Abstract
In the absence of an effective vaccine and lack of a complete cure, gene therapy approaches to control HIV infection offer feasible alternatives. Due to the chronic nature of infection, a wide window of opportunity exists to gene modify the HIV susceptible cells that continuously arise from the bone marrow source. To evaluate promising gene therapy approaches that employ various anti-HIV therapeutic molecules, an ideal animal model is necessary to generate important efficacy and preclinical data. In this regard, the humanized mouse models that harbor human hematopoietic cells susceptible to HIV infection provide a suitable in vivo system. This review summarizes the currently used humanized mouse models and different anti-HIV molecules utilized for conferring HIV resistance. Humanized mouse models are compared for their utility in this context and provide perspectives for new directions.
1. Introduction
The advent of combinatorial antiretroviral therapy proved to be highly effective in controlling HIV disease progression to full blown AIDS and thus in dramatically decreasing the mortality rates where treatment is available [1]. However, major current issues are systemic drug toxicity and generation of drug resistant viral mutants during prolonged therapy. In addition, viral latency remains an intractable problem [2,3]. Therefore, alternative innovative approaches are continually being pursued to provide an effective cure. Among these are immune-augmentation protocols to reinvigorate the immune system [4] and gene therapy strategies to render HIV susceptible cells impervious to virus infection at the entry level and/or interfere with intracellular virus replication [5,6]. These approaches show considerable promise due to the inherent nature of HIV infection and disease progression. First, unlike acute viral diseases wherein pathogenesis is rapidly occurring in a span of few days, HIV is a chronic disease, thus providing a wide window of opportunity to interfere with the slow disease progression. Second, the viral susceptible cells are of hematopoietic origin, which are generated on a continued basis from the bone marrow source wherein the precursor cells reside. Thus, therapies can be effectively directed to the newly generated hematopoietic cells, or alternatively the precursor stem cells themselves can be genetically altered such that their progeny can be made virus resistant.
For the success of HIV gene therapy strategies, a number of important criteria need to be met: (1) An effective gene therapeutic construct should be able to permanently disable a critically needed host factor for viral infection and/or potently inhibit virally-encoded messages and/or proteins. (2) Appropriate gene delivery vehicles with minimal toxicity such as gene transducing viral vectors are needed that have high efficiency of gene transduction into HIV target cells or their progenitors. (3) The anti-HIV genes should have long term efficacy and should not promote generation of viral escape mutants. (4) They should not display adverse effects on lineage specific differentiation and immunological function. (5) Finally, promising anti-HIV gene therapeutic constructs need to be evaluated in a suitable in vivo system to derive critical preclinical data necessary for subsequent human clinical trials. This review is mainly focused on currently available humanized mouse models and their utility in testing a variety of anti-HIV gene constructs.
2. An Ideal in Vivo Animal Model for HIV Gene Therapy
HIV is a human virus causing severe disease in its natural host. While chimpanzees can be infected with HIV, they rarely show severe disease. In comparative studies, non-human primate (NHP) macaque models employing related simian immunodeficiency virus (SIV) and chimeric viruses such as simian-human immunodeficiency viruses (SHIVs) have yielded important data [5]. However, their utility is somewhat limited to derive full-fledged relevant data on HIV. In this regard, humanized mice transplanted with HIV susceptible human cells currently are becoming indispensable for testing various anti-HIV constructs [7] (Figure 1). While a variety of humanized mice are currently available, an ideal model should satisfy the following criteria. (1) They should harbor HIV susceptible cells long term and permit chronic HIV infection and helper CD4 T cell loss. (2) Ideally they should continuously generate the full spectrum of HIV susceptible cells, namely CD4 T cells, macrophages and dendritic cells which are primary viral targets. (3) They should permit HIV viral latency as seen in a typical HIV patient. (4) Finally they should generate human immune responses such that immune-restoration by gene therapy strategies can be effectively evaluated.
3. Immunodeficient Strains Used to Generate Humanized Mice
Various humanized mouse models have been used to test gene therapy strategies since the concept of intracellular immunization for HIV was conceived [7,8]. A common denominator has been the utilization of immunodeficient mice which do not reject xenografts for human cell reconstitution. Among the early immunocompromized mice is the SCID mouse which lacks T and B cells which permitted creation of hu-PBL-SCID and SCID-hu mouse models [9,10,11,12]. Later improvements led to generation of NOD-SCID mice with lower levels of NK cells and innate immunity, permitting improved levels of human cell engraftments [13]. A subsequent innovation was the targeted inactivation of the murine IL-2 receptor common gamma chain (IL2-Rcγ) gene, thus nullifying the actions of native mouse cytokines IL-2, IL-4, IL-7, IL-9, IL- 15 and IL-21 [13,14]. This trait, when bred into mice harboring SCID, NOD, RAG1 or RAG2 gene mutations yielded more severe immunocompromized mice (Rag2−/− cγ−/− , Rag1−/− cγ−/− (RG), NOD/shi-scid/cγ−/− null (NOG) and NOD/SCID/cγ−/− (NSG) mice) which were far superior for human cell engraftment [7,15,16]. Transplantation with human hematopoietic stem cells (HSC) into these mice leads to generation of all the necessary human immune cell subsets, namely T, B, NK cells, macrophages and dendritic cells [17,18]. Levels of different cell sets vary in different mouse models, for example NK cells are produced in suboptimal levels [19], but can be increased with IL-15 treatment. Both humoral and cell mediated immune responses are seen [20]. Newer refinements currently underway include introduction of human HLA Class I and II immune system and cytokine genes to generate more robust human immune responses [15,21].
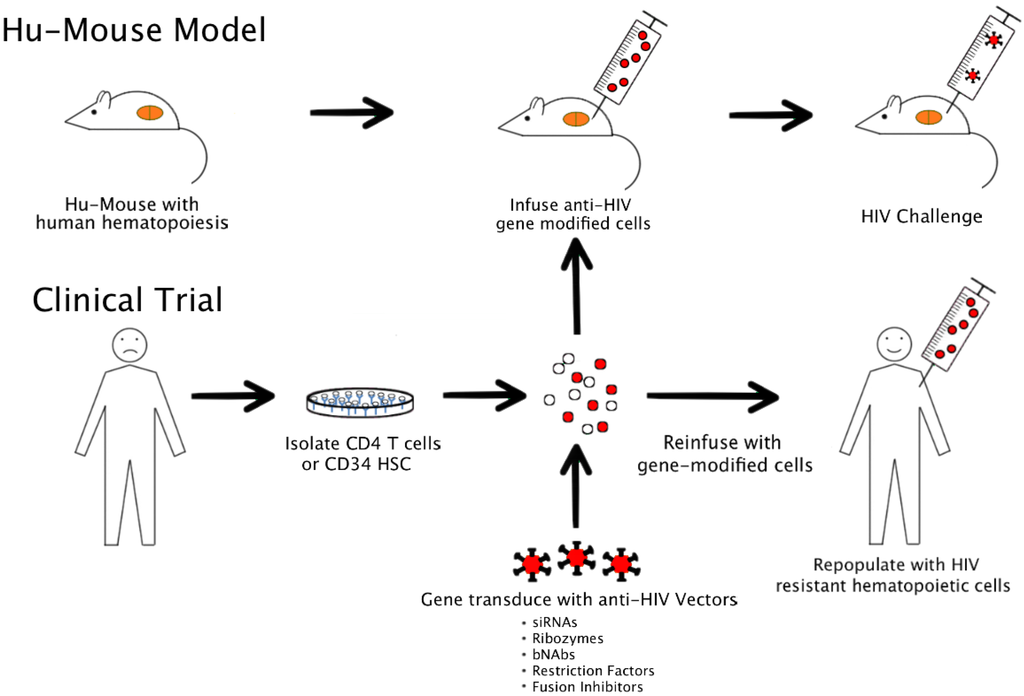
Figure 1.
Modeling HIV gene therapy in humanized mice and clinical application.
4. Currently Used Humanized Mouse Models
Different versions of humanized mice (Hu-Mice) currently exist, each with its own advantages and disadvantages [7]. A major distinguishing feature of new Hu-Mouse models with those of the earlier versions is their ability to support primary human immune responses. A general description describing various features and their utility for testing gene therapy approaches is detailed below and summarized in Table 1.

Table 1.
Current Humanized Mouse Models and Preclinical Gene Therapy Studies.
| Model | Method (Mouse Strain) | Advantages | Disadvantages | Gene Therapy Approaches Studied |
|---|---|---|---|---|
| Hu-PBL | i/p injection of human PBMC. (SCID, NOD-SCID, NSG, NOG) | Easy to produce. Immediate use. Good T cell engraftment. | Lacks multilineage hematopoiesis. Lacks primary immune response. Graft versus host disease. | CCR5 shRNA [22,23] |
| tat-rev shRNA [24] | ||||
| vif/pol shRNA [25] | ||||
| antisense env [24] | ||||
| fusion inhibitor [24] | ||||
| BNAb [26] | ||||
| TCR [27] | ||||
| TRIM5α [28] | ||||
| ZFN for CCR5 [29] | ||||
| ZFN for CXCR4 [30,31] | ||||
| LEDGF/p75 [32] | ||||
| Hu-HSC | Intrahepatic injection of CD34+ HSC into neonates. Intravenous injection of CD34+ HSC into adults. (Rag2−/− yc−/−, NSG, NOG) | Easy to produce. Multilineage hematopoiesis. Primary humoral and cellular immune responses. IgM production. Mucosal engraftment. | Weak human HLA restriction. Weak IgG production. | CCR5 shRNA [33,34] |
| gag, pol shRNA [35] | ||||
| tat-rev shRNA [35,36] | ||||
| nef shRNA [37] | ||||
| LTR shRNA [38] | ||||
| antisense env [39] | ||||
| TAR decoy [34] | ||||
| fusion inhibitor [36] | ||||
| BNAb [40] | ||||
| TRIM5α [33,34] | ||||
| ZFN for CCR5 [41] | ||||
| SCID-Hu | Co-implantation of human fetal liver and thymic tissue under kidney capsule. (SCID or NOD-SCID) | Abundant T cell lymphopoiesis. | Surgery needed, labor intensive. Requires human fetal tissue. No multilineage hematopoiesis. No primary immune response. Poor peripheral T cell engraftment. | Integrase antibody [42] |
| tat-rev shRNA [43] | ||||
| rev shRNA [44] | ||||
| TAR decoy [43,45] | ||||
| CCR5 ribozyme [43,45,46] | ||||
| tat-rev ribozyme rev aptamer [47] | ||||
| CCR5 intrabody [48] | ||||
| TRIM5α [28] | ||||
| Transgenic TCR [49] | ||||
| RevM10 [50] | ||||
| BLT | Co-implantation of human fetal liver and thymic tissue under kidney capsule with additional i/v injection of autologous CD34+ HSC. (Rag2−/− yc−/−, NOD-SCID, NSG) | Multilineage hematopoiesis. Primary humoral and cellular immune responses. IgM production. Presence of human thymus. Human HLA T cell restriction. Mucosal engraftment. | Surgery needed, labor intensive. Requires human fetal tissue. Weak IgG production. | CCR5 shRNA [51,52] LTR shRNA [52] transgenic TCR [53] BNAb [54] |
Hu-PBL mice: By far the easiest model to prepare, this model is created by engraftment of human mature PBMCs by i/p route into SCID, NSG or RG mouse strains [10]. Human immune cells persist for many weeks and HIV-1 can productively infect these mice. However, the injected cells eventually decline and no notable primary immune responses could be seen due to lack of de novo multilineage human hematopoiesis. Graft versus host disease is a problem, thus complicating the interpretation of results.
