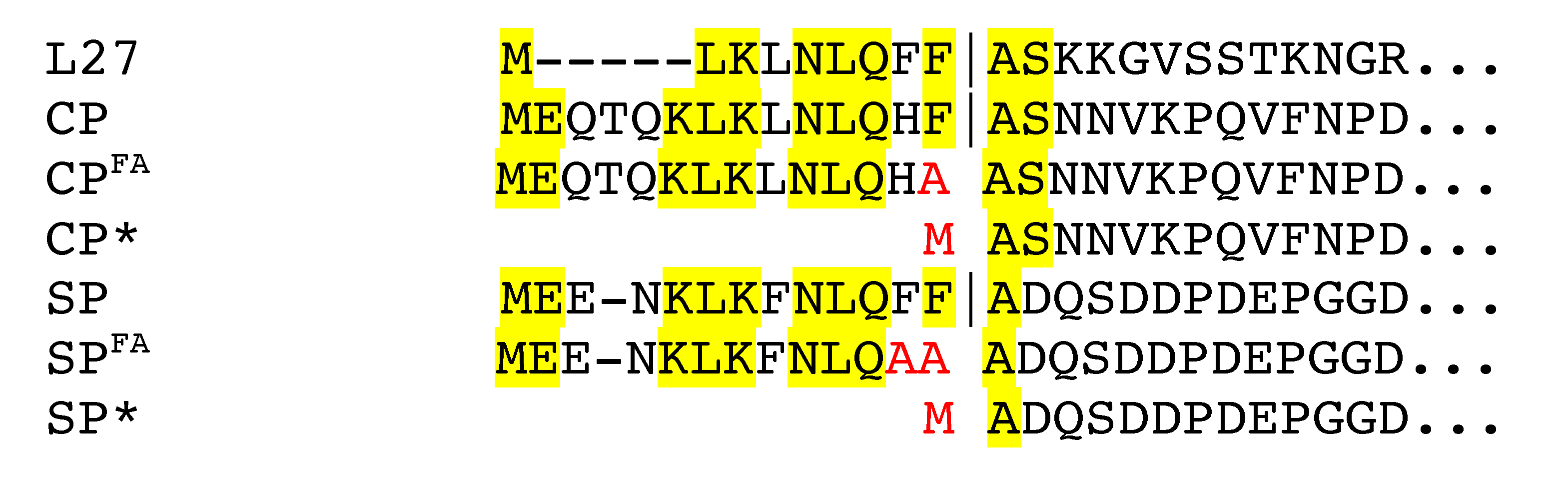
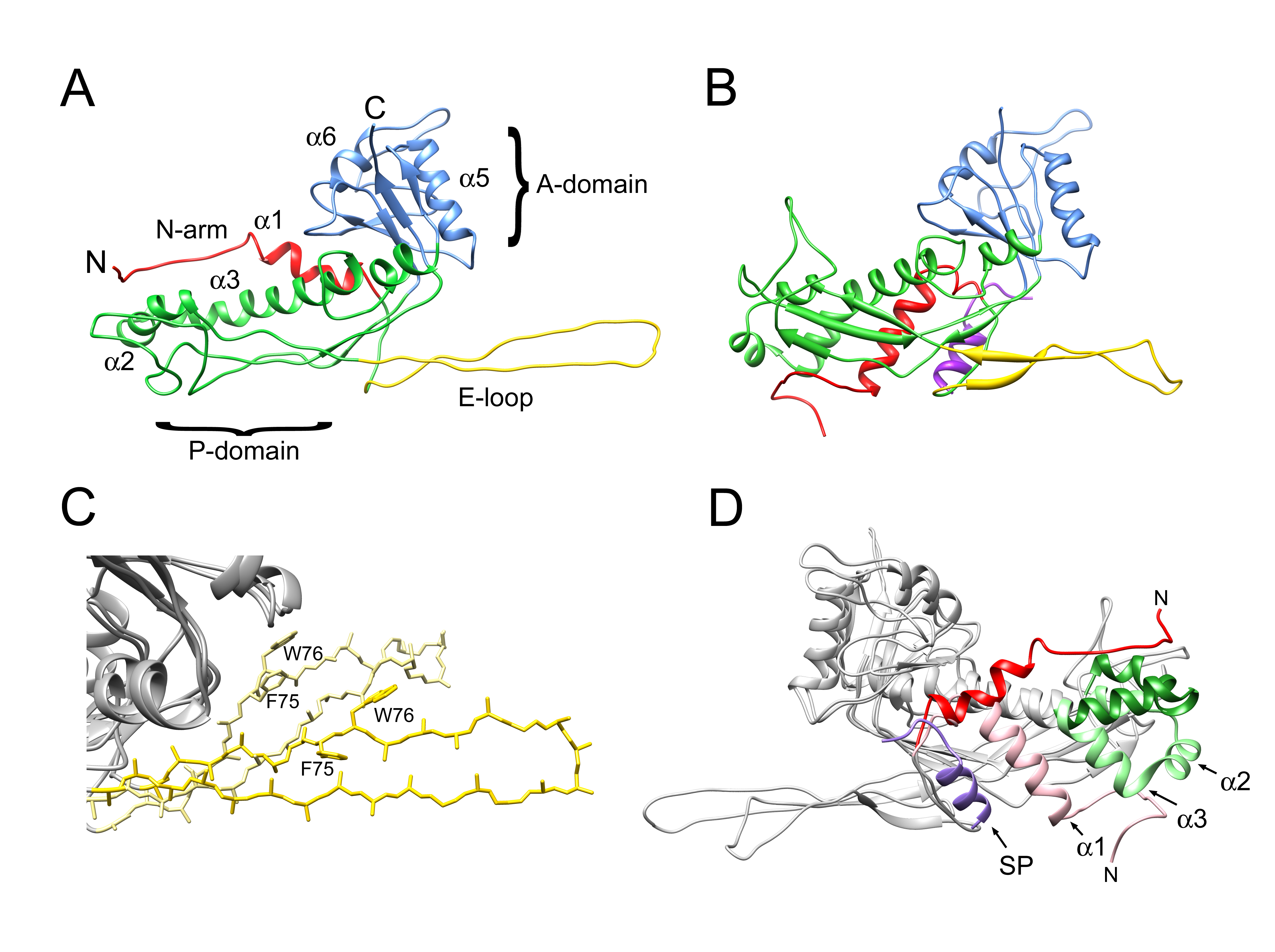
3.1. Cleavage of Capsid and Scaffolding Protein
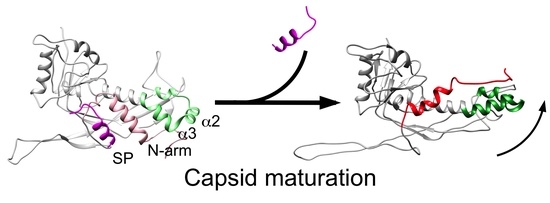
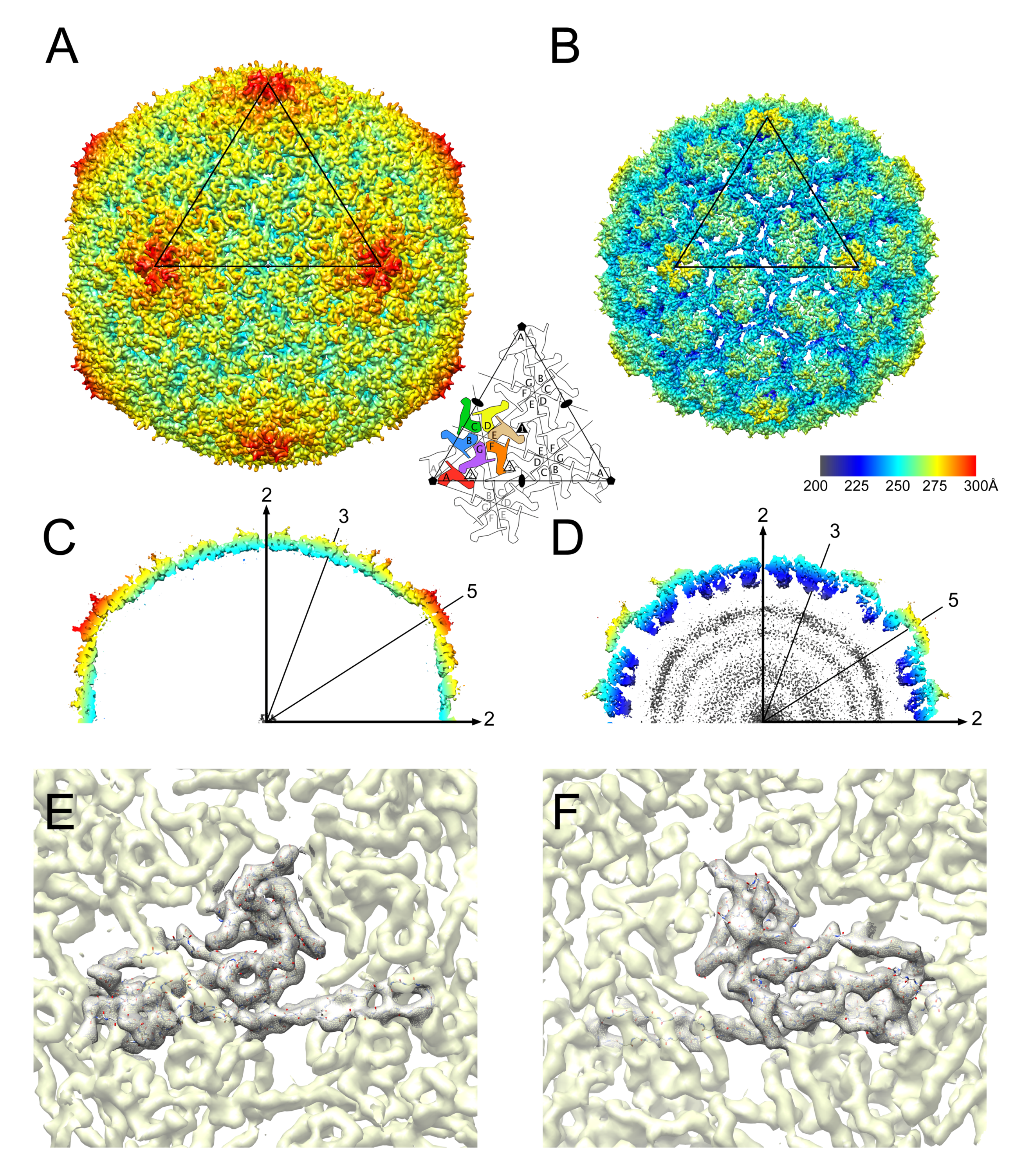
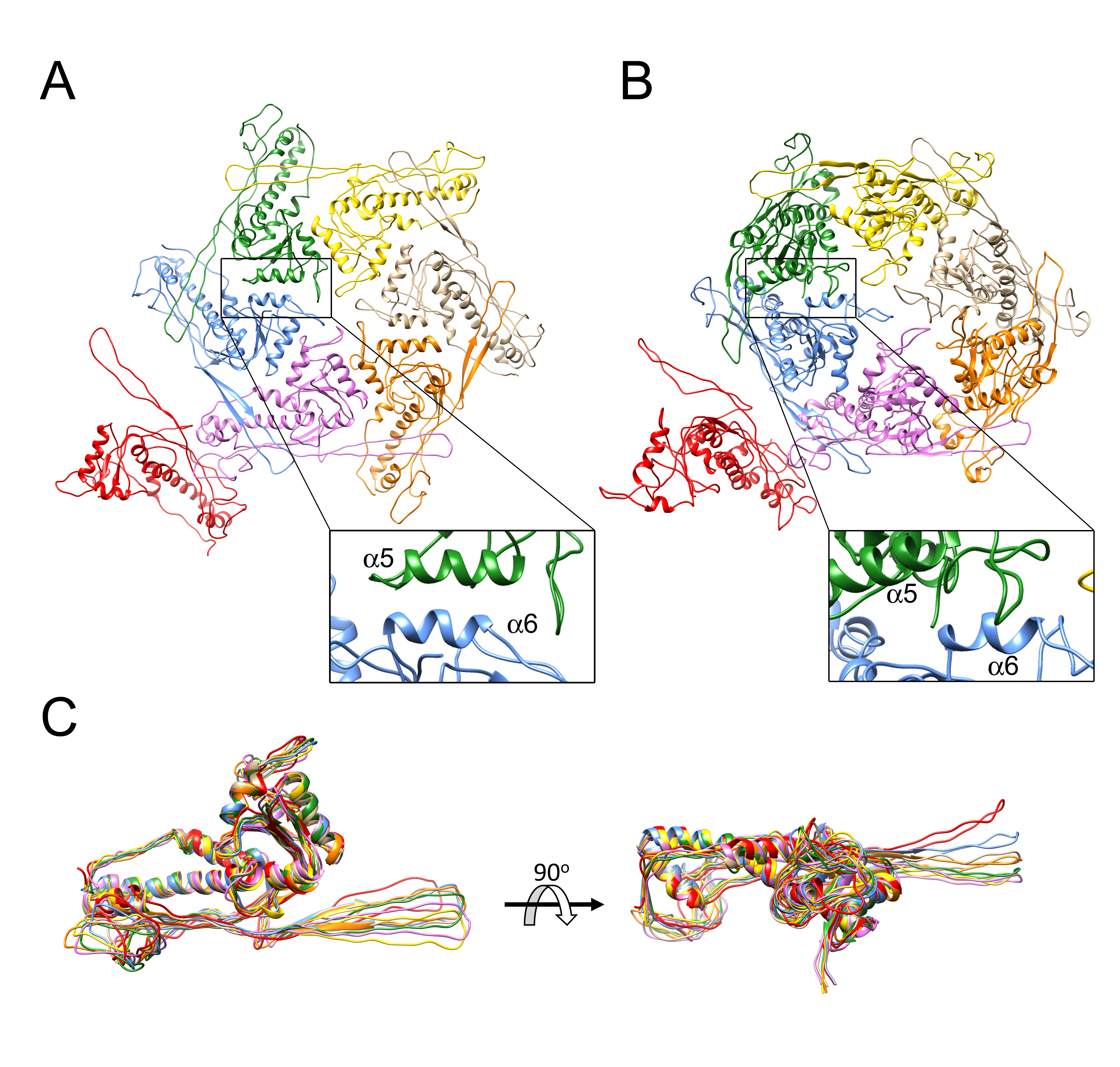
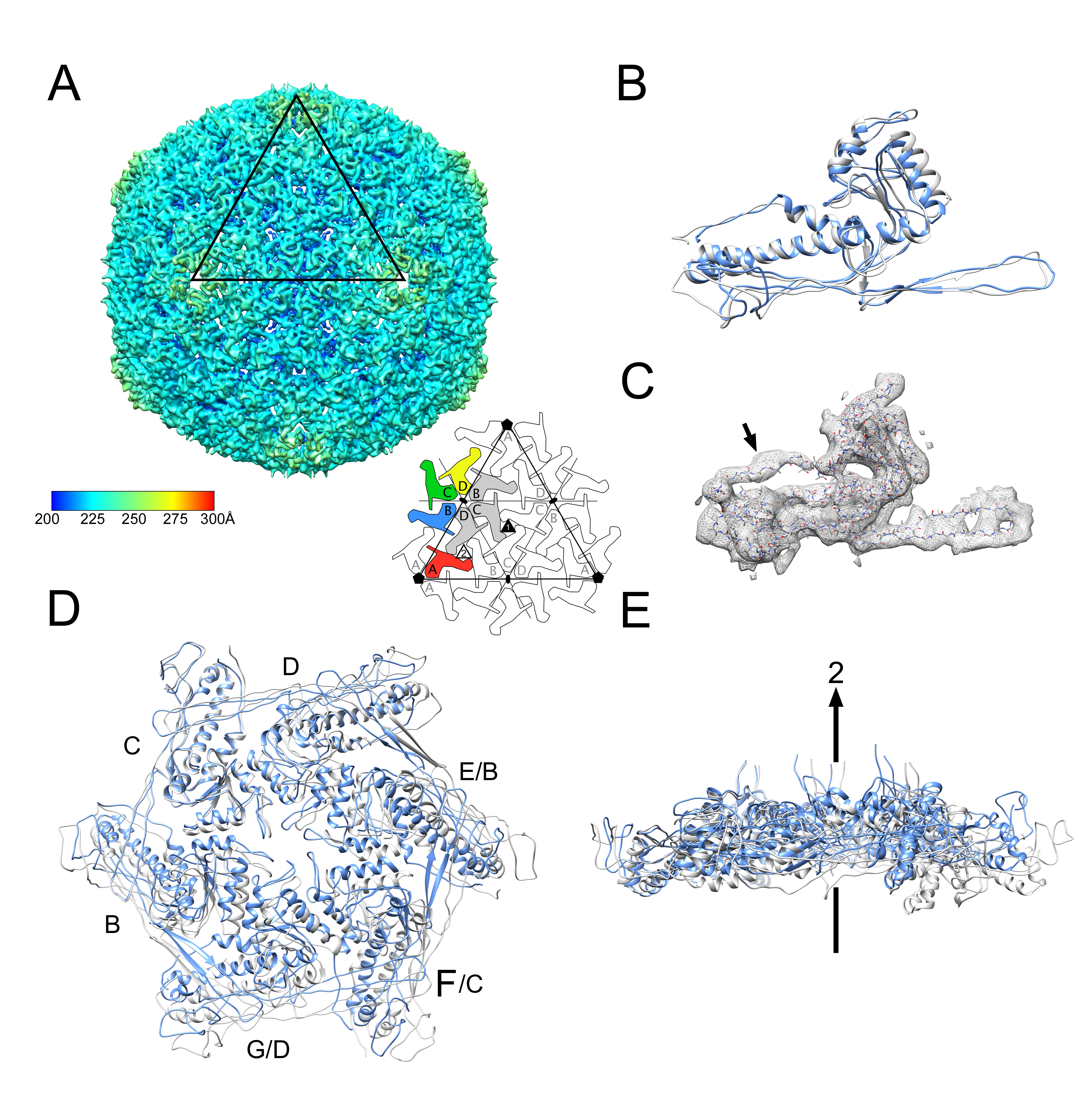
3.2. Structure of the Mature 80α Capsid
3.3. Structure of the Mature SaPI1 Capsid
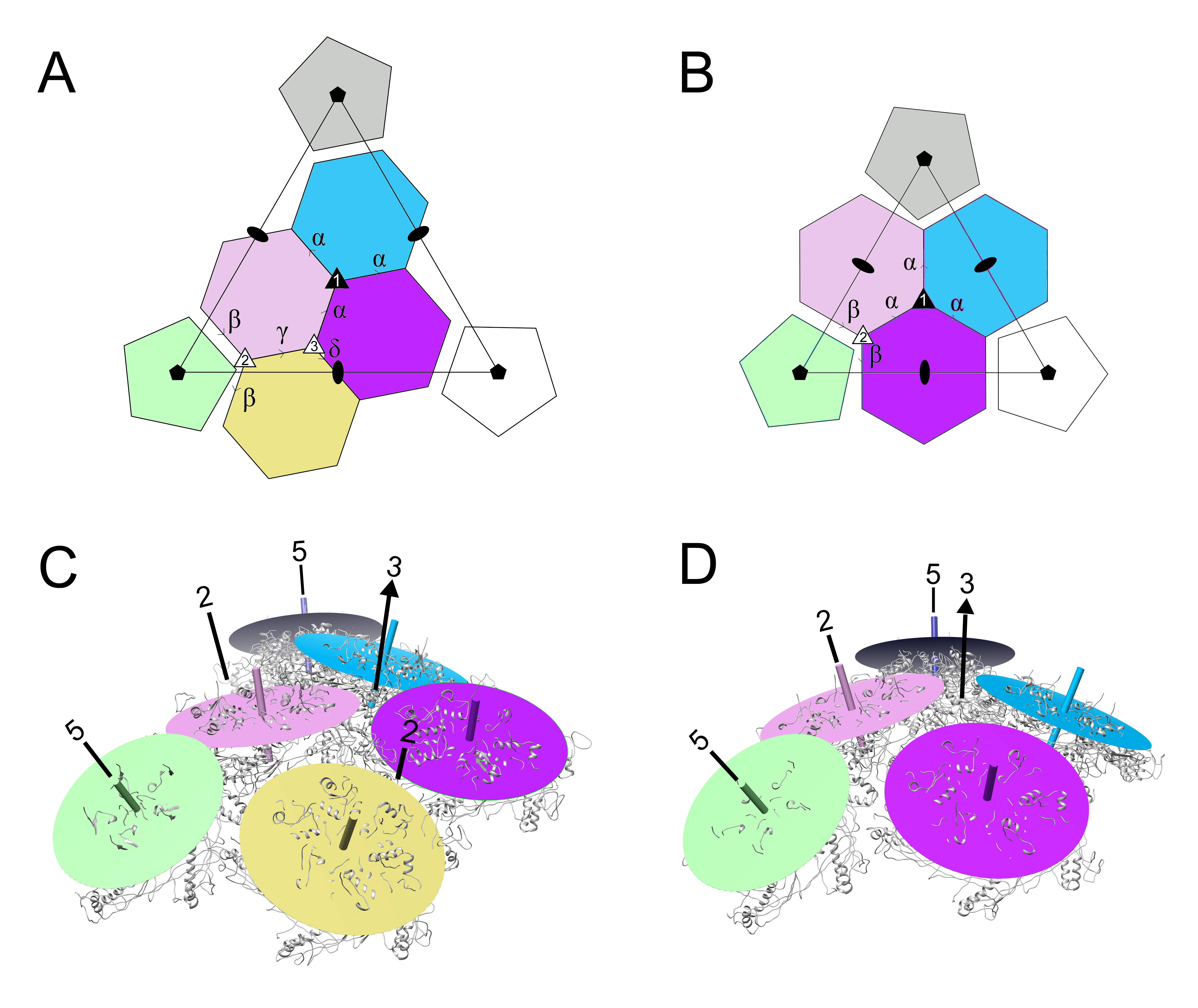
3.4. Comparison of Capsomer Angles
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Archer, G.L. Staphylococcus aureus: A well-armed pathogen. Clin. Infect. Dis. 1998, 26, 1179–1181. [Google Scholar] [CrossRef] [PubMed]
- Lowy, F.D. Staphylococcus aureus infections. N. Eng. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- DeLeo, F.R.; Otto, M.; Kreiswirth, B.N.; Chambers, H.F. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 2010, 375, 1557–1568. [Google Scholar] [CrossRef]
- Boucher, H.W.; Corey, G.R. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 2008, 46 (Suppl. 5), S344–S349. [Google Scholar] [CrossRef] [PubMed]
- Malachowa, N.; DeLeo, F.R. Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 2010, 67, 3057–3071. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu. Rev. Microbiol. 2010, 64, 143–162. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, J.A. Staphylococcus aureus genomics and the impact of horizontal gene transfer. Int. J. Med. Microbiol. 2014, 304, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Novick, R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 1967, 33, 155–166. [Google Scholar] [CrossRef]
