Construction and Segmental Reconstitution of Full-Length Infectious Clones of Milk Vetch Dwarf Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus Source and Nucleic Acid Extraction
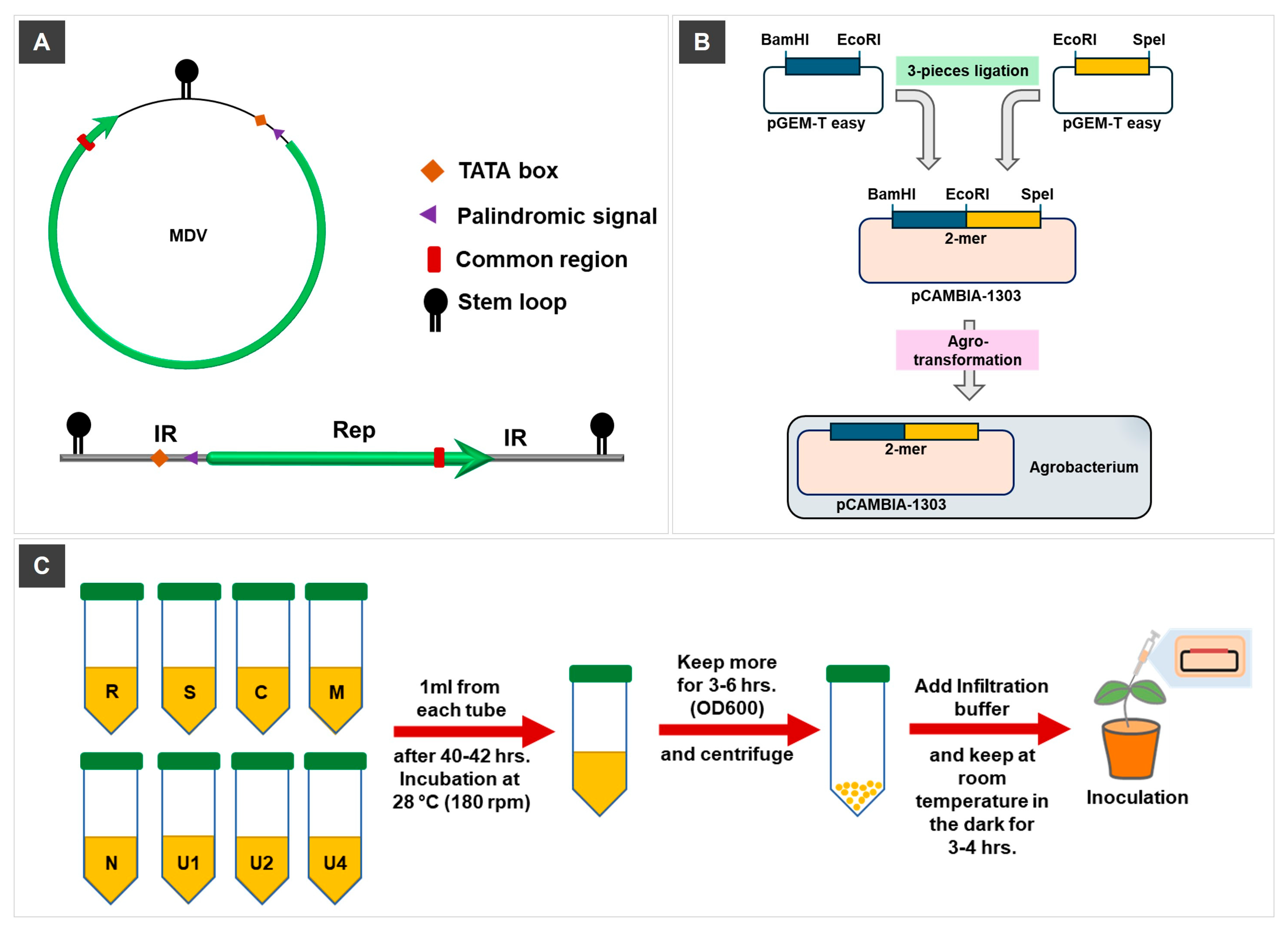
2.2. Strategy for Infectious Clone (IC) Construction of MDV
2.3. Agrobacterium Transformation and Agroinoculation Procedure
2.4. Plant Maintenance and Segment Reconstitution Analysis by PCR
2.5. Quantification of Segment Abundance by qPCR and Fold Change Analysis
3. Results
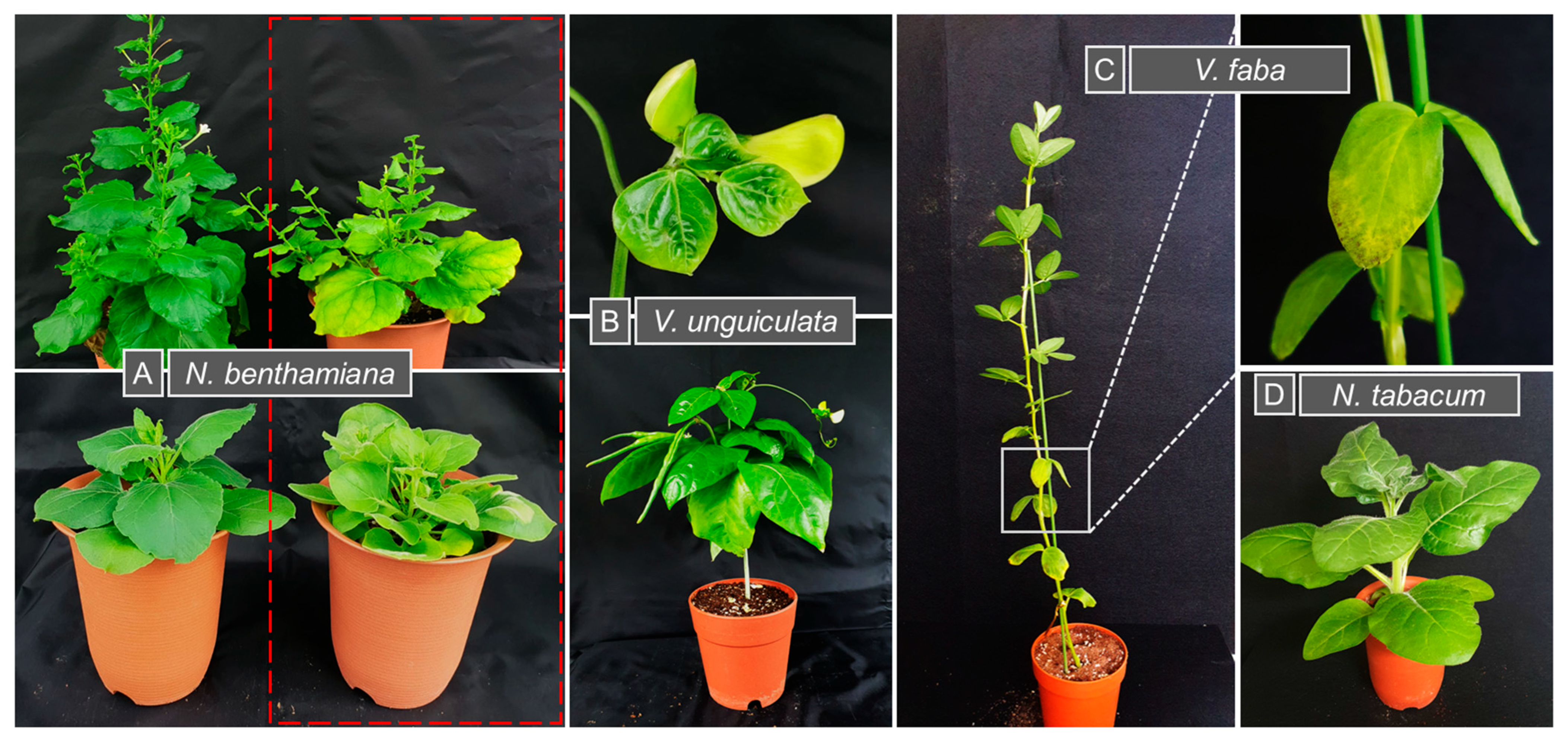
3.1. Reconstitution of MDV Segments Through PCR
3.2. Relative Accumulation of MDV Segments in Infected Plants
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Lal, A.; Vo, T.T.B.; Sanjaya, I.G.N.P.W.; Ho, P.T.; Kim, J.-K.; Kil, E.-J.; Lee, S. Nanovirus disease complexes: An emerging threat in the modern era. Front. Plant Sci. 2020, 11, 558403. [Google Scholar] [CrossRef]
- Lal, A.; Kil, E.J.; Thuy, V.T.B.; Fadhila, C.; Ho, P.T.; Byun, H.S.; Dao, H.T.; Kim, J.K.; Lee, S. Milk vetch dwarf virus infection in the Solanaceae and Caricaceae families in Southeast Asia. Plant Pathol. 2020, 69, 1026–1033. [Google Scholar] [CrossRef]
- Mandal, B. Advances in small isometric multicomponent ssDNA viruses infecting plants. Indian J. Virol. 2010, 21, 18–30. [Google Scholar] [CrossRef]
- Gronenborn, B. Nanoviruses: Genome organisation and protein function. Vet. Microbiol. 2004, 98, 103–109. [Google Scholar] [CrossRef]
- Sicard, A.; Pirolles, E.; Gallet, R.; Vernerey, M.-S.; Yvon, M.; Urbino, C.; Peterschmitt, M.; Gutierrez, S.; Michalakis, Y.; Blanc, S. A multicellular way of life for a multipartite virus. eLife 2019, 8, e43599. [Google Scholar] [CrossRef] [PubMed]
- Sano, Y.; Wada, M.; Hashimoto, Y.; Matsumoto, T.; Kojima, M. Sequences of ten circular ssDNA components associated with the milk vetch dwarf virus genome. J. Gen. Virol. 1998, 79, 3111–3118. [Google Scholar] [CrossRef] [PubMed]
- Hanley-Bowdoin, L.; Settlage, S.B.; Orozco, B.M.; Nagar, S.; Robertson, D. Geminiviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 1999, 18, 71–106. [Google Scholar] [CrossRef]
- Lal, A.; Shamim, A.; Kil, E.-J.; Vo, T.T.B.; Qureshi, M.A.; Bupi, N.; Tabassum, M.; Lee, S. Insights into the Differential Composition of Stem-Loop Structures of Nanoviruses and Their Impacts. Microbiol. Spectr. 2023, 11, e04798-22. [Google Scholar] [CrossRef]