SCID-hu mice: Surgical co-implantation of human fetal thymus and liver fragments under the SCID mouse kidney capsule generates mice harboring a functional human thymus (thy/liv organoid) [9]. There is robust thymopoiesis with generation of both CD4 and CD8 mature T cells [55]. HIV infection leads to severe depletion of both mature and immature CD4 T cells. A drawback with this model is human immune responses are lacking due to the absence of the full array of human immune cells.
Hu-HSC mice: These are generated by transplantation with hematopoietic stem cells (HSC) into either adult or neonatal mice. IL-2Rcγ−/− mice such as RG, NOG or NSG mice are commonly utilized [7,13]. Engraftment of HSC into conditioned newborn mice via routes such as intra-hepatic injection results in superior human cell engraftment with generation of a full complement of T cells, B cells, macrophages, NK cells and dendritic cells [17,18]. HIV infection leads to chronic viremia lasting more than a year with concomitant CD4 T cell loss [56]. Human antibody responses are seen [57]. While cell mediated immune responses are detected, they are not human HLA restricted [58]. Mucosal human cell engraftment permits HIV vaginal transmission, thus permitting viral challenges by the natural route of infection to assess viral resistance conferred by anti-HIV genes [54,59,60].
BLT mice: These are created by a modification and improvement of the earlier SCID-hu mouse model by additional engraftment with autologous HSC (bone marrow, liver and thymus) [61,62]. Superior human cell engraftment with multilineage generation of T cells, B cells, macrophages, NK cells and dendritic cells is seen. Presence of a functional autologous human thymus in this model permits appropriate T cell education and human HLA cell restriction. HIV infection leads to viremia and helper CD4 T cell loss. Good mucosal human cell engraftment is seen thus permitting mucosal viral challenges [63].
5. Preclinical Gene Therapy Questions that Can be Effectively Assessed in Humanized Mice
In vitro evaluation of various anti-HIV gene therapy constructs in lab adopted T cell lines and human PBMCs provides preliminary efficacy data. However, many important physiological questions cannot be answered by these. Therefore, it is essential that promising strategies be tested in vivo. Among these are, how long does the efficacy last and do escape viral mutants arise after prolonged use? What types of toxicities such as adverse cytokine production are associated with in vivo use? For stem cell-based strategies it needs to be evaluated whether a particular anti-HIV construct displays adverse effects on the stem cell lineage specific differentiation into the end stage hematopoietic cells such as T cells, B cells, macrophages and dendritic cells. Is there any gene silencing/deletion occurring during prolonged in vivo application? Do some specific gene transduced cells have preferential clonal expansions and possibly have oncogenic potential? With specific gene transducing vectors it also needs to be determined if there is a preference to integrate into selective chromosomal sites. Can different anti-viral constructs when introduced in combination work synergistically in providing anti-viral protection? Finally, are the gene transduced lineage specific differentiated cells functionally competent and work synergistically with other cell types in generating an effective immune response such that immune functions can be restored in the AIDS patient? Features that distinguish the newer generation hu-HSC and BLT humanized mouse models from those of previous hu-PBL and SCID-hu mouse models are de novo multilineage human hematopoiesis and the capacity for generating both adaptive and innate immune responses [15,20]. Therefore many of the above questions can be effectively evaluated in an experimental setting to derive important pre-clinical data.
6. Anti-HIV Gene Therapy Constructs
HIV is a highly evolved complex retrovirus with many unique features, especially a great tendency for genomic variation [64]. Antigenic variation in the infected host frustrates the immune system, thus posing a difficulty in controlling infection and in designing effective vaccines. Therefore, novel intracellular immunization strategies offer innovative avenues. Whereas mature CD4 T cells can be genetically modified to confer anti-viral resistance and/or potent effector functions, targeting these approaches to HSC will be long-lasting and therefore potentially conferring a permanent viral control [5,6]. For these strategies to be effective, vulnerable steps of the viral infection need to be targeted. To date, a great deal of data has accumulated on various stages of viral replication. Steps that are amenable to preventive/therapeutic targeting encompass both cellular and virally-encoded molecules. Among these are cellular receptors CD4, CCR5 and CXCR4, virally-encoded regulatory molecules such as tat and rev, and finally host factors that assist or restrict viral replication [5,8]. Varieties of anti-HIV constructs have been experimented both in vitro and in vivo. These fall into two broad categories, namely, nucleic acid based and protein based. A particular advantage with the nucleic acid based approaches versus protein-based is that they are not immunogenic and thus are more suitable for long-range application. The following is a brief description of the various constructs and how they have been successfully evaluated in humanized mice. For brevity, only representative examples are discussed with some historical perspective.
7. RNAi
The phenomenon of RNA interference (RNAi) is a native cellular process that can be exploited to silence any gene of choice [65]. It involves small RNAs that can perform post-transcriptional gene silencing (PTGS) as well as transcriptional gene silencing (TGS). TGS involves the prevention of transcription through epigenetic modification of DNA sequences. This approach is beginning to be harnessed to silence HIV [66]. PTGS involves either sequence specific cleavage of fully complementary target RNAs by siRNAs or translational inhibition and destruction of mRNAs that have imperfect complementary miRNAs. These pathways involve a complex set of intracellular reactions that employ a number of cellular proteins, with the final cleavage of the target RNA performed by an RNA induced silencing complex (RISC), which acts by endonuclease activity [67]. Since the discovery of RNAi, numerous reports described use of siRNAs to inhibit HIV in vitro, and some of these studies progressed to in vivo evaluation [5,8,68]. Both cellular and viral molecules essential for viral infection were targeted. For synthetic siRNA delivery into cells, most methods used a variety of transfection methods in addition to using novel molecules such as dendrimers [69,70]. For infected cell-specific delivery, ligand-based approaches such as using antibodies or aptamers were employed [71,72,73,74]. The gene therapy approach calls for constitutive endogenous expression requiring incorporation of siRNA coding sequence into the target cell genomes. In this regard lentiviral vectors are the most commonly used for gene transduction [8].
One particular complication with the use of RNAi as a gene therapy for HIV involves the ability of HIV to generate escape mutations relatively easily. Even silent mutations with no consequences on the amino acid sequence (thus not conferring a fitness cost) could be sufficient to confer resistance to shRNA targeting viral transcripts [75]. Thus, for RNAi to be fully effective against HIV, it should be deployed as part of a combinatorial approach, either targeting multiple viral genes, or alternatively acting against cellular factors necessary for viral infection and/or replication.
Among the prominent host cell targets evaluated in the RNAi context are the primary cell surface receptor CD4 and co-receptors CXCR4 and CCR5 [6,8]. Of these CCR5 received the most attention recently due to the now famous case of the “Berlin patient” who was cured of the HIV infection after allogenic bone marrow transplantation with CCR5 negative HSC [76,77]. Based on this example, many ongoing studies are directed towards silencing the CCR5 gene.
In early studies evaluating CCR5 siRNAs using gene transduced human PBMCs in vivo in humanized mice, An et al. found that high expression levels achieved under the control of the U6 promoter resulted in significant cytotoxicity [78]. However, this was avoided through the use of a weaker H1 promoter but resulting in the desirable levels of CCR5 down regulation. In later studies using a similar approach, CD34 HSCs were transduced and engrafted into BLT mice [51]. T cell development was normal and CCR5 knockdown was maintained during long term in vivo and these cells were resistant to HIV challenge ex vivo. For targeted delivery of CCR5 shRNA only into CCR5-expressing cells, a ZZ domain/monoclonal antibody conjugated Sindbis virus glycoprotein pseudotyped lentiviral vector was used [23]. In vitro results showed inhibition of HIV-1 replication, and in vivo results with NSG mice engrafted with these cells confirmed CCR5 expressing cells were targeted for transduction. Transduction of iPSCs with a lentiviral vector encoding CCR5 shRNA has also been tested, however, the in vivo efficacy of the transduced cells was not demonstrated [33]. A potential limitation of targeting only CCR5, however, is that such a strategy may promote selection of a CXCR4-tropic strain, with the possibility of a quicker progression to AIDS.
In addition to host cell molecules, a number of virally-encoded RNAs were also targeted by siRNAs. Some examples are, tat, rev, nef and env. Many of these were successfully tested in humanized mice (See Table 1). SiRNAs against the viral tat-rev genes were found to be effective at inhibiting HIV in vivo and ex vivo [24,36,43]. shRNA for rev alone [44], nef alone [37], vif/pol [25], and the HIV LTR [38,52] have been shown to have efficacy as well in humanized mice, along with the full length antisense env gene [24,39]. More recently the safety of a multi-shRNA based gene therapy for HIV-1 targeting gag, pol and tat/rev, has been evaluated in humanized mice [35]. Phase I clinical trials using this construct are planned.