- Christie, G.E.; Matthews, A.M.; King, D.G.; Lane, K.D.; Olivarez, N.P.; Tallent, S.M.; Gill, S.R.; Novick, R.P. The complete genomes of Staphylococcus aureus bacteriophages 80 and 80α—Implications for the specificity of SaPI mobilization. Virology 2010, 407, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Spilman, M.S.; Dearborn, A.D.; Chang, J.R.; Damle, P.K.; Christie, G.E.; Dokland, T. A conformational switch involved in maturation of Staphylococcus aureus bacteriophage 80α capsids. J. Mol. Biol. 2011, 405, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Poliakov, A.; Chang, J.R.; Spilman, M.S.; Damle, P.K.; Christie, G.E.; Mobley, J.A.; Dokland, T. Capsid size determination by Staphylococcus aureus pathogenicity island SaPI1 involves specific incorporation of SaPI1 proteins into procapsids. J. Mol. Biol. 2008, 380, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Wall, E.A.; Caufield, J.H.; Lyons, C.E.; Manning, K.A.; Dokland, T.; Christie, G.E. Specific N-terminal cleavage of ribosomal protein L27 in Staphylococcus aureus and related bacteria. Mol. Microbiol. 2015, 95, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Wall, E.A.; Johnson, A.L.; Peterson, D.L.; Christie, G.E. Structural modeling and functional analysis of the essential ribosomal processing protease Prp from Staphylococcus aureus. Mol. Microbiol. 