- Timchenko, T.; Katul, L.; Sano, Y.; de Kouchkovsky, F.; Vetten, H.J.; Gronenborn, B. The master rep concept in nanovirus replication: Identification of missing genome components and potential for natural genetic reassortment. Virology 2000, 274, 189–195. [Google Scholar] [CrossRef]
- Horser, C.L.; Harding, R.M.; Dale, J.L. Banana bunchy top nanovirus DNA-1 encodes the ‘master’ replication initiation protein. J. Gen. Virol. 2001, 82, 459–464. [Google Scholar] [CrossRef]
- Trapani, S.; Bhat, E.A.; Yvon, M.; Lai-Kee-Him, J.; Hoh, F.; Vernerey, M.S.; Pirolles, E.; Bonnamy, M.; Schoehn, G.; Zeddam, J.L.; et al. Structure-guided mutagenesis of the capsid protein indicates that a nanovirus requires assembled viral particles for systemic infection. PLoS Pathog. 2023, 19, e1011086. [Google Scholar] [CrossRef] [PubMed]
- Wanitchakorn, R.; Harding, R.M.; Dale, J.L. Banana bunchy top virus DNA-3encodes the viral coat protein. Arch. Virol. 1997, 142, 1673–1680. [Google Scholar] [CrossRef] [PubMed]
- Aronson, M.N.; Meyer, A.D.; Györgyey, J.; Katul, L.; Vetten, H.J.; Gronenborn, B.; Timchenko, T. Clink, a nanovirus-encoded protein, binds both pRB and SKP1. J. Virol. 2000, 74, 2967–2972. [Google Scholar] [CrossRef]
- Grigoras, I.; Vetten, H.-J.; Commandeur, U.; Ziebell, H.; Gronenborn, B.; Timchenko, T. Nanovirus DNA-N encodes a protein mandatory for aphid transmission. Virology 2018, 522, 281–291. [Google Scholar] [CrossRef]
- Yan, D.; Han, K.; Lu, Y.; Peng, J.; Rao, S.; Wu, G.; Liu, Y.; Chen, J.; Zheng, H.; Yan, F. The nanovirus U2 protein suppresses RNA silencing via three conserved cysteine residues. Mol. Plant Pathol. 2024, 25, e13394. [Google Scholar] [CrossRef]
- Taniguchi, T.; Palmieri, M.; Weissmann, C. Qβ DNA-containing hybrid plasmids giving rise to Qβ phage formation in the bacterial host. Nature 1978, 274, 223–228. [Google Scholar] [CrossRef]
- Zaitlin, M.; Palukaitis, P. Advances in understanding plant viruses and virus diseases. Annu. Rev. Phytopathol. 2000, 38, 117–143. [Google Scholar] [CrossRef]
