The Lymantria dispar IPLB-Ld652Y Cell Line Transcriptome Comprises Diverse Virus-Associated Transcripts
Abstract
:1. Introduction
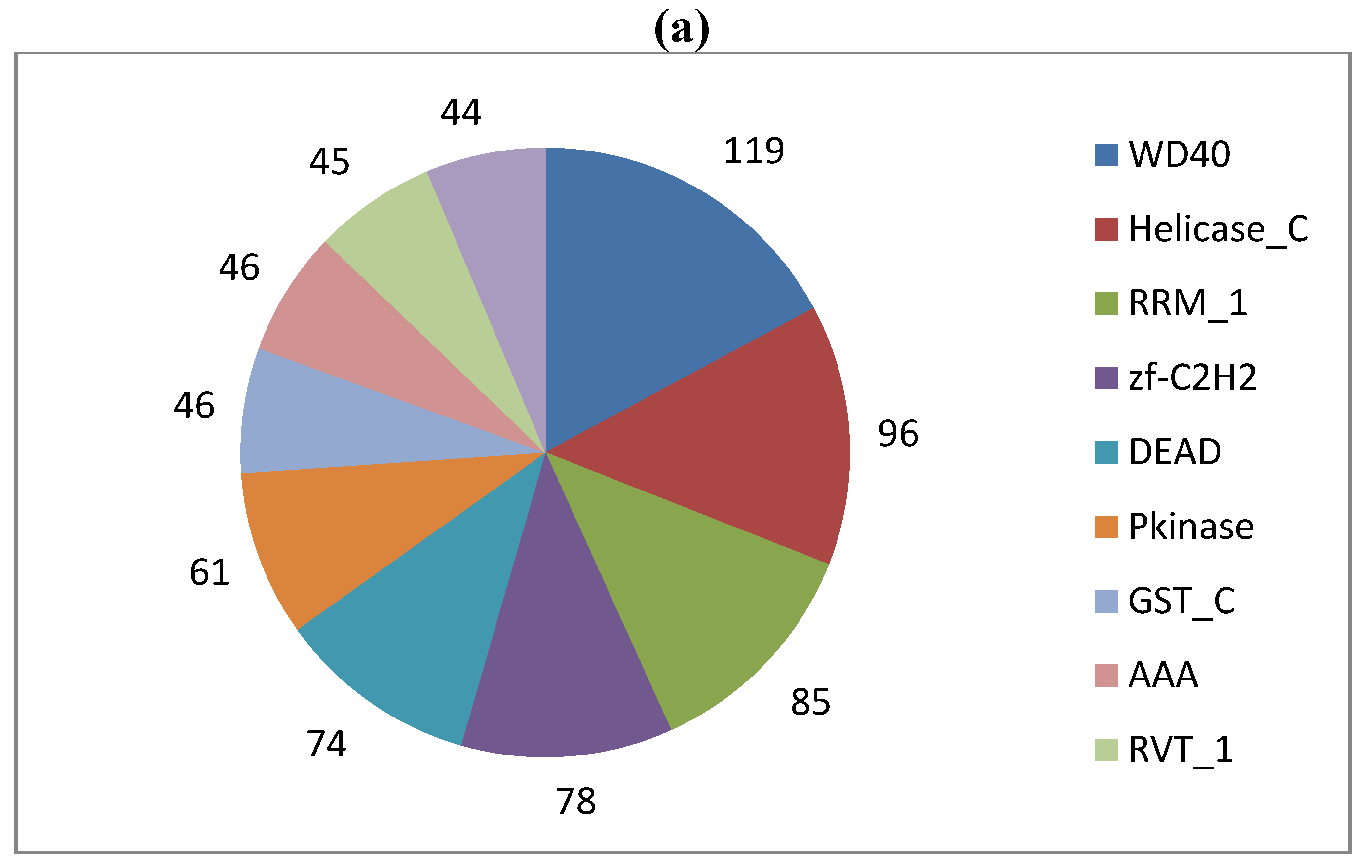
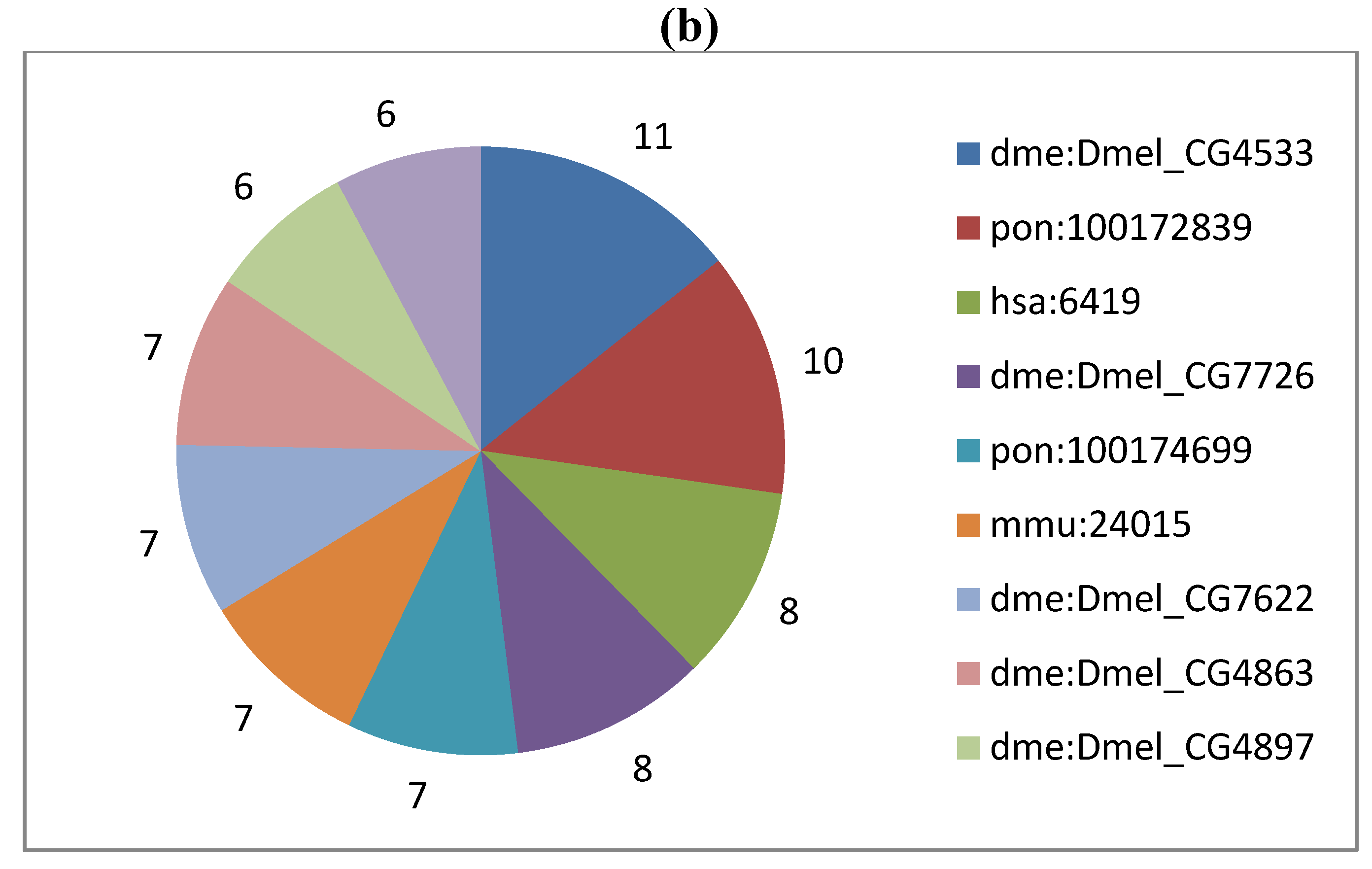
2. Results and Discussion
3. Experimental Section
3.1. EST Sequence Processing
3.2. 454 Sequence Processing
3.3. Derivation of Putative Unique Transcripts (PUTs)
3.4. Gene Content Survey and Functional Analysis
4. Conclusions
Acknowledgments
Conflict of Interest
References and Notes
- Goodwin, R.H.; Tompkins, G.J.; McCawley, P. Gypsy moth cell lines divergent in viral susceptibility. I. Culture and identification. Vitro Cell. Dev. Biol. Plant 1978, 14, 485–494. [Google Scholar] [CrossRef]
- Guzo, D.; Dougherty, E.M.; Lynn, D.E.; Braun, S.K.; Weiner, R.M. Changes in macromolecular synthesis of gypsy moth cell line IPLB-Ld652Y induced by Autographa californica nuclear polyhedrosis virus infection. J. Gen. Virol. 1991, 72, 1021–1029. [Google Scholar] [CrossRef]
- Mazzacano, C.A.; Du, X.; Thiem, S.M. Global protein synthesis shutdown in Autographa californica nucleopolyhedrovirus-infected Ld652Y cells is rescued by tRNA from uninfected cells. Virology 1999, 260, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Ikeda, M.; Yanagimoto, K.; Alves, C.A.; Katou, Y.; Laviña-Caoili, B.A.; Kobayashi, M. Induction of apoptosis in an insect cell line, IPLB-Ld652Y, infected with nucleopolyhedroviruses. J. Gen. Virol. 2003, 84, 705–714. [Google Scholar] [CrossRef] [PubMed]
- McNeil, J.; Cox-Foster, D.; Gardner, M.; Slavicek, J.; Thiem, S.; Hoover, K. Pathogenesis of Lymantria dispar multiple nucleopolyhedrovirus in L. dispar and mechanisms of developmental resistance. J. Gen. Virol. 2010, 91, 1590–1600. [Google Scholar] [CrossRef] [PubMed]
- McNeil, J.; Cox-Foster, D.; Slavicek, J.; Hoover, K. Contributions of immune responses to developmental resistance in Lymantria dispar challenged with baculovirus. J. Insect Physiol. 2010, 56, 1167–1177. [Google Scholar] [CrossRef]