2017, 104, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P.; Christie, G.E.; Penades, J.R. The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 2010, 8, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Christie, G.E.; Dokland, T. Pirates of the Caudovirales. Virology 2012, 434, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Tormo-Mas, M.A.; Mir, I.; Shrestha, A.; Tallent, S.M.; Campoy, S.; Lasa, I.; Barbe, J.; Novick, R.P.; Christie, G.E.; Penades, J.R. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature 2010, 465, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Dearborn, A.D.; Dokland, T. Mobilization of pathogenicity islands by Staphylococcus aureus strain Newman bacteriophages. Bacteriophage 2012, 2, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.L.; Dokland, T. The type 2 dUTPase of bacteriophage φNM1 initiates mobilization of Staphylococcus aureus bovine pathogenicity island 1. J. Mol. Biol. 2016, 428, 142–152. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bowring, J.; Neamah, M.M.; Donderis, J.; Mir-Sanchis, I.; Alite, C.; Ciges-Tomas, J.R.; Maiques, E.; Medmedov, I.; Marina, A.; Penadés, J.R. Pirating conserved phage mechanisms promotes promiscuous staphylococcal pathogenicity island transfer. eLife 2017, 6, e26487. [Google Scholar] [CrossRef] [PubMed]
- Damle, P.K.; Wall, E.A.; Spilman, M.S.; Dearborn, A.D.; Ram, G.; Novick, R.P.; Dokland, T.; Christie, G.E. The roles of SaPI1 proteins gp7 (CpmA) and gp6 (CpmB) in capsid size determination and helper phage interference. Virology 2012, 432, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Dearborn, A.D.; Spilman, M.S.; Damle, P.K.; Chang, J.R.; Monroe, E.B.; Saad, J.S.; Christie, G.E.; Dokland, T. The Staphylococcus aureus pathogenicity island protein gp6 functions as an internal scaffold during capsid size determination. J. Mol. Biol. 2011, 412, 710–722. [Google Scholar] [CrossRef] [PubMed]