- Shakir, S.; Zaidi, S.S.-e.-A.; Hashemi, F.S.G.; Nyirakanani, C.; Vanderschuren, H. Harnessing plant viruses in the metagenomics era: From the development of infectious clones to applications. Trends Plant Sci. 2023, 28, 297–311. [Google Scholar] [CrossRef]
- Brewer, H.C.; Hird, D.L.; Bailey, A.M.; Seal, S.E.; Foster, G.D. A guide to the contained use of plant virus infectious clones. Plant Biotechnol. J. 2018, 16, 832–843. [Google Scholar] [CrossRef]
- Howell, S.H.; Walker, L.L.; Walden, R.M. Rescue of in vitro generated mutants of cloned cauliflower mosaic virus genome in infected plants. Nature 1981, 293, 483–486. [Google Scholar] [CrossRef]
- Gronenborn, B.; Gardner, R.C.; Schaefer, S.; Shepherd, R.J. Propagation of foreign DNA in plants using cauliflower mosaic virus as vector. Nature 1981, 294, 773–776. [Google Scholar] [CrossRef]
- Stenger, D.C.; Revington, G.N.; Stevenson, M.C.; Bisaro, D.M. Replicational release of geminivirus genomes from tandemly repeated copies: Evidence for rolling-circle replication of a plant viral DNA. Proc. Natl. Acad. Sci. USA 1991, 88, 8029–8033. [Google Scholar] [CrossRef]
- Massart, S.; Candresse, T.; Gil, J.; Lacomme, C.; Predajna, L.; Ravnikar, M.; Reynard, J.S.; Rumbou, A.; Saldarelli, P.; Škorić, D.; et al. A Framework for the Evaluation of Biosecurity, Commercial, Regulatory, and Scientific Impacts of Plant Viruses and Viroids Identified by NGS Technologies. Front. Microbiol. 2017, 8, 45. [Google Scholar] [CrossRef]
- Nagyová, A.; Subr, Z. Infectious full-length clones of plant viruses and their use for construction of viral vectors. Acta Virol. 2007, 51, 223–237. [Google Scholar] [PubMed]
- Timchenko, T.; Katul, L.; Aronson, M.; Vega-Arreguin, J.C.; Ramirez, B.C.; Vetten, H.J.; Gronenborn, B. Infectivity of nanovirus DNAs: Induction of disease by cloned genome components of Faba bean necrotic yellows virus. J. Gen. Virol. 2006, 87, 1735–1743. [Google Scholar] [CrossRef] [PubMed]
- Grigoras, I.; Timchenko, T.; Katul, L.; Grande-Pérez, A.; Vetten, H.-J.; Gronenborn, B. Reconstitution of authentic nanovirus from multiple cloned DNAs. J. Virol. 2009, 83, 10778–10787. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, M.; Heydarnejad, J.; Hasanvand, V.; Massumi, H.; Kvarnheden, A.; Varsani, A. Genome characterization of Iranian sophora yellow stunt virus isolates and construction of infectious clones. Plant Pathol. 2023, 72, 1283–1292. [Google Scholar] [CrossRef]
