Combined DECS Analysis and Next-Generation Sequencing Enable Efficient Detection of Novel Plant RNA Viruses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Infected BSSV Plants
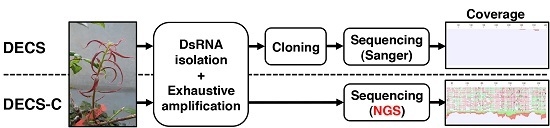
2.2. Library Preparation for NGS
2.3. NGS Sequencing and Data Analysis
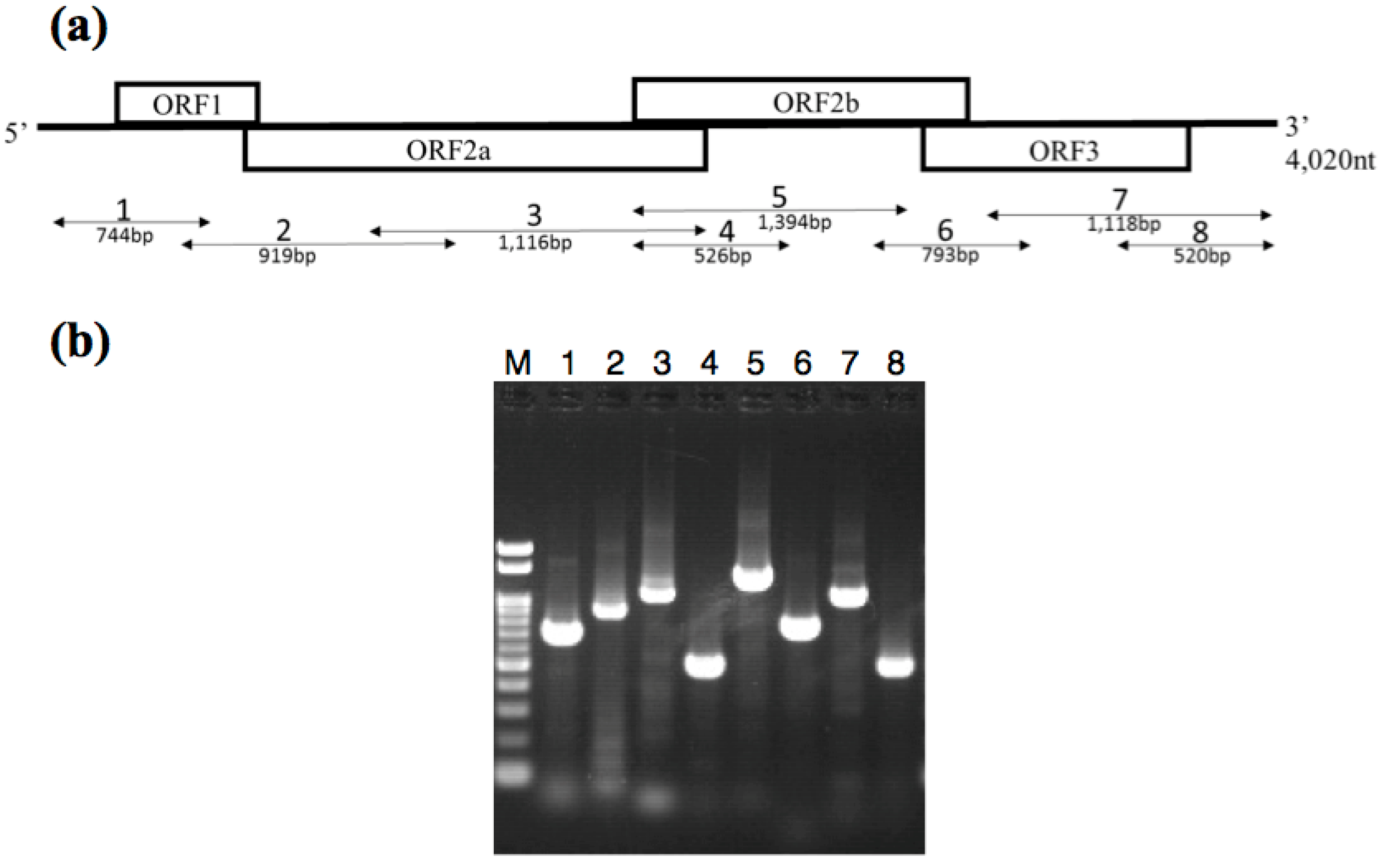
2.4. RT-PCR
2.5. Generation of a Recombinant BSSV Coat Protein and Its Detection by Immunoblotting
3. Results
3.1. Detection of the BSSV Genomic Sequence in NGS Data Analysis
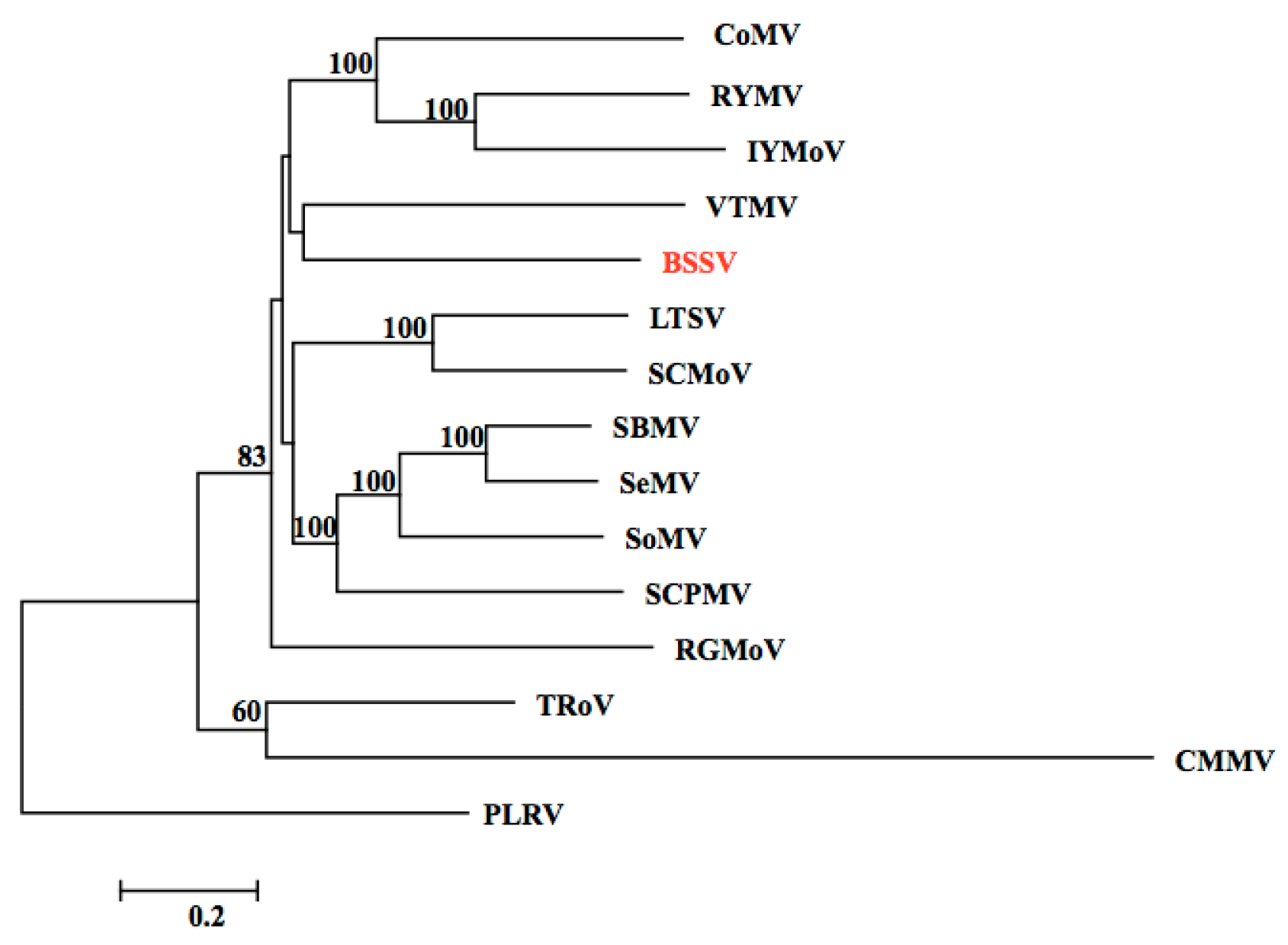
3.2. Identification of the BSSV Genome
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Okada, R.; Kiyota, E.; Moriyama, H.; Fukuhara, T.; Natsuaki, T. A simple and rapid method to purify viral dsRNA from plant and fungal tissue. J. Gen. Plant Pathol. 2015, 81, 103–107. [Google Scholar] [CrossRef]
- Castillo, A.; Cottet, L.; Castro, M.; Sepúlveda, F. Rapid isolation of mycoviral double-stranded RNA from Botrytis cinerea and Saccharomyces cerevisiae. Virol. J. 2011, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Diaz-ruiz, J.R.; Kaper, J.M. Isoletion of viral double-stranded RNAs using a LiCl fractionation procedure. Prep. Biochm. 1978, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Tomita, R.; Sakamoto, M. Recombinant plant dsRNA-binding protein as an effecttive tool for the isolation of viral replicative form dsRNA and universal detection of RNA viruses. J. Gen. Plant Pathol. 2009, 75, 87–91. [Google Scholar] [CrossRef]
- Atsumi, G.; Sekine, K.T.; Kobayashi, K. A New Method to Isolated Total dsRNA. Plant Virol. Protoc. 2015, 1236, 27–37. [Google Scholar]
- Kobayashi, K.; Atsumi, G.; Iwadate, Y.; Tomita, R.; Chiba, K.; Akasaka, S.; Nishihara, M.; Takahashi, H.; Yamaoka, N.; Nishiguchi, M.; et al. Gentian Kobu-sho-associated virus: A tentative, novel double-strand RNA virus that is relevant to gentian Kibu-sho syndrome. J. Gen. Plant Pathol. 2013, 79, 56–63. [Google Scholar] [CrossRef]