8. Ribozymes
Ribozymes are RNA molecules with catalytic activity which can cleave phosphodiester bonds present in target RNA sequences, and thus can be exploited for gene silencing. They can be custom designed to achieve site specific cleavage of desired target RNA molecules [78]. There was much initial excitement in using ribozymes for clinical application, although later discovery of RNAi provided more potent molecules for similar applications. For HIV gene therapy, ribozymes were employed to target either host or virally-encoded RNAs, some of which were later evaluated in clinical trials [79,80]. Modest decreases in viral load and increases in frequencies of CD4 T cells were seen in treated individuals [80]. Similar to siRNAs, ribozymes are also non-immunogenic, thus avoiding the risk of immune rejection of gene transduced cells.
In early in vivo studies of CCR5, specific ribozymes were incorporated into retroviral vectors and transduced into HSCs for gene therapy application [6]. Vector delivered ribozymes have played a key role in setting the stage for deriving preclinical data on transgene effects on hematopoietic lineage specific differentiation and expression in terminally differentiated cells such as T cells [42]. In these studies, the SCID-hu mouse model was used. Work of Bai et al. that tested a CCR5 ribozyme showed no adverse effects on thymocyte differentiation from retroviral gene transduced HSC [46]. Subsequent studies using a lentiviral vector that gave much higher transduction efficiencies showed similar results [45]. In vivo derived transgenic cells were found to resist ex vivo HIV challenge confirming the efficacy of the ribozyme in down regulating the CCR5 co-receptor in differentiated cells. Later studies in SCID-hu mice have also successfully tested the efficacy of anti-HIV tat-rev and env ribozymes [47]. In a combinatorial approach, CCR5 ribozymes were incorporated into a lentiviral vector that also harbored a TAR decoy and a tat-rev siRNA and tested in humanized mice [43] (see below).
9. RNA Decoys and Aptamers
RNA decoys consist of the protein binding sequences of parent native RNA molecules whereas RNA aptamers are derived by in vitro selection for high affinity binding [69]. These molecules sequester the cognate viral regulatory or essential proteins and neutralize their function. TAR decoys inhibit HIV by neutralizing viral tat protein and rev aptamers interfere with rev function. Both TAR decoys [45] and a rev aptamer [47] were evaluated in SCID-hu mice. Transgenic T cells derived in vivo were found to show anti-HIV resistance. Later studies used the TAR decoy in combination with anti-CCR5 ribozymes and tat-rev siRNAs [43], and a TAR decoy in combination with CCR5 shRNA and a chimeric human-rhesus macaque TRIM5α gene was shown in hu-HSC mice to have in vivo efficacy against HIV [34]. Many other aptamers to different viral or cellular targets were developed for use in gene therapy approaches and await in vivo testing in a gene therapy setting [81]. With regard to immune rejection, RNA decoys and aptamers are also considered to be safe due to lack of immunogenicity.
10. Antibodies
While antibody responses are typically generated in response to HIV-1 infection, they are found to be inadequate in control of viral replication due to the propensity of the virus for constant antigenic variation [82]. This feature of the virus has also been frustrating in the development of an effective vaccine. Therefore, novel gene therapy efforts have also been directed at boosting host humoral immunity by delivering anti-HIV antibody coding genes into human cells in vivo. A first report utilizing this strategy employed an SV40 vector encoding a variable antibody fragment against the viral integrase protein [42]. SCID-hu mice thy/liv grafts were injected with the vector leading to the production of antibody by the transduced thymocytes. HIV challenge in mice showed marked viral resistance, supporting the value of this approach. Broadly neutralizing anti-HIV antibodies (bNAbs) are seen in a subset of HIV infected individuals [82]. These bNAbs are superior due to their high potency against a broad range of HIV strains, and therefore vaccines that can elicit these will have obvious advantages. However, induction of these in immunized individuals has not been accomplished with the current experimental vaccines. Therefore, supplementation of these via what is recently termed as vectored immunoprophylaxis shows considerable promise.
Such an approach was first tested by using bNAb 2G12 in humanized mice [40]. An antibody encoding lentiviral vector was employed to transduce human HSC, which were then engrafted into NSG mice. Systemic 2G12 antibody production was seen, and there was marked reduction in viral loads upon HIV challenge compared to non-treated mice. A separate approach used a tumor cell “backpack” constitutively producing a dimeric 2G12 bNAb. Hu-Mice harboring this “backpack” showed reduced viral loads and preserved CD4 T cell levels [83]. Using a different vector, AAV coding for bNABs b12, 2G12, 4E10 and VRC01, humanized mice (NSG and RG mice backgrounds) were vaccinated intramuscularly, leading to systemic antibody production lasting as long as 52 weeks [84]. Full protection from HIV challenge via i/v route was seen with the B12 and VRC01 antibodies, whereas partial protection was seen with 2G12, 4E10, and 2F5 antibodies. Since HIV is transmitted primarily by sexual transmission by the mucosal routes and IgA is the predominant mucosal antibody, another study evaluated the efficacy of lentiviral vector delivered IgA [54]. In humanized NSG, BLT, and hu-HSC RG mice prepared by transplantation with antibody gene transduced HSC, the transgenes were expressed by B cells and plasma cells in both lymphoid organs and mucosal sites. While there was no protection from HIV infection following HIV-1 vaginal challenge, CD4 T cell depletion was drastically reduced in mucosal sites.
Antibodies directed against host proteins involved in viral infection have also been tested in humanized mice in a different approach. CD34+ HSCs were transduced with a lentiviral vector encoding a single chain CCR5 antibody (termed intrabody). During intracellular synthesis the intrabody prevents export of CCR5 protein to the cell surface. SCID-hu mice engrafted with the intrabody transduced HSC were found to be resistant to R5 tropic HIV infection [48].
11. Transgenic T cell Receptors
Well chosen viral specific T cell receptors (TCRs) genetically introduced into primary T cells or HSCs have the potential to mediate potent effector T cell functions against infected cells expressing conserved viral antigens [8]. Therefore their exploitation for HIV gene therapy holds considerable promise. In vitro studies have documented the efficacy of such gene transduced CD8 T cells bearing transgenic TCRs in inhibiting HIV replication [84]. In vivo efficacy was evaluated by the use of a lentiviral vector containing a gag-SL9 (SLYNTVATL) specific TCR. Vector transduced human PBMC were injected into the spleens of SCID mice along with HIV infected PBMC [27]. Significantly lower viral titers were seen, suggesting that the engineered CTLs were able to exert some control over viral replication in vivo. In a stem cell based approach, human HSC transduced with a similar gag-SL9 TCR and allowed to mature in SCID-hu mice thy/liv grafts [49]. Functional HIV specific CTLs were generated which recognized SL9 epitope in the context of proper HLA type. Later, this same group demonstrated in vivo suppression of HIV in NSG BLT mice prepared by injection of HSC transduced with gag-SL9 TCR [53]. Higher numbers of SL9-specific CD8 T cells in vivo correlated with lower plasma viral loads. These data demonstrated the feasibility of cell mediated immune engineering against HIV. However, for a broad and long lasting protection it is necessary that multiple TCRs must be chosen carefully to target the most conserved HIV epitopes, since escape from CTL responses is common [85].
12. Transdominant Proteins
Transdominant proteins exert their effect by interfering with the action of their counterpart native proteins involved in critical functions essential for viral replication. Some of these with anti-HIV properties have been evaluated in humanized mice to determine protection from HIV-1. An example is RevM10 that interferes with HIV rev protein action [50]. RevM10 encoding retroviral vector transduced CD34 HSC when engrafted into SCID-hu thymic grafts gave rise to normal T cells which upon ex vivo challenge showed HIV resistance. However, a disadvantage in a clinical setting is the immunogenicity of the protein leading to eventual elimination of gene transduced cells in vivo. Indeed in clinical trials this has not fared well, resulting in only a modest survival of CD4 T cells containing the RevM10 gene [86,87,88].
Another target of HIV-1 gene therapy tested in humanized mice is Lens Epithelium Derived Growth Factor (LEDGF) which is an essential cellular cofactor for viral integration [89]. Its inactivation by siRNAs or over expression of its C-terminal domain was shown to inhibit HIV-1 in vitro. This effect was also confirmed in vivo in humanized mice using gene transduced CD4 T cells [32]. Lower viral loads and protection from CD4 T cell decline were observed.
13. Zinc finger, TALENS and CRISPR Nucleases
Novel gene disruption methods employ gene targeted nucleases which encompass ZFN (zinc finger nucleases), TALENS (transcription activator-like effector nucleases), and CRISPR (clustered regulatory interspaced short palindromic repeats) systems [90,91,92,93]. ZFNs were among the nucleases initially exploited to silence the CCR5 gene to render HIV susceptible cells impervious to infection with the R5 tropic HIV, the predominant viral strain involved in natural viral transmission. Engineered ZFNs, which are an array of DNA-binding zinc fingers tethered covalently to a nonspecific FokI restriction nuclease, act by causing double strand DNA breaks at specific recognition sites leading to gene deletions and insertions thus permanently disabling a targeted gene [94]. An advantage with these is that their transient expression can lead to permanent gene disruption. In some of the first studies with CCR5 specific ZFNs, Perez et al. showed that human CD4 T cells that had been modified by ZFNs showed efficacy in humanized mice [95], with lower viral loads and preserved CD4 T cell levels. Later studies of Holt et al. also evaluated this approach in vivo in humanized mice using ZFN modified HSCs [41]. There was multilineage human hematopoiesis and generation of cells lacking the CCR5 receptor. HIV infection of these mice resulted in selection of CCR5 negative cells, lower viral loads and protection from CD4 T cell loss. More recently, an adenoviral vector encoding CCR5-ZFNs was used to modify human CD4 T cells, which were then engrafted into NSG mice to evaluate potential toxicity [29]. No detectable ZFN-specific toxicity or T-cell transformation was observed.