- Sicard, A.; Zeddam, J.-L.; Yvon, M.; Michalakis, Y.; Gutiérrez, S.; Blanc, S. Circulative Nonpropagative Aphid Transmission of Nanoviruses: An Oversimplified View. J. Virol. 2015, 89, 9719–9726. [Google Scholar] [CrossRef]
- Di Mattia, J.; Vernerey, M.-S.; Yvon, M.; Pirolles, E.; Villegas, M.; Gaafar, Y.; Ziebell, H.; Michalakis, Y.; Zeddam, J.-L.; Blanc, S. Route of a multipartite nanovirus across the body of its aphid vector. J. Virol. 2020, 94, 10–1128. [Google Scholar] [CrossRef]
- Lal, A.; Kil, E.J.; Byun, H.S.; Zarghami, D.S.; Kim, J.K.; Lee, S. First report of milk vetch dwarf virus associated with dwarfism in papaya in Korea. Plant Dis. 2018, 102, 2666. [Google Scholar] [CrossRef]
- Choi, H.; Jo, Y.; Zhou, Y.; Cho, W.K. First report of milk vetch dwarf virus infecting lily in Korea. Plant Dis. 2019, 103, 2144. [Google Scholar] [CrossRef]
- Yang, J.G.; Wang, S.P.; Liu, W.; Li, Y.; Shen, L.L.; Qian, Y.M.; Wang, F.L.; Du, Z.G. First report of milk vetch dwarf virus associated with a disease of Nicotiana tabacum in China. Plant Dis. 2016, 100, 1255. [Google Scholar] [CrossRef]
- Kumari, S.G.; Rodoni, B.; Vetten, H.J.; Loh, M.H.; Freeman, A.; Van Leur, J.; Bao, S.; Wang, X. Detection and partial characterization of Milk vetch dwarf virus isolates from faba bean (Vicia faba L.) in Yunnan Province, China. J. Phytopathol. 2010, 158, 35–39. [Google Scholar] [CrossRef]
- Seol, E.; Jung, Y.; Lee, J.; Cho, C.; Kim, T.; Rhee, Y.; Lee, S. In planta transformation of Notocactus scopa cv. Soonjung by Agrobacterium tumefaciens. Plant Cell Rep. 2008, 27, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ahlquist, P.; French, R.; Janda, M.; Loesch-Fries, L.S. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc. Natl. Acad. Sci. USA 1984, 81, 7066–7070. [Google Scholar] [CrossRef]
- Scholthof, H.B.; Scholthof, K.-B.G.; Jackson, A.O. Plant virus gene vectors for transient expression of foreign proteins in plants. Annu. Rev. Phytopathol. 1996, 34, 299–323. [Google Scholar] [CrossRef]
- Buragohain, A.K.; Sung, Y.K.; Coffin, R.S.; Coutts, R.H.A. The infectivity of dimeric potato yellow mosaic geminivirus clones in different hosts. J. Gen. Virol. 1994, 75, 2857–2861. [Google Scholar] [CrossRef]