- Atsumi, G.; Tomota, R.; Yamashita, T.; Sekine, K.T. A novel virus transmitted through pollination causes ring-spot diseasae on gentian (Gentiana triflora) ovaries. J. Gen. Virol. 2015, 96 Pt 2, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Shimomoto, Y.; Kobayashi, K.; Okuda, M. Identification and characterization of Lisianthus necrotic ringspot virus, a novel distinct tospovirus species causing necrotic disease of lisianthus (Eustoma grandiflorum). J. Gen. Plant Pathol. 2014, 80, 169–175. [Google Scholar] [CrossRef]
- Elbeaino, T.; Giampetruzzi, A.; Stradis, A.D.; Digiaro, M. Deep-sequencing analysis of an apricot tree with vein clearing symptoms reveals the presence of a novel betaflexivirus. Virus Res. 2014, 181, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Kishigami, R.; Yamagishi, N.; Ito, T.; Yoshikawa, N. Detection of apple latent spherical virus in seeds and seedlings from infected apple trees by reverse transcription quantitative PCR and deep sequencing: Evidence for lack of transmission of the virus to most progeny seedlings. J. Gen. Plant Pathol. 2014, 80, 490–498. [Google Scholar] [CrossRef]
- Thekke-Veetil, T.; Polashock, J.J.; Marn, M.V.; Plesko, I.M.; Schilder, A.C.; Keller, K.E.; Martin, R.R.; Tzanetakis, I.E. Population structure of blueberry mosaic associated virus: Evidence of reassortment in geographically distinct isolates. Virus Res. 2015, 201, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Al Rwahnih, M.; Daubert, S.; Golino, D.; Rowhani, A. Deep sequencing analysis of RNAs from a grapevine showing Syrah decline symptoms reveals a multiple virus infection that includes a novel virus. Virology 2009, 387, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Villacreses, J.; Rojas-Herrera, M.; Sánchez, C.; Hewstone, N.; Undurraga, S.F.; Alzate, J.F.; Manque, P.; Maracaja-Coutinho, V.; Polanco, V. Deep Sequencing Reveals the Complete Genome and Evidence for Transcriptional Activity of the First Virus-Like Sequences Identified in Aristotelia chilensis (Maqui Berry). Viruses 2015, 7, 1685–1699. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Jaramillo, J.L.; Ruiz-Medrano, R.; Rojas-Morales, L.; López-Buenfil, J.A.; Morales-Galván, O.; Chavarín-Palacio, C.; Ramírez-Pool, J.A.; Xoconostle-Cázares, B. Characterization of a Proposed Dichorhavirus Associated with the Citrus Leprosis Disease and Analysis of the Host Response. Viruses 2014, 6, 2602–2633. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Suzaki, K.; Nakano, M.; Sato, A. Characterization of a new apscaviroid from American persimmon. Arch. Virol. 2013, 158, 2629–2631. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, K.M.; Ramsdell, D.C.; Gillett, J.M.; Chaney, W.G. Acquisition and transmission of blueberry shoestring virus by its aphid vector Illinoia pepperi. Phytopathology 1985, 75, 709–712. [Google Scholar] [CrossRef]
- Lsaacs, R.; Schilder, A.; Miles, T.; Longstroth, M. Blueberry Aphid and Blueberry Shoestring Virus, Michigan Blueberry Facts, 2008, Michigen State University. Available online: https://www.oakgov.com/msu/Documents/publications/e3050_blueberry_aphid.pdf (accessed on 27 June 2015).
- Urban, L.A.; Ramsdell, D.C.; Klomparens, K.L.; Lynck, T.; Hancock, J.F. Detection of blueberry shoestring virus in xylem and phloem tissues of highbush blueberry. Phytopathology 1989, 79, 488–493. [Google Scholar] [CrossRef]
- Ramsdell, D.C. Physical and chemical properties of blueberry shoestring virus. Phytopathology 1979, 69, 1087–1091. [Google Scholar] [CrossRef]
- Sekine, K.T.; Shirakawa, A.S.; Yanagisawa, H.; Tomita, R.; Atsumi, G. Modification of “DECS analysis”, an exhaustive RNA virus diagnostic system, to extend the applicability to various plant species. Virus and other Graft Transmissible Diseases of Fruit Crops. In Proceedings of the 23rd International Council for the Study of Virus and other Graft Transmissible Diseases of Fruit Crops, Morioka, Japan, 8–12 June 2015.
- Atsumi, G.; Tomita, R.; Kobayashi, K.; Sekine, K.T. Prevalence and genetic diversity of an unusual virus associated with Kobu-sho disease of gentian in Japan. J. Gen. Virol. 2013, 94, 2360–2365. [Google Scholar] [CrossRef] [PubMed]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A Plant DNA Minipreparation, Version. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Tomita, R.; Sekine, K.T.; Mizumoto, H.; Sakamoto, M.; Murai, J.; Kiba, A.; Hikichi, Y.; Suzuki, K.; Kobayashi, K. Genetic basis for the hierarchical interaction between tobamoviruses and allelic L resistance genes from different pepper species. Mol. Plant Microbe Interact. 2011, 24, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Sekine, K.T.; Tomita, R.; Takeuchi, S.; Atsumi, G.; Saitoh, H.; Mizumoto, H.; Kiba, A.; Yamaoka, N.; Nishiguchi, M.; Hikichi, Y.; et al. Functional differentiation in the LRR domains of closely related plant virus resistance proteins that recognize common Avr proteins. Mol. Plant Microbe Interact. 2012, 25, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- Isogai, M.; Muramatu, S.; Watanabe, M.; Yoshikawa, N. Complete nucleotide sequence and latency of a novel blueberry-infecting closterovirus. J. Gen. Plant Pathol. 2013, 79, 123–127. [Google Scholar] [CrossRef]
- Martin, R.R.; Polashockr, J.J.; Tzanetakis, I.E. New and Emerging Viruses of Blueberry and Cranberry. Viruses 2012, 4, 2831–2852. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.; Tzanetakis, I.E. Development of a virus detection and discovery pipeline using next generation sequencing. Virology 2014, 471–473, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Coetzee, B.; Freeborough, M.J.; Maree, H.J.; Celton, J.M.; Rees, D.J.G.; Burger, J.T. Deep sequencing analysis of viruses infecting grapevines: Virome of a vineyard. Virology 2010, 400, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Suzaki, K.; Nakano, M. Genetic characterization of novel putative rhabdovirus and dsRNA virus from Japanese persimmon. J. Gen. Virol. 2013, 94, 1917–1921. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Atsumi, G.; Yamaoka, N.; Sekine, K.T. Sequencing-based virus hunting and virus detection. Jpn. Agric. Res. Q. 2012, 46, 123–128. [Google Scholar] [CrossRef]
- Jenkins, G.M.; Rambaut, A.; Pybus, O.G.; Holmes, E.C. Rates of molecular evolution in RNA viruses: A quantitative phylogenetic analysis. J. Mol. Evol. 2002, 54, 152–161. [Google Scholar] [CrossRef] [PubMed]