- Dearborn, A.D.; Wall, E.A.; Kizziah, J.L.; Klenow, L.; Parker, L.K.; Manning, K.A.; Spilman, M.S.; Spear, J.M.; Christie, G.E.; Dokland, T. Competing scaffolding proteins determine capsid size during mobilization of Staphylococcus aureus pathogenicity islands. eLife 2017, 6, e30822. [Google Scholar] [CrossRef] [PubMed]
- Bento, J.C.; Lane, K.D.; Read, E.K.; Cerca, N.; Christie, G.E. Sequence determinants for DNA packaging specificity in the S. aureus pathogenicity island SaPI1. Plasmid 2014, 71, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, M.; Chastanet, A.; Debarbouille, M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 2004, 70, 6887–6891. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P. Genetic systems in Staphylococci. Methods Enzymol. 1991, 204, 587–636. [Google Scholar] [PubMed]
- Kreiswirth, B.N.; Lofdahl, S.; Betley, M.J.; O’Reilly, M.; Schlievert, P.M.; Bergdoll, M.S.; Novick, R.P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 1983, 305, 709–712. [Google Scholar] [CrossRef] [PubMed]
- Ubeda, C.; Olivarez, N.P.; Barry, P.; Wang, H.; Kong, X.; Matthews, A.; Tallent, S.M.; Christie, G.E.; Novick, R.P. Specificity of staphylococcal phage and SaPI DNA packaging as revealed by integrase and terminase mutations. Mol. Microbiol. 2009, 72, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Tormo, M.A.; Ferrer, M.D.; Maiques, E.; Ubeda, C.; Selva, L.; Lasa, I.; Calvete, J.J.; Novick, R.P.; Penades, J.R. SaPI DNA is packaged in particles composed of phage proteins. J. Bacteriol. 2008, 190, 2434–2440. [Google Scholar] [CrossRef] [PubMed]