- de Oliveira Ferreira, P.d.T.; Lemos, T.O.; Nagata, T.; Inoue-Nagata, A.K. One-step cloning approach for construction of agroinfectious begomovirus clones. J. Virol. Methods 2008, 147, 351–354. [Google Scholar] [CrossRef]
- Bally, J.; Nakasugi, K.; Jia, F.; Jung, H.; Ho, S.Y.W.; Wong, M.; Paul, C.M.; Naim, F.; Wood, C.C.; Crowhurst, R.N. The extremophile Nicotiana benthamiana has traded viral defence for early vigour. Nat. Plants 2015, 1, 15165. [Google Scholar] [CrossRef]
- Goodin, M.M.; Zaitlin, D.; Naidu, R.A.; Lommel, S.A. Nicotiana benthamiana: Its History and Future as a Model for Plant–Pathogen Interactions. Mol. Plant Microbe Interact. 2008, 21, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zheng, H.; Yan, D.; Han, K.; Song, X.; Liu, Y.; Zhang, D.; Chen, J.; Yan, F. Complete genomic characterization of milk vetch dwarf virus isolates from cowpea and broad bean in Anhui province, China. Arch. Virol. 2017, 162, 2437–2440. [Google Scholar] [CrossRef] [PubMed]
- Sicard, A.; Yvon, M.; Timchenko, T.; Gronenborn, B.; Michalakis, Y.; Gutierrez, S.; Blanc, S. Gene copy number is differentially regulated in a multipartite virus. Nat. Commun. 2013, 4, 2248. [Google Scholar] [CrossRef] [PubMed]
- Grigoras, I.; Ginzo, A.; Martin, D.P.; Varsani, A.; Romero, J.; Mammadov, A.C.; Huseynova, I.M.; Aliyev, J.A.; Kheyr-Pour, A.; Huss, H.; et al. Genome diversity and evidence of recombination and reassortment in nanoviruses from Europe. J. Gen. Virol. 2014, 95, 1178–1191. [Google Scholar] [CrossRef]

| Segment | Primer Name | Restriction Site | Sequences (5′ → 3′) | Primer Name | Restriction Site | Sequences (5′ → 3′) | Product Size (bp) |
|---|---|---|---|---|---|---|---|
| MDV-C | IC1-F | BamHI | GGATCCTATATTAAGTTG TTATCTGAGAAATCTATT | IC1-R | EcoRI | GAATTCAACTCAGCAGGTGAAG | 634 |
| IC2-F | EcoRI | GAATTCGTTAAGTA AGTTTTTAAATGCTs | IC2-R | SpeI | ACTAGTTTTCGTTGTA AGAACAACGAAGAAA | 810 | |
| MDV-R | IC1-F | KpnI | GGTACCCGTCATAT GATCCCGTGCT | IC1-R | EcoRI | GAATTCGAACTCCCT TAGAGTACTTGC | 557 |
| IC2-F | EcoRI | GAATTCAGAGGTG AGTTGAAGAAGA | IC2-R | SpeI | ACTAGTATTTTATTGATGAATG ATAAAATATTACAACTTG | 584 | |
| MDV-M | IC1-F | KpnI | GGTACCAGAATGATTATA GATTGTAATTAGTTATTC | IC1-R | EcoRV | GATATCGCCGTCGTCTTGATA | 698 |