- Yamada, H.; Shibuya, M.; Kobayashi, M.; Ikeda, M. Identification of a novel apoptosis suppressor gene from the baculovirus Lymantria dispar multicapsid nucleopolyhedrovirus. J. Virol. 2011, 85, 5237–5242. [Google Scholar] [CrossRef]
- Lu, H.; Burand, J.P. Replication of the gonad-specific virus Hz-2V in Ld652Y cells mimics replication in vivo. J. Invertebr. Pathol. 2001, 77, 44–50. [Google Scholar] [CrossRef]
- Boguski, M.S.; Tolstoshev, C.M.; Bassett, D.E. Gene discovery in dbEST. Science 1994, 265, 1993–1994. [Google Scholar] [CrossRef]
- Leinonen, R.; Sugawara, H.; Shumway, M. The sequence read archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef]
- Landais, I.; Ogliastro, M.; Mita, K.; Nohata, J.; López-Ferber, M.; Duonor-Cérutti, M.; Shimada, T.; Fournier, P.; Devauchelle, G. Annotation pattern of ESTs from Spodoptera frugiperda Sf9 cells and analysis of the ribosomal protein genes reveal insect-specific features and unexpectedly low codon usage bias. Bioinformatics 2003, 19, 2343–2350. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Dong, Y.; Thodima, V.; Clem, R.J.; Passarelli, A.L. Analysis and functional annotation of expressed sequence tags from the fall armyworm Spodoptera frugiperda. BMC Genomics 2006, 7, 264. [Google Scholar] [CrossRef] [PubMed]
- Okano, K.; Shimada, T.; Mita, K.; Maeda, S. Comparative expressed-sequence-tag analysis of differential gene expression profiles in BmNPV-infected BmN cells. Virology 2001, 282, 348–356. [Google Scholar] [CrossRef]
- Braaten, D.; Franke, E.K.; Luban, J. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIV(CPZ)GAB but not group O HIV-1 or other primate immunodeficiency viruses. J. Virol. 1996, 70, 4220–4227. [Google Scholar] [CrossRef] [PubMed]
- Sherry, B.; Zybarth, G.; Alfano, M.; Dubrovsky, L.; Mitchell, R.; Rich, D.; Ulrich, P.; Bucala, R.; Cerami, A.; Bukrinsky, M. Role of cyclophilin A in the uptake of HIV-1 by macrophages and T lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 1758–1763. [Google Scholar] [CrossRef]
- Kaneko, Y.; Takaki, K.; Iwami, M.; Sakurai, S. Developmental profile of annexin IX and its possible role in programmed cell death of the Bombyx mori anterior silk gland. Zool. Sci. 2006, 23, 533–542. [Google Scholar] [CrossRef]