- Spilman, M.S.; Damle, P.K.; Dearborn, A.D.; Rodenburg, C.M.; Chang, J.R.; Wall, E.A.; Christie, G.E.; Dokland, T. Assembly of bacteriophage 80α capsids in a Staphylococcus aureus expression system. Virology 2012, 434, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Suloway, C.; Pulokas, J.; Fellmann, D.; Cheng, A.; Guerra, F.; Quispe, J.; Stagg, S.; Potter, C.S.; Carragher, B. Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 2005, 151, 41–60. [Google Scholar] [CrossRef] [PubMed]
- Lander, G.C.; Stagg, S.M.; Voss, N.R.; Cheng, A.; Fellmann, D.; Pulokas, J.; Yoshioka, C.; Irving, C.; Mulder, A.; Lau, P.W.; et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 2009, 166, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Voss, N.R.; Yoshioka, C.K.; Radermacher, M.; Potter, C.S.; Carragher, B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 2009, 166, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Roseman, A.M. FindEM—A fast, efficient program for automatic selection of particles from electron micrographs. J. Struct. Biol. 2004, 145, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Mallick, S.P.; Carragher, B.; Potter, C.S.; Kriegman, D.J. ACE: Automated CTF estimation. Ultramicroscopy 2005, 104, 8–29. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Jiang, W. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol. Biol. 2014, 1117, 401–443. [Google Scholar] [PubMed]