ZFNs targeting the second coreceptor CXCR4 have also been evaluated in humanized mice [30,31]. When engrafted with zinc-finger modified CD4 T cells and challenged with X4 tropic virus, these mice were shown to be resistant to in vivo challenge. However, disabling CXCR4 in HSCs has disadvantages since this receptor is indispensable for normal homing patterns and hematopoietic homeostasis. Potential downsides to the nuclease-based gene disruption approaches include the relatively modest bi-allelic gene modification and off target effects [96], resulting in unwanted disruption of important regulatory genes, possibly leading to malignancy. While no short term toxicities have been observed so far in ongoing clinical trials [29], long term follow up for several years is required to assess potential negative effects.
TALENs, like ZFNs, bind to specific DNA sequences via transcription activator-like (TAL) proteins originally isolated from plant-pathogenic bacteria of the Xanthomonas species [97]. They create double stranded breaks through the action of covalently bound FokI nuclease domain [94]. They have an advantage of higher sequence specificity than ZFNs and relatively simple retargeting, with the disadvantage of a larger size and subsequent greater difficulty in delivery to target cells.
Originally identified as elements of an adaptive immune system found in bacteria and archea, CRISPR, in constrast to ZFN and TALENs, relies on the use of guide RNA (gRNA) to deliver CRISPR-associated (Cas) nucleases [98]. When the gRNA bound to a codon-optimized Cas9 protein binds to a complementary sequence on DNA, it creates double strand DNA breaks at the unique target sequences. This has the advantages of even greater sequence specificity and is relatively easy to retarget to new sequences [99]. Potential off-target effects remain to be evaluated in future in depth studies.
There has been considerable excitement in the use of TALENs or CRISPR to target the latent viral reservoir through disruption of the proviral genome. One in vitro study involving the CRISPR/Cas9 system demonstrated inhibition of an HIV LTR [100]. In vivo studies on the use of TALENs and CRISPR for HIV prevention and therapies are the next step to establish their future clinical application.
14. Host Restriction Factors
As a host defense, mammalian cells encode many dominant acting proteins that help suppress viral replication. These are termed host restriction factors [101]. With regard to HIV restriction, a number of such factors were identified. Prominent ones are TRIM5α, TRIMcyp, APOBEC 3F and G, SAMHD1 and tetherin, which can potentially be harnessed for HIV gene therapy. Many in vitro studies evaluated their anti-HIV effects. With regard to in vivo testing in humanized mice, early reports used a chimeric human-rhesus TRIM5α [28], which was humanized to reduce potential immunogenicity while retaining the critical rhesus macaque motif responsible for HIV restriction. HSCs were transduced with a lentiviral vector containing this construct and introduced into SCID-hu grafts. The transgenic T cells matured in these mice were found to be HIV resistant. In a different study, a human version of the monkey AoTRIM5Cyp (hT5Cyp) was transduced into human CD4 T cells and evaluated in hu-PBL mice [102]. Viral challenge experiments demonstrated decreased HIV viral loads and protection from CD4 T cell decline. Chimeric versions of TRIM5α in combination with other anti-HIV genes were also tested in vivo in humanized mice with encouraging results [34] (see below). A note of caution with the use of modified restriction factors for long-range gene therapy, however, is the potential for generating unwanted human immune responses, thus mitigating the expansion and survival of these transgenic cells in vivo.
15. Fusion Inhibitors
Clinically licensed peptide drugs such as enfuvirtide (T20) inhibit HIV entry by interfering with viral fusion with the cell membranes [103]. A fusion inhibitor C46 (derived from a 46 amino acid sequence of gp41) with a similar structure to T20 can be stably expressed as a membrane anchored peptide (maC46) in retro or lentiviral vector transduced cells thus enabling its use as a gene therapeutic construct. C46 was tested in hu-PBL mice by introducing gene transduced CD4 T cells which showed efficacy [24]. When compared with two other anti-HIV constructs, namely a tat-rev siRNA and an antisense RNA VRX494, it was found to confer higher selective survival advantage in vivo. These studies did not address potential immunogenicity of the peptide, however, which remains a concern for long term efficacy.
16. Combinatorial Approaches
Due to the ability of HIV to rapidly mutate and escape under selective pressure, the early mono-antiretroviral therapies were not fully effective during long-term use. Use of combinatorial ART (HAART) has overcome this problem to a large extent [1]. Similarly, use of a single anti-HIV gene therapeutic construct will invariably lead to generation of viral escape mutants thus requiring the design and utilization of an ideal combinatorial construct for effective viral control. Based on this premise, many studies investigated this approach using viral vectors that harbor multiple genes [5]. Ideally, a combination of anti-HIV constructs with different mechanisms of action targeted to different stages of viral life cycle is desirable. A number of different constructs that include ribozymes and siRNAs targeted to both cellular and viral RNA targets, fusion proteins and restriction factors in different combinations were evaluated, some of which have been tested in vivo in humanized mice. Among these, one study employed a triple anti-HIV gene encoding lentiviral vector harboring a TAR decoy, CCR5 ribozyme and a tat-rev siRNA [43]. HSCs transduced with this combinatorial vector gave rise to HIV resistant cells when engrafted into SCID-hu mouse thy/liv grafts, providing data for a subsequent human clinical trial. In another study, a triple combination of CCR5 siRNA, chimeric human/rhesus TRIM5a and TAR decoy was evaluated in hu-HSC mice [34]. These studies, in addition to showing anti-viral efficacy, also demonstrated no apparent toxicity with the combination of genes, although neither study directly compared the effectiveness of combination therapy versus their respective single therapies. An in vitro study, however, showed that while HIV-1 can escape from a single shRNA, this was not the case when four anti-HIV shRNAs were expressed in the same cell [104]. As can be seen, humanized mice have been instrumental for deriving important pre-clinical data.
17. Clinical Studies and Future Directions
While numerous in vitro and in vivo studies laid the groundwork by identifying a large variety of anti-HIV constructs for testing gene therapy strategies, only a few of these reached clinical trials [105] (summarized in Table 2). Most were phase I studies aimed at safety and feasibility. Previous stem cell based trials involved retroviral vectors harboring RevM10 transdominant protein [88], RRE decoy [106,107] or anti-HIV ribozymes [80,108,109,110]. No adverse effects were seen in patients receiving the treatment, thus showing safety. While there were detectable levels of transgene expressing cells, the gene marking levels were too low, however, to provide any durable clinical benefit. In a recent combinatorial approach using HSC, the triple lentiviral vector described above containing a CCR5 ribozyme, TAR decoy and a tat-rev siRNA was tested in AIDS lymphoma patients [79]. Again, levels of gene marking were low and there was no clinical benefit. Many phase I/II clinical trials also evaluated gene transduced T cells, again showing less than ideal levels of gene marking and persistence of gene modified cells [105]. Therefore, it is clear that a number of important hurdles need to be overcome to bring HIV gene therapy to a clinical reality. Among the main ones are “making space” for gene transduced HSC by myeloablation methods to allow durable engraftment, protocols to enable selection of gene modified cells, and high level gene transduction of the true and long lasting subpopulation of HSCs. In this regard, humanized mice can be exploited to evaluate new innovative experimental strategies to realize the full potential of gene therapy approaches in a clinical setting.

Table 2.
HIV Gene Therapy Clinical Trials.