- Thiem, S.M.; Chejanovsky, N. The role of baculovirus apoptotic suppressors in AcMNPV-mediated translation arrest in Ld652Y cells. Virology 2004, 319, 292–305. [Google Scholar] [CrossRef]
- Clem, R.J. Baculoviruses and apoptosis: A diversity of genes and responses. Curr. Drug Targets 2007, 8, 1069–1074. [Google Scholar] [CrossRef]
- Bao, Y.-Y.; Tang, X.D.; Lv, Z.Y.; Wang, X.Y.; Tian, C.H.; Xu, Y.P.; Zhang, C.X. Gene expression profiling of resistant and susceptible Bombyx mori strains reveals nucleopolyhedrovirus-associated variations in host gene transcript levels. Genomics 2009, 94, 138–145. [Google Scholar] [CrossRef]
- Nobiron, I.; O’Reilly, D.R.; Olszewski, J.A. Autographa californica nucleopolyhedrovirus infection of Spodoptera frugiperda cells: A global analysis of host gene regulation during infection, using a differential display approach. J. Gen. Virol. 2003, 84, 3029–3039. [Google Scholar] [CrossRef]
- Salem, T.Z.; Zhang, F.; Xie, Y.; Thiem, S.M. Comprehensive analysis of host gene expression in Autographa californica nucleopolyhedrovirus-infected Spodoptera frugiperda cells. Virology 2011, 412, 167–178. [Google Scholar] [CrossRef]
- Breitenbach, J.E.; Shelby, K.S.; Popham, H.J.R. Baculovirus induced transcripts in hemocytes from the larvae of Heliothis virescens. Viruses 2011, 3, 2047–2064. [Google Scholar] [CrossRef] [PubMed]
- Menzel, T.; Rohrmann, G.F. Diversity of errantivirus (retrovirus) sequences in two cell lines used for baculovirus expression, Spodoptera frugiperda and Trichoplusia ni. Virus Genes 2008, 36, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; Yutin, N.; Shabalina, S.A.; Robert, C.; Fournous, G.; La Scola, B.; Raoult, D.; Koonin, E.V. Viruses with More Than 1,000 Genes: Mamavirus, a new Acanthamoeba polyphaga mimivirus Strain, and Reannotation of Mimivirus Genes. Genome Biol. Evol. 2011, 3, 737–742. [Google Scholar] [CrossRef]
- Colson, P.; Gimenez, G.; Boyer, M.; Fournous, G.; Raoult, D. The giant Cafeteria roenbergensis virus that infects a widespread marine phagocytic protist is a new member of the fourth domain of life. PLoS One 2011, 6, e18935. [Google Scholar] [CrossRef]