- Brown, A.; Long, F.; Nicholls, R.A.; Toots, J.; Emsley, P.; Murshudov, G. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 136–153. [Google Scholar] [CrossRef] [PubMed]
- Ludtke, S.J.; Baldwin, P.R.; Chiu, W. EMAN: Semiautomated software for high-resolution single-partice reconstructions. J. Struct. Biol. 1999, 128, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Sinkovits, R.S.; Baker, T.S. AUTO3DEM—An automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 2007, 157, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Heymann, J.B.; Belnap, D.M. Bsoft: Image processing and molecular modeling for electron microscopy. J. Struct. Biol. 2007, 157, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Hohn, T.; Katsura, I. Structure and assembly of bacteriophage lambda. Curr. Top. Microbiol. Immunol. 1977, 78, 69–110. [Google Scholar] [PubMed]
- King, J.; Lenk, E.V.; Botstein, D. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. II. Morphogenetic pathway. J. Mol. Biol. 1973, 80, 697–731. [Google Scholar] [CrossRef]
- Conway, J.F.; Duda, R.L.; Cheng, N.; Hendrix, R.W.; Steven, A.C. Proteolytic and conformational control of virus capsid maturation: The bacteriophage HK97 system. J. Mol. Biol. 1995, 253, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.E. Virus particle maturation: Insights into elegantly programmed nanomachines. Curr. Opin. Struct. Biol. 2010, 20, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Wyckoff, E.; Casjens, S. Autoregulation of the bacteriophage P22 scaffolding protein gene. J. Virol. 1985, 53, 192–197. [Google Scholar] [PubMed]