| IC2-F | EcoRV | GATATCGATGCCCAGAAGAG | IC2-R | SpeI | ACTAGTGAAATTTCAAT GGACAATAAAAACAC | 953 | |
| MDV-S | IC1-F | KpnI | GGTACCTGTAATGAAGAACAC TATGAAATAATGAAACC | IC1-R | PstI | CTGCAGACCAATTAACAA TGGGAGAA | 767 |
| IC2-F | PstI | CTGCAGCTTTTACCGCTCC | IC2-R | BamHI | GGATCCTTTCGTTGTG AGTACAACGAATAC | 720 | |
| MDV-U1 | IC1-F | BamHI | GGATCCTAATGAATATTTGTTT CAGGATCAAACA | IC1-R | PsiI | TTATAAAAAACATTCTAATA CCTATCAAATAA | 888 |
| IC2-F | PsiI | TTATAAATATTAATCAGTTG ATTAATACTTGT | IC2-R | SpeI | ACTAGTGACCTCAATA GAAGCTTTAGTTTG | 655 | |
| MDV-N | IC1-F | BamHI | GGATCCTAATCATAATTATTGT AAGATTATGCAATTG | IC1-R | ScaI | AGTACTGGGATTC AATATCAAGGT | 839 |
| IC2-F | ScaI | AGTACTTGAAGAAG GACGAAGAC | IC2-R | SpeI | ACTAGTTTTTTGCAGTTGCA GAAAATGATGAC | 621 | |
| MDV-U2 | IC1-F | BamHI | GGATCCTGTGATATATGAAAAC AATTTGTTGTTTTTTCCATTG | IC1-R | EcoRV | GATATCTATAATTACCTGAA TCGTACAAATCTTTCAAG | 930 |
| IC2-F | EcoRV | GATATCAAGTGTATTATTC TTCGTCATGTAAAAGAG | IC2-R | SpeI | ACTAGTAGAACAAGA ACGAGACTAACGC | 464 | |
| MDV-U4 | IC1-F | BamHI | GAGCAATAACAAGAATAAATA AGGATCCAAATGCAA | IC1-R | SalI | GTCGACATCTTCA AAGGGATTCTT | 876 |
| IC2-F | SalI | GTCGACCCTGATGTTACC | IC2-R | SpeI | GTGGGGACCATACTAGT TTCTCACTTATTA | 551 |
| Experiment | Segment | Primer Name | Sequences (5′ → 3′) | Product Size (bp) |
|---|---|---|---|---|
| Reconstitution after agro-inoculation | MDV-C | C-Forward | CCTGCTGAATTGAATTCTCTGAGTA | 295 |
| C-Reverse | AAACTATCTGAATACCTAGCGACTTAAAC | |||
| MDV-S | S-Forward | CTGCTTTGTTGAAGAAAGATGAAGTC | 488 | |
| S-Reverse | AAACACGGAAACATACCGCTAC | |||
| MDV-N | N-Forward | GAAGCTTCTTCGTTGCTCTATAAATACAAG | 673 | |
| N-Reverse | TCAGATGACGTCATATTCATTTGGG | |||
| MDV-M | M-Forward | CCTGAGCCGCTATTGTCAT | 672 | |
| M-Reverse | TTCCTCATTGGCTACTGAATTGG | |||
| MDV-R | R-Forward | GAGATGAAGAAACGCACGTCT | 526 | |
| R-Reverse | GCACTAACTCTTGGTGGTC | |||
| MDV-U1 | U1-Forward | CGTCTGAGAGGAAATTGATAGCC | 486 | |
| U1-Reverse | GGGCCTAGACATATAGCTTCG | |||
| MDV-U2 | U2-Forward | CGAGCGTTAGTCTCGTTCTTG | 516 | |
| U2-Reverse | TGTTATCAATTGTAGTTGTCTTCCACC | |||
| MDV-U4 | U4-Forward | CCACGCACTATATGAACCTTGC | 516 | |
| U4-Reverse | GCAAATATTGAAGGTCTTCACCATC |
| Segment | Primer Name | Sequences (5′ → 3′) | Product Size (bp) |
|---|---|---|---|
| MDV-M | MDVQ-M-F | GCCCAGAAGAGACATCAAGC | 196 |