- Thézé, J.; Bézier, A.; Periquet, G.; Drezen, J.-M.; Herniou, E.A. Paleozoic origin of insect large dsDNA viruses. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 15931–15935. [Google Scholar] [CrossRef] [PubMed]
- Drezen, J.-M.; Bézier, A.; Lesobre, J.; Huguet, E.; Cattolico, L.; Periquet, G.; Dupuy, C. The few virus-like genes of Cotesia congregata bracovirus. Arch. Insect Biochem. Physiol. 2006, 61, 110–122. [Google Scholar] [CrossRef]
- Katsuma, S.; Tanaka, S.; Omuro, N.; Takabuchi, L.; Daimon, T.; Imanishi, S.; Yamashita, S.; Iwanaga, M.; Mita, K.; Maeda, S.; et al. Novel macula-like virus identified in Bombyx mori cultured cells. J. Virol. 2005, 79, 5577–5584. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Vijayendran, D.; Bonning, B.C. Next generation sequencing technologies for insect virus discovery. Viruses 2011, 3, 1849–1869. [Google Scholar] [CrossRef]
- Wu, Q.; Luo, Y.; Lu, R.; Lau, N.; Lai, E.C.; Li, W.X.; Ding, S.W. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 1606–1611. [Google Scholar] [CrossRef]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V.; Lee, R.C. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol. Biol. 2004, 265, 131–158. [Google Scholar] [PubMed]
- Finn, R.D.; Mistry, J.; Tate, J.; Coggill, P.; Heger, A.; Pollington, J.E.; Gavin, O.L.; Gunasekaran, P.; Ceric, G.; Forslund, K.; et al. The Pfam protein families database. Nucleic Acids Res. 2010, 38, D211–D222. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Furumichi, M.; Tanabe, M.; Hirakawa, M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010, 38, D355–D360. [Google Scholar] [CrossRef]
- Ewing, B.; Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998, 8, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Chou, H.-H.; Holmes, M.H. DNA sequence quality trimming and vector removal. Bioinformatics 2001, 17, 1093–1104. [Google Scholar] [CrossRef]
- Smit, A.F.A.; Hubley, R.; Green, P. RepeatMasker Open-3.0; 1996–2010. Available online: http://www.repeatmasker.org (accessed on 21 July 2011).