- Dokland, T.; Lindqvist, B.H.; Fuller, S.D. Image reconstruction from cryo-electron micrographs reveals the morphopoietic mechanism in the P2–P4 bacteriophage system. EMBO J. 1992, 11, 839–846. [Google Scholar] [PubMed]
- Dearborn, A.D.; Laurinmaki, P.; Chandramouli, P.; Rodenburg, C.M.; Wang, S.; Butcher, S.J.; Dokland, T. Structure and size determination of bacteriophage P2 and P4 procapsids: Function of size responsiveness mutations. J. Struct. Biol. 2012, 178, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Marzec, C.J.; Day, L.A. Pattern formation in icosahedral virus capsids: The papova viruses and Nudaurelia capensis β virus. Biophys. J. 1993, 65, 2559–2577. [Google Scholar] [CrossRef]
| Strain | Genotype/description | Alias | Reference |
|---|---|---|---|
| RN450 | Phage cured version of reference strain NCTC 8535 | – | [8] |
| RN4220 | Phage cured NCTC 8535 mutated to accept foreign DNA | – | [26] |
| RN10616 | RN4220 (80α) | WT | [27] |
| JP3569 | RN450 (80α ΔCP) | ΔCP | [28] |
| ST91 | RN4220 (80α ΔSP) | ΔSP | [29] |
| ST65 | RN4220 (80α Δorf4 ) SaPI1 tst::tetM ) SaPI1 tst::tetM | SaPI1 | [21] |
| ST209 | RN450 (80α CP Δ1-14) | CP* | This work |
| ST208 | RN450 (80α CP::F14A) | CPFA | This work |
| ST247 | RN4220 (80α SP Δ1-13) | SP* | This work |
| ST379 | RN4220 (80α SP::F12A,F13A) | SPFA | This work |
| Strain | Phage | Titer (pfu/mL) |
|---|---|---|
| RN10616 | 80α WT | 3.0 × 1010 |
| ST208 | 80α CPFA | 2.7 × 1010 |
| ST209 | 80α CP* | 1.8 × 1010 |
| ST379 | 80α SPFA | 4.2 × 1010 |
| ST247 | 80α SP* | <10 |
| Angle | 80α | SaPI1 | ||||
|---|---|---|---|---|---|---|