| MDVQ-M-R | CGAAGGGTGTGCGTGTTATAG | ||
| MDV-U4 | MDVQ-U4-F | ATGGA ACCCAGGTTCCTTCTT | 163 |
| MDVQ-U4-R | TCCTCTGGTTGTTCAAACGTAT | ||
| MDV-N | MDVQ-N-F | GAAGGTCAGAAGACATTCAACCT | 159 |
| MDVQ-N-R | ACACTTTGATCCTAAGAGCATG | ||
| MDV-R | MDVQ-R-F | GGCTTAGTATTACCCCCGCC | 137 |
| MDVQ-R-R | GCACCAGCATATAACTTGCCG | ||
| MDV-C | MDVQ-C-F | AATACGCGTGGACGATCAGG | 177 |
| MDVQ-C-R | CGGGAAGAAGCAAAGACAGC | ||
| MDV-S | MDVQ-S-F | CCGGTATCAGCCAAACCCAA | 164 |
| MDVQ-S-R | ATACCGCTACGCGGAGTTTT | ||
| MDV-U1 | MDVQ-U1-F | CTTCGTCTCGAAGCAAAGGAC | 145 |
| MDVQ-U1-R | TCGTTCGCAGACATAACCTCAA | ||
| MDV-U2 | MDVQ-U2-F | AAGGAAGAACAAGATGCTTTCTGG | 150 |
| MDVQ-U2-R | TCTAAGAACCCACCGTGCAG |
| Host Species | Symptoms Observed * | DNA-R | DNA-S | DNA-C | DNA-M | DNA-N | DNA-U1 | DNA-U2 | DNA-U4 |
|---|---|---|---|---|---|---|---|---|---|
| N. benthamiana | Bushy growth Stunting and leaf yellowing | 36/36 | 33/36 | 35/36 | 31/36 | 36/36 | 32/36 | 33/36 | 34/36 |
| V. unguiculata | Leaf yellowing | 18/18 | 18/18 | 15/18 | 14/18 | 18/18 | 14/18 | 16/18 | 12/18 |
| V. faba | Necrosis, leaf yellowing, curling | 36/36 | 33/36 | 35/36 | 23/36 | 36/36 | 30/36 | 33/36 | 34/36 |
| N. tabacum | Mild stunting | 18/18 | 18/18 | 15/18 | 07/18 | 18/18 | 03/18 | 16/18 | 12/18 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lal, A.; Qureshi, M.A.; Son, M.-C.; Lee, S.; Kil, E.-J. Construction and Segmental Reconstitution of Full-Length Infectious Clones of Milk Vetch Dwarf Virus. Viruses 2025, 17, 1213. https://doi.org/10.3390/v17091213
Lal A, Qureshi MA, Son M-C, Lee S, Kil E-J. Construction and Segmental Reconstitution of Full-Length Infectious Clones of Milk Vetch Dwarf Virus. Viruses. 2025; 17(9):1213. https://doi.org/10.3390/v17091213
Chicago/Turabian StyleLal, Aamir, Muhammad Amir Qureshi, Man-Cheol Son, Sukchan Lee, and Eui-Joon Kil. 2025. "Construction and Segmental Reconstitution of Full-Length Infectious Clones of Milk Vetch Dwarf Virus" Viruses 17, no. 9: 1213. https://doi.org/10.3390/v17091213
APA StyleLal, A., Qureshi, M. A., Son, M.-C., Lee, S., & Kil, E.-J. (2025). Construction and Segmental Reconstitution of Full-Length Infectious Clones of Milk Vetch Dwarf Virus. Viruses, 17(9), 1213. https://doi.org/10.3390/v17091213