| Gene therapy construct (viral or cellular target) | Proprietary name | Gene modified cells | Delivery method | Phase, status | Reference(s) |
|---|---|---|---|---|---|
| Antisense (env mRNA) | VRX496 | Autologous CD4+ T cells | Lentiviral vector | I-II, Ongoing | [111,112,113] |
| *NCT00622232 | |||||
| *NCT00295477 | |||||
| *NCT00131560 | |||||
| ZFN (CCR5 gene) | SB 728T | Autologous CD4+ T cells | Adenoviral vector | I-II, Ongoing | [114] |
| *NCT01543152 | |||||
| *NCT01252641 | |||||
| *NCT01044654 | |||||
| *NCT00842634 | |||||
| shRNA (CCR5 mRNA) Fusion inhibitor C46 (env protein) | Cal-1 | Autologous CD34+ HSCs and CD4+ T cells | Lentiviral vector | I-II, Ongoing | [115] |
| *NCT01734850 | |||||
| Fusion inhibitor C46 (env protein) | M87o | Autologous of Allogeneic CD34+ HSCs | Retroviral vector | I-II, Ongoing | [116] |
| *NCT00858793 | |||||
| Endoribonuclease (ACA sequences) | MazF-T | Autologous CD4+ T cells | Retroviral vector | I, Ongoing | *NCT01787994 |
| Transgenic TCR (gag epitope) | Autologous CD8+ T cells | Lentiviral vector | I, Ongoing | *NCT0091224 | |
| Chimeric antigen receptor (gp120 protein) | Autologous CD4+ and CD8+ T cells | Retroviral vector | I-II, Completed | [117,118,119,120] | |
| *NCT00001409 | |||||
| *NCT01013415 | |||||
| Antisense (TAR, tat/rev mRNA) | HGTV43 | Autologous CD34+ HSCs | Retroviral vector | I-II, Ongoing | [121] |
| Ribozyme (tat/vpr mRNA) | OZ1 | Autologous CD34+ HSCs | Retroviral vector | II, Ongoing | [80,122] |
| *NCT01177059 | |||||
| *NCT00074997 | |||||
| Ribozyme (tat/vpr mRNA) | Rz2 | Syngeneic CD4+ T cells | Retroviral vector | I, Completed | [108,123,124,125] |
| Ribozyme (tat/rev mRNA) | Autologous CD34+ HSCs | Retroviral vector | II, Completed | *NCT00002221 | |
| shRNA (tat/rev mRNA) TAR decoy (tat protein) Ribozyme (CCR5 mRNA) | Autologous CD34+ HSCs | Lentiviral vector | Pilot, Ongoing | [79] | |
| *NCT01153464 | |||||
| *NCT00569985 | |||||
| Ribozyme (U5/pol mRNA) | MY-2 | Autologous CD4+ T cells | Retroviral vector | I, Completed | [110] |
| RRE decoy (rev protein) | Autologous CD34+ HSCs | Retroviral vector | Pilot, Completed | [107] | |
| Transdominant rev (rev protein) | Autologous CD34+ HSCs | Retroviral vector | I, Completed | [126,127] | |
| Transdominant rev (rev protein) | Autologous CD4+ T cells | Gold particles | I, Completed | [128] | |
| Transdominant rev (rev protein) | Autologous CD4+ T cells | Retroviral vector | I, Completed | [88] | |
| Transdominant rev (rev protein) Antisense (pol mRNA) | Autologous CD34+ HSCs | Retroviral vector | I/II, Completed | *NCT00003942 |
* clinicaltrials.gov reference number
Acknowledgements
We would like to thank Jonathan LeCureux for assistance with this review and Alex Harvey for art work. Members of Akkina lab are gratefully acknowledged for their contributions toward progress in Hu-Mouse work. Research in Akkina laboratory is supported by NIH grants AI073255, AI095101, AI099783, AI100845, HL074704, HL94257 and U54AI065357.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Richman, D.D.; Margolis, D.M.; Delaney, M.; Greene, W.C.; Hazuda, D.; Pomerantz, R.J. The challenge of finding a cure for HIV infection. Science 2009, 323, 1304–1307. [Google Scholar] [CrossRef]
- Blankson, J.N.; Persaud, D.; Siliciano, R.F. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 2002, 53, 557–593. [Google Scholar] [CrossRef]
- Chun, T.W.; Fauci, A.S. Latent reservoirs of HIV: Obstacles to the eradication of virus. Proc. Natl. Acad. Sci. USA 1999, 96, 10958–10961. [Google Scholar] [CrossRef]
- Palmer, B.E.; Neff, C.P.; Lecureux, J.; Ehler, A.; Dsouza, M.; Remling-Mulder, L.; Korman, A.J.; Fontenot, A.P.; Akkina, R. In vivo blockade of the PD-1 receptor suppresses HIV-1 viral loads and improves CD4+ T cell levels in humanized mice. J. Immunol. 2013, 190, 211–219. [Google Scholar] [CrossRef]
- Kiem, H.P.; Jerome, K.R.; Deeks, S.G.; McCune, J.M. Hematopoietic-stem-cell-based gene therapy for HIV disease. Cell Stem Cell 2012, 10, 137–147. [Google Scholar] [CrossRef]
- Strayer, D.S.; Akkina, R.; Bunnell, B.A.; Dropulic, B.; Planelles, V.; Pomerantz, R.J.; Rossi, J.J.; Zaia, J.A. Current status of gene therapy strategies to treat HIV/AIDS. Mol. Ther. 2005, 11, 823–842. [Google Scholar] [CrossRef]
- Akkina, R. New generation humanized mice for virus research: Comparative aspects and future prospects. Virology 2013, 435, 14–28. [Google Scholar] [CrossRef]
- Kitchen, S.G.; Shimizu, S.; An, D.S. Stem cell-based anti-HIV gene therapy. Virology 2011, 411, 260–272. [Google Scholar] [CrossRef]
- Jamieson, B.D.; Zack, J.A. Murine models for HIV disease. AIDS 1999, 13, S5–S11. [Google Scholar]
- Mosier, D.E. Human xenograft models for virus infection. Virology 2000, 271, 215–219. [Google Scholar] [CrossRef]
- Namikawa, R.; Kaneshima, H.; Lieberman, M.; Weissman, I.L.; McCune, J.M. Infection of the SCID-hu mouse by HIV-1. Science 1988, 242, 1684–1686. [Google Scholar]
- Mosier, D.E.; Gulizia, R.J.; Baird, S.M.; Wilson, D.B. Transfer of a functional human immune system to mice with severe combined immunodeficiency. Nature 1988, 335, 256–259. [Google Scholar] [CrossRef]
- Shultz, L.D.; Ishikawa, F.; Greiner, D.L. Humanized mice in translational biomedical research. Nat. Rev. Immunol. 2007, 7, 118–130. [Google Scholar] [CrossRef]
- Ito, M.; Hiramatsu, H.; Kobayashi, K.; Suzue, K.; Kawahata, M.; Hioki, K.; Ueyama, Y.; Koyanagi, Y.; Sugamura, K.; Tsuji, K.; et al. NOD/SCID/gamma(c)(null) mouse: An excellent recipient mouse model for engraftment of human cells. Blood 2002, 100, 3175–3182. [Google Scholar] [CrossRef]
- Rongvaux, A.; Takizawa, H.; Strowig, T.; Willinger, T.; Eynon, E.E.; Flavell, R.A.; Manz, M.G. Human hemato-lymphoid system mice: Current use and future potential for medicine. Annu. Rev. Immunol. 2013, 31, 635–674. [Google Scholar] [CrossRef]
- Ishikawa, F.; Yasukawa, M.; Lyons, B.; Yoshida, S.; Miyamoto, T.; Yoshimoto, G.; Watanabe, T.; Akashi, K.; Shultz, L.D.; Harada, M. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain(null) mice. Blood 2005, 106, 1565–1573. [Google Scholar] [CrossRef]
- Berges, B.K.; Rowan, M.R. The utility of the new generation of humanized mice to study HIV-1 infection: Transmission, prevention, pathogenesis, and treatment. Retrovirology 2011, 8. [Google Scholar] [CrossRef]
- Shultz, L.D.; Brehm, M.A.; Bavari, S.; Greiner, D.L. Humanized mice as a preclinical tool for infectious disease and biomedical research. Ann. NY Acad. Sci. 2011, 1245, 50–54. [Google Scholar] [CrossRef]
- Ramer, P.C.; Chijioke, O.; Meixlsperger, S.; Leung, C.S.; Munz, C. Mice with human immune system components as in vivo models for infections with human pathogens. Immunol. Cell Biol. 2011, 89, 408–416. [Google Scholar] [CrossRef]
- Akkina, R. Human immune responses and potential for vaccine assessment in humanized mice. Curr. Opin. Immunol. 2013, 25, 403–409. [Google Scholar] [CrossRef]
- Willinger, T.; Rongvaux, A.; Strowig, T.; Manz, M.G.; Flavell, R.A. Improving human hemato-lymphoid-system mice by cytokine knock-in gene replacement. Trends Immunol. 2011, 32, 321–327. [Google Scholar] [CrossRef]
- An, D.S.; Qin, F.X.; Auyeung, V.C.; Mao, S.H.; Kung, S.K.; Baltimore, D.; Chen, I.S. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol. Ther. 2006, 14, 494–504. [Google Scholar] [CrossRef]
- Anderson, J.S.; Walker, J.; Nolta, J.A.; Bauer, G. Specific transduction of HIV-susceptible cells for CCR5 knockdown and resistance to HIV infection: A novel method for targeted gene therapy and intracellular immunization. J. Acquir. Immune Defic. Syndr. 2009, 52, 152–161. [Google Scholar] [CrossRef]
- Kimpel, J.; Braun, S.E.; Qiu, G.; Wong, F.E.; Conolle, M.; Schmitz, J.E.; Brendel, C.; Humeau, L.M.; Dropulic, B.; Rossi, J.J.; et al. Survival of the fittest: Positive selection of CD4+ T cells expressing a membrane-bound fusion inhibitor following HIV-1 infection. PLoS One 2010, 5, e12357. [Google Scholar] [CrossRef]
- Gu, Y.; Hou, W.; Xu, C.; Li, S.; Shih, J.W.; Xia, N. The enhancement of RNAi against HIV in vitro and in vivo using H-2K(k) protein as a sorting method. J. Virol. Methods 2012, 182, 9–17. [Google Scholar] [CrossRef]
- Balazs, A.B.; Chen, J.; Hong, C.M.; Rao, D.S.; Yang, L.; Baltimore, D. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 2012, 481, 81–84. [Google Scholar]