| Virus Name | Reference Length (nt) | Mapped Region (nt) | Total Mapped Reads | Average of Coverage | Accession No |
|---|---|---|---|---|---|
| Southern bean mosaic virus | 4132 | 339 | 117 | 4.12 | NC_004060.2 |
| Sesbania mosaic virus | 4148 | 186 | 1 | 0.05 | NC_002568.2 |
| Southern cowpea mosaic virus | 4193 | 228 | 12 | 0.3 | NC_001625.2 |
| Velvet tobacco mottle virus | 4247 | 201 | 2 | 0.09 | NC_014509.2 |
| Rice yellow mottle virus | 4449 | 189 | 25 | 0.77 | NC_001575.2 |
| Imperata yellow mottle virus | 4547 | 258 | 94 | 3.60 | NC_011536.1 |
| Sample Name | Sample No. | Value of Ct a |
|---|---|---|
| BSSV-infected scion-grafted seedling | 1 | 13.184 |
| 2 | 21.951 | |
| 3 | 13.485 | |
| 4 | 12.619 | |
| BSSV-infected original scion | 1 | 18.555 |
| 2 | 13.635 | |
| 3 | 18.922 | |
| 4 | 16.898 | |
| Healthy seedling | 1~20 | N.D. b |
| NTC c | - | N.D. |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yanagisawa, H.; Tomita, R.; Katsu, K.; Uehara, T.; Atsumi, G.; Tateda, C.; Kobayashi, K.; Sekine, K.-T. Combined DECS Analysis and Next-Generation Sequencing Enable Efficient Detection of Novel Plant RNA Viruses. Viruses 2016, 8, 70. https://doi.org/10.3390/v8030070
Yanagisawa H, Tomita R, Katsu K, Uehara T, Atsumi G, Tateda C, Kobayashi K, Sekine K-T. Combined DECS Analysis and Next-Generation Sequencing Enable Efficient Detection of Novel Plant RNA Viruses. Viruses. 2016; 8(3):70. https://doi.org/10.3390/v8030070
Chicago/Turabian StyleYanagisawa, Hironobu, Reiko Tomita, Koji Katsu, Takuya Uehara, Go Atsumi, Chika Tateda, Kappei Kobayashi, and Ken-Taro Sekine. 2016. "Combined DECS Analysis and Next-Generation Sequencing Enable Efficient Detection of Novel Plant RNA Viruses" Viruses 8, no. 3: 70. https://doi.org/10.3390/v8030070
APA StyleYanagisawa, H., Tomita, R., Katsu, K., Uehara, T., Atsumi, G., Tateda, C., Kobayashi, K., & Sekine, K.-T. (2016). Combined DECS Analysis and Next-Generation Sequencing Enable Efficient Detection of Novel Plant RNA Viruses. Viruses, 8(3), 70. https://doi.org/10.3390/v8030070