| Procapsid | Mature Capsid | Δ | Procapsid | Mature Capsid | Δ | |
| α | 157.1 | 157.5 | 0.4 | 144.0 | 144.0 | 0.0 |
| β | 152.9 | 145.8 | −7.1 | 148.3 | 148.3 | 0.0 |
| γ | 149.0 | 141.5 | −7.5 | - | - | - |
| δ | 158.3 | 163.7 | 5.4 | - | - | - |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kizziah, J.L.; Manning, K.A.; Dearborn, A.D.; Wall, E.A.; Klenow, L.; Hill, R.L.L.; Spilman, M.S.; Stagg, S.M.; Christie, G.E.; Dokland, T. Cleavage and Structural Transitions during Maturation of Staphylococcus aureus Bacteriophage 80α and SaPI1 Capsids. Viruses 2017, 9, 384. https://doi.org/10.3390/v9120384
Kizziah JL, Manning KA, Dearborn AD, Wall EA, Klenow L, Hill RLL, Spilman MS, Stagg SM, Christie GE, Dokland T. Cleavage and Structural Transitions during Maturation of Staphylococcus aureus Bacteriophage 80α and SaPI1 Capsids. Viruses. 2017; 9(12):384. https://doi.org/10.3390/v9120384
Chicago/Turabian StyleKizziah, James L., Keith A. Manning, Altaira D. Dearborn, Erin A. Wall, Laura Klenow, Rosanne L. L. Hill, Michael S. Spilman, Scott M. Stagg, Gail E. Christie, and Terje Dokland. 2017. "Cleavage and Structural Transitions during Maturation of Staphylococcus aureus Bacteriophage 80α and SaPI1 Capsids" Viruses 9, no. 12: 384. https://doi.org/10.3390/v9120384
APA StyleKizziah, J. L., Manning, K. A., Dearborn, A. D., Wall, E. A., Klenow, L., Hill, R. L. L., Spilman, M. S., Stagg, S. M., Christie, G. E., & Dokland, T. (2017). Cleavage and Structural Transitions during Maturation of Staphylococcus aureus Bacteriophage 80α and SaPI1 Capsids. Viruses, 9(12), 384. https://doi.org/10.3390/v9120384