- Joseph, A.; Zheng, J.H.; Follenzi, A.; Dilorenzo, T.; Sango, K.; Hyman, J.; Chen, K.; Piechocka-Trocha, A.; Brander, C.; Hooijberg, E.; et al. Lentiviral vectors encoding human immunodeficiency virus type 1 (HIV-1)-specific T-cell receptor genes efficiently convert peripheral blood CD8 T lymphocytes into cytotoxic T lymphocytes with potent in vitro and in vivo HIV-1-specific inhibitory activity. J. Virol. 2008, 82, 3078–3089. [Google Scholar] [CrossRef]
- Anderson, J.; Akkina, R. Human immunodeficiency virus type 1 restriction by human-rhesus chimeric tripartite motif 5alpha (TRIM 5alpha) in CD34+ cell-derived macrophages in vitro and in T cells in vivo in severe combined immunodeficient (SCID-hu) mice transplanted with human fetal tissue. Hum. Gene Ther. 2008, 19, 217–228. [Google Scholar] [CrossRef]
- Maier, D.A.; Brennan, A.L.; Jiang, S.; Binder-Scholl, G.K.; Lee, G.; Plesa, G.; Zheng, Z.; Cotte, J.; Carpenito, C.; Wood, T.; et al. Efficient clinical scale gene modification via zinc finger nuclease-targeted disruption of the HIV co-receptor CCR5. Hum. Gene Ther. 2013, 24, 245–258. [Google Scholar] [CrossRef]
- Wilen, C.B.; Wang, J.; Tilton, J.C.; Miller, J.C.; Kim, K.A.; Rebar, E.J.; Sherrill-Mix, S.A.; Patro, S.C.; Secreto, A.J.; Jordan, A.P.; et al. Engineering HIV-resistant human CD4+ T cells with CXCR4-specific zinc-finger nucleases. PLoS Pathog. 2011, 7, e1002020. [Google Scholar] [CrossRef]
- Yuan, J.; Wang, J.; Crain, K.; Fearns, C.; Kim, K.A.; Hua, K.L.; Gregory, P.D.; Holmes, M.C.; Torbett, B.E. Zinc-finger nuclease editing of human cxcr4 promotes HIV-1 CD4+ T cell resistance and enrichment. Mol. Ther. 2012, 20, 849–859. [Google Scholar] [CrossRef]
- Vets, S.; Kimpel, J.; Volk, A.; de Rijck, J.; Schrijvers, R.; Verbinnen, B.; Maes, W.; von Laer, D.; Debyser, Z.; Gijsbers, R. Lens epithelium-derived growth factor/p75 qualifies as a target for HIV gene therapy in the NSG mouse model. Mol. Ther. 2012, 20, 908–917. [Google Scholar] [CrossRef]
- Kambal, A.; Mitchell, G.; Cary, W.; Gruenloh, W.; Jung, Y.; Kalomoiris, S.; Nacey, C.; McGee, J.; Lindsey, M.; Fury, B.; et al. Generation of HIV-1 resistant and functional macrophages from hematopoietic stem cell-derived induced pluripotent stem cells. Mol. Ther. 2011, 19, 584–593. [Google Scholar] [CrossRef]
- Walker, J.E.; Chen, R.X.; McGee, J.; Nacey, C.; Pollard, R.B.; Abedi, M.; Bauer, G.; Nolta, J.A.; Anderson, J.S. Generation of an HIV-1-resistant immune system with CD34+ hematopoietic stem cells transduced with a triple-combination anti-HIV lentiviral vector. J. Virol. 2012, 86, 5719–5729. [Google Scholar] [CrossRef]
- Centlivre, M.; Legrand, N.; Klamer, S.; Liu, Y.P.; Eije, K.J.; Bohne, M.; Rijnstra, E.S.; Weijer, K.; Blom, B.; Voermans, C.; et al. Preclinical in vivo evaluation of the safety of a multi-shRNA-based gene therapy against HIV-1. Mol. Ther. Nucleic Acids 2013, 2, e120. [Google Scholar] [CrossRef]
- Kiem, H.P.; Wu, R.A.; Sun, G.; von Laer, D.; Rossi, J.J.; Trobridge, G.D. Foamy combinatorial anti-HIV vectors with MGMTP140K potently inhibit HIV-1 and SHIV replication and mediate selection in vivo. Gene Ther. 2010, 17, 37–49. [Google Scholar] [CrossRef]
- Ter Brake, O.; Legrand, N.; von Eije, K.J.; Centlivre, M.; Spits, H.; Weijer, K.; Blom, B.; Berkhout, B. Evaluation of safety and efficacy of RNAi against HIV-1 in the human immune system (Rag-2−/−gammac−/−) mouse model. Gene Ther. 2009, 16, 148–153. [Google Scholar] [CrossRef]
- Santat, L.; Paz, H.; Wong, C.; Li, L.; Macer, J.; Forman, S.; Wong, K.K.; Chatterjee, S. Recombinant AAV2 transduction of primitive human hematopoietic stem cells capable of serial engraftment in immune-deficient mice. Proc. Natl. Acad. Sci. USA 2005, 102, 11053–11058. [Google Scholar] [CrossRef]
- Davis, B.M.; Humeau, L.; Dropulic, B. In vivo selection for human and murine hematopoietic cells transduced with a therapeutic MGMT lentiviral vector that inhibits HIV replication. Mol. Ther. 2004, 9, 160–172. [Google Scholar] [CrossRef]
- Joseph, A.; Zheng, J.H.; Chen, K.; Dutta, M.; Chen, C.; Stiegler, G.; Kunert, R.; Follenzi, A.; Goldstein, H. Inhibition of in vivo HIV infection in humanized mice by gene therapy of human hematopoietic stem cells with a lentiviral vector encoding a broadly neutralizing anti-HIV antibody. J. Virol. 2010, 84, 6645–6653. [Google Scholar] [CrossRef]
- Holt, N.; Wang, J.; Kim, K.; Friedman, G.; Wang, X.; Taupin, V.; Crooks, G.M.; Kohn, D.B.; Gregory, P.D.; Holmes, M.C.; et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 2010, 28, 839–847. [Google Scholar] [CrossRef]
- Goldstein, H.; Pettoello-Mantovani, M.; Anderson, C.M.; Cordelier, P.; Pomerantz, R.J.; Strayer, D.S. Gene therapy using a simian virus 40-derived vector inhibits the development of in vivo human immunodeficiency virus type 1 infection of severe combined immunodeficiency mice implanted with human fetal thymic and liver tissue. J. Infect. Dis. 2002, 185, 1425–1430. [Google Scholar] [CrossRef]
- Anderson, J.; Li, M.J.; Palmer, B.; Remling, L.; Li, S.; Yam, P.; Yee, J.K.; Rossi, J.; Zaia, J.; Akkina, R. Safety and efficacy of a lentiviral vector containing three anti-HIV genes—CCR5 ribozyme, tat-rev siRNA, and TAR decoy—In SCID-hu mouse-derived T cells. Mol. Ther. 2007, 15, 1182–1188. [Google Scholar]
- Banerjea, A.; Li, M.J.; Bauer, G.; Remling, L.; Lee, N.S.; Rossi, J.; Akkina, R. Inhibition of HIV-1 by lentiviral vector-transduced siRNAs in T lymphocytes differentiated in SCID-hu mice and CD34+ progenitor cell-derived macrophages. Mol. Ther. 2003, 8, 62–71. [Google Scholar] [CrossRef]
- Banerjea, A.; Li, M.J.; Remling, L.; Rossi, J.; Akkina, R. Lentiviral transduction of Tar Decoy and CCR5 ribozyme into CD34+ progenitor cells and derivation of HIV-1 resistant T cells and macrophages. AIDS Res. Ther. 2004, 1. [Google Scholar] [CrossRef]
- Bai, J.; Gorantla, S.; Banda, N.; Cagnon, L.; Rossi, J.; Akkina, R. Characterization of anti-CCR5 ribozyme-transduced CD34+ hematopoietic progenitor cells in vitro and in a SCID-hu mouse model in vivo. Mol. Ther. 2000, 1, 244–254. [Google Scholar] [CrossRef]
- Bai, J.; Banda, N.; Lee, N.S.; Rossi, J.; Akkina, R. RNA-based anti-HIV-1 gene therapeutic constructs in SCID-hu mouse model. Mol. Ther. 2002, 6, 770–782. [Google Scholar] [CrossRef]
- Swan, C.H.; Buhler, B.; Steinberger, P.; Tschan, M.P.; Barbas, C.F., 3rd; Torbett, B.E. T-cell protection and enrichment through lentiviral CCR5 intrabody gene delivery. Gene Ther. 2006, 13, 1480–1492. [Google Scholar] [CrossRef]
- Kitchen, S.G.; Bennett, M.; Galic, Z.; Kim, J.; Xu, Q.; Young, A.; Lieberman, A.; Joseph, A.; Goldstein, H.; Ng, H.; et al. Engineering antigen-specific T cells from genetically modified human hematopoietic stem cells in immunodeficient mice. PLoS One 2009, 4, e8208. [Google Scholar] [CrossRef]
- Bonyhadi, M.L.; Moss, K.; Voytovich, A.; Auten, J.; Kalfoglou, C.; Plavec, I.; Forestell, S.; Su, L.; Bohnlein, E.; Kaneshima, H. RevM10-expressing T cells derived in vivo from transduced human hematopoietic stem-progenitor cells inhibit human immunodeficiency virus replication. J. Virol. 1997, 71, 4707–4716. [Google Scholar]
- Shimizu, S.; Hong, P.; Arumugam, B.; Pokomo, L.; Boyer, J.; Koizumi, N.; Kittipongdaja, P.; Chen, A.; Bristol, G.; Galic, Z.; et al. A highly efficient short hairpin RNA potently down-regulates CCR5 expression in systemic lymphoid organs in the hu-BLT mouse model. Blood 2010, 115, 1534–1544. [Google Scholar] [CrossRef]
- Ringpis, G.E.; Shimizu, S.; Arokium, H.; Camba-Colon, J.; Carroll, M.V.; Cortado, R.; Xie, Y.; Kim, P.Y.; Sahakyan, A.; Lowe, E.L.; et al. Engineering HIV-1-resistant T-cells from short-hairpin RNA-expressing hematopoietic stem/progenitor cells in humanized BLT mice. PLoS One 2012, 7, e53492. [Google Scholar]
- Kitchen, S.G.; Levin, B.R.; Bristol, G.; Rezek, V.; Kim, S.; Aguilera-Sandoval, C.; Balamurugan, A.; Yang, O.O.; Zack, J.A. In vivo suppression of HIV by antigen specific T cells derived from engineered hematopoietic stem cells. PLoS Pathog. 2012, 8, e1002649. [Google Scholar] [CrossRef]
- Hur, E.M.; Patel, S.N.; Shimizu, S.; Rao, D.S.; Gnanapragasam, P.N.; An, D.S.; Yang, L.; Baltimore, D. Inhibitory effect of HIV-specific neutralizing IgA on mucosal transmission of HIV in humanized mice. Blood 2012, 120, 4571–4582. [Google Scholar] [CrossRef]
- McCune, J.M.; Namikawa, R.; Kaneshima, H.; Shultz, L.D.; Lieberman, M.; Weissman, I.L. The SCID-hu mouse: Murine model for the analysis of human hematolymphoid differentiation and function. Science 1988, 241, 1632–1639. [Google Scholar]