- Jurka, J.; Kapitonov, V.V.; Pavlicek, A.; Klonowski, P.; Kohany, O.; Walichiewicz, J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005, 110, 462–467. [Google Scholar] [CrossRef]
- Morgulis, A.; Gertz, E.M.; Schäffer, A.A.; Agarwala, R. A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J. Comput. Biol. 2006, 13, 1028–1040. [Google Scholar] [CrossRef]
- Beldade, P.; Rudd, S.; Gruber, J.D.; Long, A.D. A wing expressed sequence tag resource for Bicyclus anynana butterflies, an evo-devo model. BMC Genomics 2006, 7, 130. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Schlicker, A.; Huthmacher, C.; Ramírez, F.; Lengauer, T.; Albrecht, M. Functional evaluation of domain–domain interactions and human protein interaction networks. Bioinformatics 2007, 23, 859–865. [Google Scholar] [CrossRef] [PubMed]

| Subject Count | PUT Count | Virus |
|---|---|---|
| 444 | 31 | Human immunodeficiency virus 1 |
| 111 | 2 | Porcine reproductive and respiratory syndrome virus |
| 89 | 25 | Cowpox virus |
| 63 | 5 | Human herpesvirus 5 |
| 60 | 23 | Vaccinia virus |
| 57 | 54 | Acanthamoeba polyphaga mimivirus |
| 37 | 3 | Equine infectious anemia virus |
| 36 | 1 | Beak and feather disease virus |
| 35 | 13 | Variola virus |
| 31 | 2 | Cassava brown streak virus |
| 27 | 2 | Human herpesvirus 1 |
| 25 | 30 | Amsacta moorei entomopoxvirus ’L’ |
| 24 | 30 | Cafeteria roenbergensis virus BV-PW1 |
| 21 | 9 | Simian immunodeficiency virus |
| 20 | 28 | Autographa californica nucleopolyhedrovirus |
| 17 | 17 | Monkeypox virus |
| 17 | 33 | Melanoplus sanguinipes entomopoxvirus |
| 17 | 18 | Fowlpox virus |
| 16 | 6 | Hyposoter fugitivus ichnovirus |
| 15 | 8 | Human herpesvirus 8 |
| Count | GO Id. | Description |
|---|---|---|
| 399 | GO:0006412 | Translation |
| 340 | GO:0006351 | transcription, DNA-dependent |
| 287 | GO:0006355 | regulation of transcription, DNA-dependent |
| 140 | GO:0006397 | mRNA processing |
| 128 | GO:0015031 | protein transport |
| 128 | GO:0006457 | protein folding |
| 124 | GO:0006810 | Transport |
| 110 | GO:0008380 | RNA splicing |
| 106 | GO:0007275 | multicellular organismal development |
| 105 | GO:0006281 | DNA repair |
| 103 | GO:0006508 | Proteolysis |
| 95 | GO:0051301 | cell division |
| 95 | GO:0007067 | Mitosis |
| 95 | GO:0006364 | rRNA processing |
| 92 | GO:0006915 | Apoptosis |
| 90 | GO:0022900 | electron transport chain |
| 64 | GO:0000022 | mitotic spindle elongation |
| 63 | GO:0055085 | transmembrane transport |
| 63 | GO:0006260 | DNA replication |
| 56 | GO:0007049 | cell cycle |
| 55 | GO:0030154 | cell differentiation |
| 51 | GO:0006886 | intracellular protein transport |
| 49 | GO:0045454 | cell redox homeostasis |
| 48 | GO:0007264 | small GTPase mediated signal transduction |
| 48 | GO:0006511 | ubiquitin-dependent protein catabolic process |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2011 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sparks, M.E.; Gundersen-Rindal, D.E. The Lymantria dispar IPLB-Ld652Y Cell Line Transcriptome Comprises Diverse Virus-Associated Transcripts. Viruses 2011, 3, 2339-2350. https://doi.org/10.3390/v3112339
Sparks ME, Gundersen-Rindal DE. The Lymantria dispar IPLB-Ld652Y Cell Line Transcriptome Comprises Diverse Virus-Associated Transcripts. Viruses. 2011; 3(11):2339-2350. https://doi.org/10.3390/v3112339
Chicago/Turabian StyleSparks, Michael E., and Dawn E. Gundersen-Rindal. 2011. "The Lymantria dispar IPLB-Ld652Y Cell Line Transcriptome Comprises Diverse Virus-Associated Transcripts" Viruses 3, no. 11: 2339-2350. https://doi.org/10.3390/v3112339
APA StyleSparks, M. E., & Gundersen-Rindal, D. E. (2011). The Lymantria dispar IPLB-Ld652Y Cell Line Transcriptome Comprises Diverse Virus-Associated Transcripts. Viruses, 3(11), 2339-2350. https://doi.org/10.3390/v3112339