- Berges, B.K.; Wheat, W.H.; Palmer, B.E.; Connick, E.; Akkina, R. HIV-1 infection and CD4 T cell depletion in the humanized Rag2−/−gamma c−/− (RAG-hu) mouse model. Retrovirology 2006, 3. [Google Scholar] [CrossRef]
- Chang, H.; Biswas, S.; Tallarico, A.S.; Sarkis, P.T.; Geng, S.; Panditrao, M.M.; Zhu, Q.; Marasco, W.A. Human B-cell ontogeny in humanized NOD/SCID gammac(null) mice generates a diverse yet auto/poly- and HIV-1-reactive antibody repertoire. Genes Immun. 2012, 13, 399–410. [Google Scholar] [CrossRef]
- Strowig, T.; Gurer, C.; Ploss, A.; Liu, Y.F.; Arrey, F.; Sashihara, J.; Koo, G.; Rice, C.M.; Young, J.W.; Chadburn, A.; et al. Priming of protective T cell responses against virus-induced tumors in mice with human immune system components. J. Exp. Med. 2009, 206, 1423–1434. [Google Scholar] [CrossRef]
- Berges, B.K.; Akkina, S.R.; Folkvord, J.M.; Connick, E.; Akkina, R. Mucosal transmission of R5 and X4 tropic HIV-1 via vaginal and rectal routes in humanized Rag2−/− gammac−/− (RAG-hu) mice. Virology 2008, 373, 342–351. [Google Scholar] [CrossRef]
- Veselinovic, M.; Neff, C.P.; Mulder, L.R.; Akkina, R. Topical gel formulation of broadly neutralizing anti-HIV-1 monoclonal antibody VRC01 confers protection against HIV-1 vaginal challenge in a humanized mouse model. Virology 2012, 432, 505–510. [Google Scholar] [CrossRef]
- Lan, P.; Tonomura, N.; Shimizu, A.; Wang, S.; Yang, Y.G. Reconstitution of a functional human immune system in immunodeficient mice through combined human fetal thymus/liver and CD34+ cell transplantation. Blood 2006, 108, 487–492. [Google Scholar] [CrossRef]
- Melkus, M.W.; Estes, J.D.; Padgett-Thomas, A.; Gatlin, J.; Denton, P.W.; Othieno, F.A.; Wege, A.K.; Haase, A.T.; Garcia, J.V. Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat. Med. 2006, 12, 1316–1322. [Google Scholar] [CrossRef]
- Denton, P.W.; Garcia, J.V. Mucosal HIV-1 transmission and prevention strategies in BLT humanized mice. Trends Microbiol. 2012, 20, 268–274. [Google Scholar] [CrossRef]
- Swanstrom, R.; Coffin, J. HIV-1 pathogenesis: The virus. Cold Spring Harb. Perspect. Med. 2012, 2. [Google Scholar] [CrossRef]
- Castanotto, D.; Rossi, J.J. The promises and pitfalls of RNA-interference-based therapeutics. Nature 2009, 457, 426–433. [Google Scholar] [CrossRef]
- Turner, A.M.; Ackley, A.M.; Matrone, M.A.; Morris, K.V. Characterization of an HIV-targeted transcriptional gene-silencing RNA in primary cells. Hum. Gene Ther. 2012, 23, 473–483. [Google Scholar] [CrossRef]
- Pratt, A.J.; MacRae, I.J. The RNA-induced silencing complex: A versatile gene-silencing machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef]
- Burnett, J.C.; Rossi, J.J. RNA-based therapeutics: Current progress and future prospects. Chem. Biol. 2012, 19, 60–71. [Google Scholar] [CrossRef]
- Thiel, K.W.; Giangrande, P.H. Intracellular delivery of RNA-based therapeutics using aptamers. Ther. Deliv. 2010, 1, 849–861. [Google Scholar] [CrossRef]
- Zhou, J.; Neff, C.P.; Liu, X.; Zhang, J.; Li, H.; Smith, D.D.; Swiderski, P.; Aboellail, T.; Huang, Y.; Du, Q.; et al. Systemic administration of combinatorial dsiRNAs via nanoparticles efficiently suppresses HIV-1 infection in humanized mice. Mol. Ther. 2011, 19, 2228–2238. [Google Scholar] [CrossRef]
- Kumar, P.; Ban, H.S.; Kim, S.S.; Wu, H.; Pearson, T.; Greiner, D.L.; Laouar, A.; Yao, J.; Haridas, V.; Habiro, K.; et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell 2008, 134, 577–586. [Google Scholar] [CrossRef]
- Neff, C.P.; Zhou, J.; Remling, L.; Kuruvilla, J.; Zhang, J.; Li, H.; Smith, D.D.; Swiderski, P.; Rossi, J.J.; Akkina, R. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4+ T cell decline in humanized mice. Sci. Transl. Med. 2011, 3. [Google Scholar] [CrossRef]
- Peer, D.; Lieberman, J. Special delivery: Targeted therapy with small RNAs. Gene Ther. 2011, 18, 1127–1133. [Google Scholar] [CrossRef]
- Zhou, J.; Neff, C.P.; Swiderski, P.; Li, H.; Smith, D.D.; Aboellail, T.; Remling-Mulder, L.; Akkina, R.; Rossi, J.J. Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a chemically synthesized aptamer with a sticky bridge. Mol. Ther. 2013, 21, 192–200. [Google Scholar] [CrossRef]
- Westerhout, E.M.; Ooms, M.; Vink, M.; Das, A.T.; Berkhout, B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005, 33, 796–804. [Google Scholar] [CrossRef]
- Allers, K.; Hutter, G.; Hofmann, J.; Loddenkemper, C.; Rieger, K.; Thiel, E.; Schneider, T. Evidence for the cure of HIV infection by CCR5Delta32/Delta32 stem cell transplantation. Blood 2011, 117, 2791–2799. [Google Scholar] [CrossRef]
- Hutter, G.; Nowak, D.; Mossner, M.; Ganepola, S.; Mussig, A.; Allers, K.; Schneider, T.; Hofmann, J.; Kucherer, C.; Blau, O.; et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 2009, 360, 692–698. [Google Scholar] [CrossRef]
- Rossi, J.J. Ribozyme therapy for HIV infection. Adv. Drug Deliv. Rev. 2000, 44, 71–78. [Google Scholar] [CrossRef]
- DiGiusto, D.L.; Krishnan, A.; Li, L.; Li, H.; Li, S.; Rao, A.; Mi, S.; Yam, P.; Stinson, S.; Kalos, M.; et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34+ cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2010, 2. [Google Scholar] [CrossRef]
- Mitsuyasu, R.T.; Merigan, T.C.; Carr, A.; Zack, J.A.; Winters, M.A.; Workman, C.; Bloch, M.; Lalezari, J.; Becker, S.; Thornton, L.; et al. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat. Med. 2009, 15, 285–292. [Google Scholar] [CrossRef]
- Zhou, J.; Bobbin, M.L.; Burnett, J.C.; Rossi, J.J. Current progress of RNA aptamer-based therapeutics. Front. Genet. 2012, 3. [Google Scholar] [CrossRef]
- Burton, D.R.; Poignard, P.; Stanfield, R.L.; Wilson, I.A. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 2012, 337, 183–186. [Google Scholar] [CrossRef]
- Luo, X.M.; Lei, M.Y.; Feidi, R.A.; West, A.P., Jr.; Balazs, A.B.; Bjorkman, P.J.; Yang, L.; Baltimore, D. Dimeric 2G12 as a potent protection against HIV-1. PLoS Pathog. 2010, 6, e1001225. [Google Scholar] [CrossRef]
- Bennett, M.S.; Joseph, A.; Ng, H.L.; Goldstein, H.; Yang, O.O. Fine-tuning of T-cell receptor avidity to increase HIV epitope variant recognition by cytotoxic T lymphocytes. AIDS 2010, 24, 2619–2628. [Google Scholar] [CrossRef]
- Leslie, A.J.; Pfafferott, K.J.; Chetty, P.; Draenert, R.; Addo, M.M.; Feeney, M.; Tang, Y.; Holmes, E.C.; Allen, T.; Prado, J.G.; et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 2004, 10, 282–289. [Google Scholar] [CrossRef]
- Morgan, R.A.; Walker, R.; Carter, C.S.; Natarajan, V.; Tavel, J.A.; Bechtel, C.; Herpin, B.; Muul, L.; Zheng, Z.; Jagannatha, S.; et al. Preferential survival of CD4+ T lymphocytes engineered with anti-human immunodeficiency virus (HIV) genes in HIV-infected individuals. Hum. Gene Ther. 2005, 16, 1065–1074. [Google Scholar] [CrossRef]
- Podsakoff, G.M.; Engel, B.C.; Carbonaro, D.A.; Choi, C.; Smogorzewska, E.M.; Bauer, G.; Selander, D.; Csik, S.; Wilson, K.; Betts, M.R.; et al. Selective survival of peripheral blood lymphocytes in children with HIV-1 following delivery of an anti-HIV gene to bone marrow CD34+ cells. Mol. Ther. 2005, 12, 77–86. [Google Scholar] [CrossRef]
- Ranga, U.; Woffendin, C.; Verma, S.; Xu, L.; June, C.H.; Bishop, D.K.; Nabel, G.J. Enhanced T cell engraftment after retroviral delivery of an antiviral gene in HIV-infected individuals. Proc. Natl. Acad. Sci. USA 1998, 95, 1201–1206. [Google Scholar] [CrossRef]
- Cherepanov, P.; Maertens, G.; Proost, P.; Devreese, B.; van Beeumen, J.; Engelborghs, Y.; de Clercq, E.; Debyser, Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003, 278, 372–381. [Google Scholar]
- Jao, L.E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef]
- Urnov, F.D.; Miller, J.C.; Lee, Y.L.; Beausejour, C.M.; Rock, J.M.; Augustus, S.; Jamieson, A.C.; Porteus, M.H.; Gregory, P.D.; Holmes, M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 2005, 435, 646–651. [Google Scholar] [CrossRef]
- Schiffer, J.T.; Aubert, M.; Weber, N.D.; Mintzer, E.; Stone, D.; Jerome, K.R. Targeted DNA mutagenesis for the cure of chronic viral infections. J. Virol. 2012, 86, 8920–8936. [Google Scholar] [CrossRef]
- Perez, E.E.; Wang, J.; Miller, J.C.; Jouvenot, Y.; Kim, K.A.; Liu, O.; Wang, N.; Lee, G.; Bartsevich, V.V.; Lee, Y.L.; et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 808–816. [Google Scholar] [CrossRef]
- Pattanayak, V.; Ramirez, C.L.; Joung, J.K.; Liu, D.R. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods 2011, 8, 765–770. [Google Scholar] [CrossRef]
- Bogdanove, A.J.; Schornack, S.; Lahaye, T. TAL effectors: Finding plant genes for disease and defense. Curr. Opin. Plant Biol. 2010, 13, 394–401. [Google Scholar] [CrossRef]
- Wiedenheft, B.; Sternberg, S.H.; Doudna, J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012, 482, 331–338. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef]
- Ebina, H.; Misawa, N.; Kanemura, Y.; Koyanagi, Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef]
- Malim, M.H.; Bieniasz, P.D. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harb. Perspect. Med. 2012, 2. [Google Scholar] [CrossRef]
- Neagu, M.R.; Ziegler, P.; Pertel, T.; Strambio-De-Castillia, C.; Grutter, C.; Martinetti, G.; Mazzucchelli, L.; Grutter, M.; Manz, M.G.; Luban, J. Potent inhibition of HIV-1 by TRIM5-cyclophilin fusion proteins engineered from human components. J. Clin. Invest. 2009, 119, 3035–3047. [Google Scholar] [CrossRef]
- Kuritzkes, D.R. HIV-1 entry inhibitors: An overview. Curr. Opin. HIV AIDS 2009, 4, 82–87. [Google Scholar] [CrossRef]
- Ter Brake, O.; Hooft, K.; Liu, Y.P.; Centlivre, M.; von Eije, K.J.; Berkhout, B. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol. Ther. 2008, 16, 557–564. [Google Scholar] [CrossRef]
- Hoxie, J.A.; June, C.H. Novel cell and gene therapies for HIV. Cold Spring Harb. Perspect. Med. 2012, 2. [Google Scholar] [CrossRef]
- Bauer, G.; Selander, D.; Engel, B.; Carbonaro, D.; Csik, S.; Rawlings, S.; Church, J.; Kohn, D.B. Gene therapy for pediatric AIDS. Ann. NY Acad. Sci. 2000, 918, 318–329. [Google Scholar]
- Kohn, D.B.; Bauer, G.; Rice, C.R.; Rothschild, J.C.; Carbonaro, D.A.; Valdez, P.; Hao, Q.; Zhou, C.; Bahner, I.; Kearns, K.; et al. A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34+ cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood 1999, 94, 368–371. [Google Scholar]
- Amado, R.G.; Mitsuyasu, R.T.; Rosenblatt, J.D.; Ngok, F.K.; Bakker, A.; Cole, S.; Chorn, N.; Lin, L.S.; Bristol, G.; Boyd, M.P.; et al. Anti-human immunodeficiency virus hematopoietic progenitor cell-delivered ribozyme in a phase I study: Myeloid and lymphoid reconstitution in human immunodeficiency virus type-1-infected patients. Hum. Gene Ther. 2004, 15, 251–262. [Google Scholar] [CrossRef]
- Michienzi, A.; Castanotto, D.; Lee, N.; Li, S.; Zaia, J.A.; Rossi, J.J. RNA-mediated inhibition of HIV in a gene therapy setting. Ann. NY Acad. Sci. 2003, 1002, 63–71. [Google Scholar]
- Wong-Staal, F.; Poeschla, E.M.; Looney, D.J. A controlled, Phase 1 clinical trial to evaluate the safety and effects in HIV-1 infected humans of autologous lymphocytes transduced with a ribozyme that cleaves HIV-1 RNA. Hum. Gene Ther. 1998, 9, 2407–2425. [Google Scholar] [CrossRef]
- Levine, B.L.; Humeau, L.M.; Boyer, J.; MacGregor, R.R.; Rebello, T.; Lu, X.; Binder, G.K.; Slepushkin, V.; Lemiale, F.; Mascola, J.R.; et al. Gene transfer in humans using a conditionally replicating lentiviral vector. Proc. Natl. Acad. Sci. USA 2006, 103, 17372–17377. [Google Scholar] [CrossRef]
- McGarrity, G.J.; Hoyah, G.; Winemiller, A.; Andre, K.; Stein, D.; Blick, G.; Greenberg, R.N.; Kinder, C.; Zolopa, A.; Binder-Scholl, G.; et al. Patient monitoring and follow-up in lentiviral clinical trials. J. Gene Med. 2013, 15, 78–82. [Google Scholar] [CrossRef]
- Tebas, P.; Stein, D.; Binder-Scholl, G.; Mukherjee, R.; Brady, T.; Rebello, T.; Humeau, L.; Kalos, M.; Papasavvas, E.; Montaner, L.J.; et al. Antiviral effects of autologous CD4 T cells genetically modified with a conditionally replicating lentiviral vector expressing long antisense to HIV. Blood 2013, 121, 1524–1533. [Google Scholar] [CrossRef]
- Lee, G.K.; Zeidan, J.; Lalezari, J.; Mitsuyasu, R.; Wang, S.; Giedlin, M.; Nichol, G.; Tang, W.S.; Ando, D.; Sekaly, R.P. Long term CD4 reconstitution in HIV subjects receiving ZFN CCR5 modified CD4 T-cells (SB-728-T) may be attributed to the sustained durability of the central memory T-cell subset. In Proceedings of the 16th Annual Meeting of the American Society of Gene and Cell Therapy, Salt Lake City, UT, USA, 15–18 May 2013. Abstract No. 58.
- Burke, B.P.; Boyd, M.P.; Millington, M.L.; Impey, H.; Zhang, J.; Carroll, M.V.; Camba-Colon, J.; Keech, N.; Delebecque, F.; Wolstein, O.; et al. Engineering resistance to HIV-1 infection with a dual therapeutic lentiviral vector. In Proceedings of the 16th Annual Meeting of the American Society of Gene and Cell Therapy, Salt Lake City, UT, USA, 15–18 May 2013. Abstract No. 48.
- Van Lunzen, J.; Glaunsinger, T.; Stahmer, I.; von Baehr, V.; Baum, C.; Schilz, A.; Kuehlcke, K.; Naundorf, S.; Martinius, H.; Hermann, F.; et al. Transfer of autologous gene-modified T cells in HIV-infected patients with advanced immunodeficiency and drug-resistant virus. Mol. Ther. 2007, 15, 1024–1033. [Google Scholar]
- Deeks, S.G.; Wagner, B.; Anton, P.A.; Mitsuyasu, R.T.; Scadden, D.T.; Huang, C.; Macken, C.; Richman, D.D.; Christopherson, C.; June, C.H.; et al. A phase II randomized study of HIV-specific T-cell gene therapy in subjects with undetectable plasma viremia on combination antiretroviral therapy. Mol. Ther. 2002, 5, 788–797. [Google Scholar] [CrossRef]
- Mitsuyasu, R.T.; Anton, P.A.; Deeks, S.G.; Scadden, D.T.; Connick, E.; Downs, M.T.; Bakker, A.; Roberts, M.R.; June, C.H.; Jalali, S.; et al. Prolonged survival and tissue trafficking following adoptive transfer of CD4zeta gene-modified autologous CD4+ and CD8+ T cells in human immunodeficiency virus-infected subjects. Blood 2000, 96, 785–793. [Google Scholar]
- Scholler, J.; Brady, T.L.; Binder-Scholl, G.; Hwang, W.T.; Plesa, G.; Hege, K.M.; Vogel, A.N.; Kalos, M.; Riley, J.L.; Deeks, S.G.; et al. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci. Transl. Med. 2012, 4. [Google Scholar] [CrossRef]
- Walker, R.E.; Bechtel, C.M.; Natarajan, V.; Baseler, M.; Hege, K.M.; Metcalf, J.A.; Stevens, R.; Hazen, A.; Blaese, R.M.; Chen, C.C.; et al. Long-term in vivo survival of receptor-modified syngeneic T cells in patients with human immunodeficiency virus infection. Blood 2000, 96, 467–474. [Google Scholar]
- Liu, D.; Conant, M.A.; Cowan, M.J.; Laurence, J.; Eden, C.; Dunn, E.; Thalenfeld, B.E.; Engelhardt, D.L. Engraftment and development of HGTV43-transduced CD34+ PBSC in HIV-1 seropositive individuals. In Proceedings of the XIV International AIDS Conference, Barcelona, Spain, 7–12 July 2002. Abstract No. A10139.
- Savkovic, B.; Macpherson, J.L.; Zaunders, J.; Kelleher, A.D.; Knop, A.E.; Pond, S.; Evans, L.; Symonds, G.; Murray, J.M. T-lymphocyte perturbation following large-scale apheresis and hematopoietic stem cell transplantation in HIV-infected individuals. Clin. Immunol. 2012, 144, 159–171. [Google Scholar] [CrossRef]
- Amado, R.G.; Mitsuyasu, R.T.; Symonds, G.; Rosenblatt, J.D.; Zack, J.; Sun, L.Q.; Miller, M.; Ely, J.; Gerlach, W. A phase I trial of autologous CD34+ hematopoietic progenitor cells transduced with an anti-HIV ribozyme. Hum. Gene Ther. 1999, 10, 2255–2270. [Google Scholar] [CrossRef]
- Cooper, D.; Penny, R.; Symonds, G.; Carr, A.; Gerlach, W.; Sun, L.Q.; Ely, J. A marker study of therapeutically transduced CD4+ peripheral blood lymphocytes in HIV discordant identical twins. Hum. Gene Ther. 1999, 10, 1401–1421. [Google Scholar] [CrossRef]
- Macpherson, J.L.; Boyd, M.P.; Arndt, A.J.; Todd, A.V.; Fanning, G.C.; Ely, J.A.; Elliott, F.; Knop, A.; Raponi, M.; Murray, J.; et al. Long-term survival and concomitant gene expression of ribozyme-transduced CD4+ T-lymphocytes in HIV-infected patients. J. Gene Med. 2005, 7, 552–564. [Google Scholar] [CrossRef]
- Hayakawa, J.; Washington, K.; Uchida, N.; Phang, O.; Kang, E.M.; Hsieh, M.M.; Tisdale, J.F. Long-term vector integration site analysis following retroviral mediated gene transfer to hematopoietic stem cells for the treatment of HIV infection. PLoS One 2009, 4, e4211. [Google Scholar]
- Kang, E.M.; de Witte, M.; Malech, H.; Morgan, R.A.; Phang, S.; Carter, C.; Leitman, S.F.; Childs, R.; Barrett, A.J.; Little, R.; et al. Nonmyeloablative conditioning followed by transplantation of genetically modified HLA-matched peripheral blood progenitor cells for hematologic malignancies in patients with acquired immunodeficiency syndrome. Blood 2002, 99, 698–701. [Google Scholar] [CrossRef]
- Woffendin, C.; Ranga, U.; Yang, Z.; Xu, L.; Nabel, G.J. Expression of a protective gene-prolongs survival of T cells in human immunodeficiency virus-infected patients. Proc. Natl. Acad. Sci. USA 1996, 93, 2889–2894. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).