A Single-Cell Perspective on the Effects of Dopamine in the Regulation of HIV Latency Phenotypes in a Myeloid Cell Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Promocytic Cell Lines and Culture Conditions
2.2. Cell Culture Treatments
2.3. Single-Cell RNAseq Samples Preparation
2.4. Single-Cell RNAseq Data Analysis
- (i)
- Alignment and Quality filtering—Sequencing data processing were performed using the Cloud-based Cell Ranger 7.2.0 platform and visualized in the Loupe 7.0 browser (10× Genomics). Specifically, the scRNA data was demultiplexed and aligned to the customized hg38 Homo sapiens reference genome (NCBI RefSeq assembly GCF_000001405.40) which was combined with a chromosome representing the HIV-1 genome (AF033819). The generated sequencing libraries were then aggregated using the function Cell Ranger Aggr (v3.1.0); low-quality single-cell transcriptomes were filtered based on the UMI count [500 to 30,000], gene count [300 to 5000], and mitochondrial percentage [less than 15%]. Table 1 shows the Cell Ranger-derived quality controls.
- (ii)
- Data normalization, batch correction, and clustering analysis. Following data normalization in Seurat v4 [31], batch correction was performed using Harmony. Using the 2000 most variable genes (default parameters), and 50 most significant principal components, we performed linear dimensional reduction and built a neighborhood graph using the t-SNE low-dimension visualization tools in Seurat. Cluster-specific pathways and biological processes were analyzed using iPathwayGuide (AdvaitaBio, Ann Arbor, MI, USA) [32], as well as the Database for Annotation, Visualization and Integrated Discovery (DAVID) [33], and visualized using GeneMania [34] in Cytoscape 3.10.2 [35,36].
- (iii)
- Differential gene expression (DGE) analysis was conducted with the DESeq2 R Package [37]. Cell Ranger was integrated to calculate DGEs and generate volcano plots for the visualization of differential gene expression upon different stimulations: non-treated (NT) vs. dopamine (DA), within and between U937 and U1 cells (Table 1). Statistical significance was tested using the Wilcoxon matched-pair signed-rank test. Cell Ranger data were converted to Seurat for aggregation and cluster analysis.
- (iv)
- All raw and processed data were deposited in GEO with the assessment number GSE278043.
2.5. Systems Biology and Visualizations
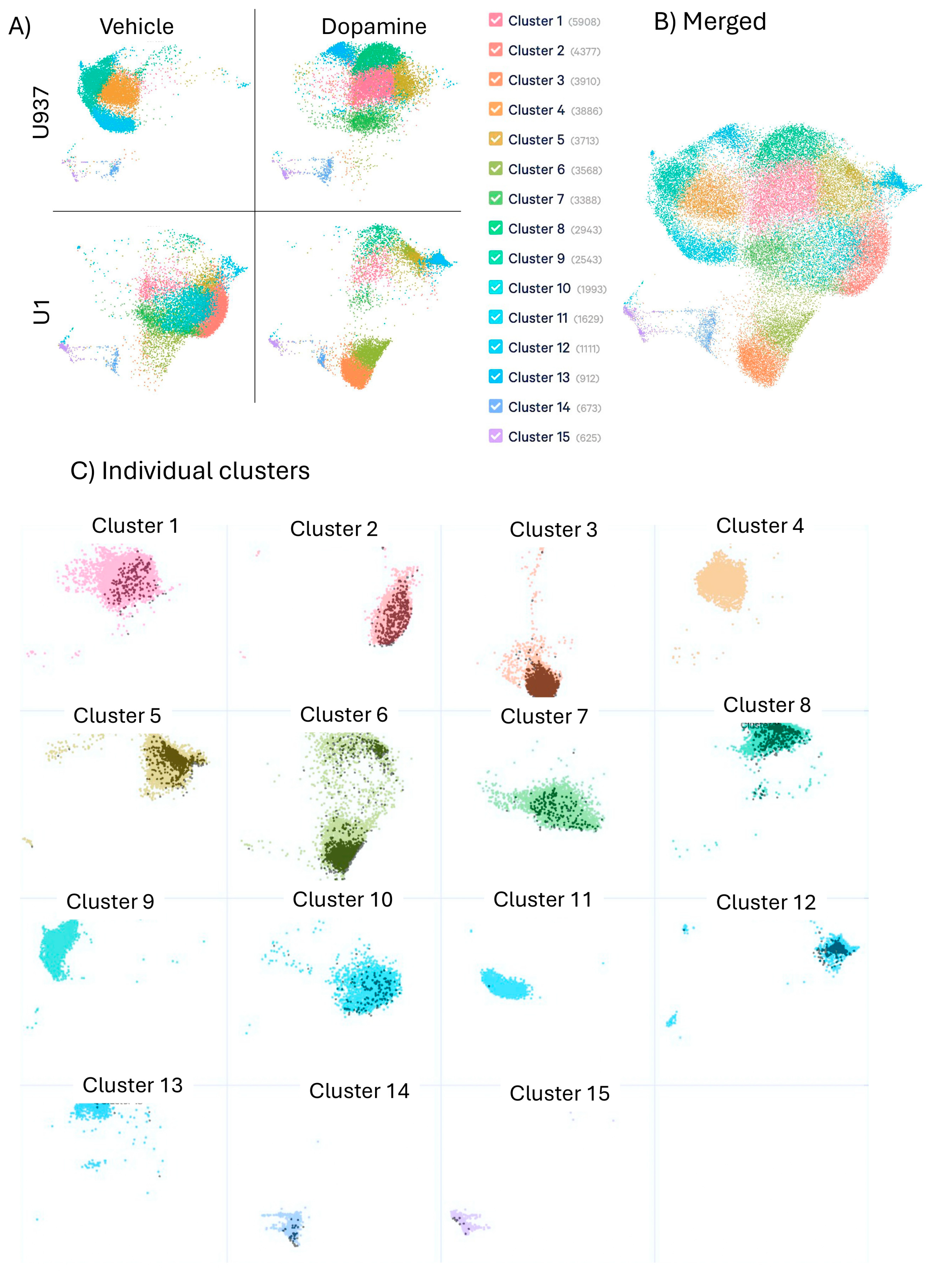
3. Results
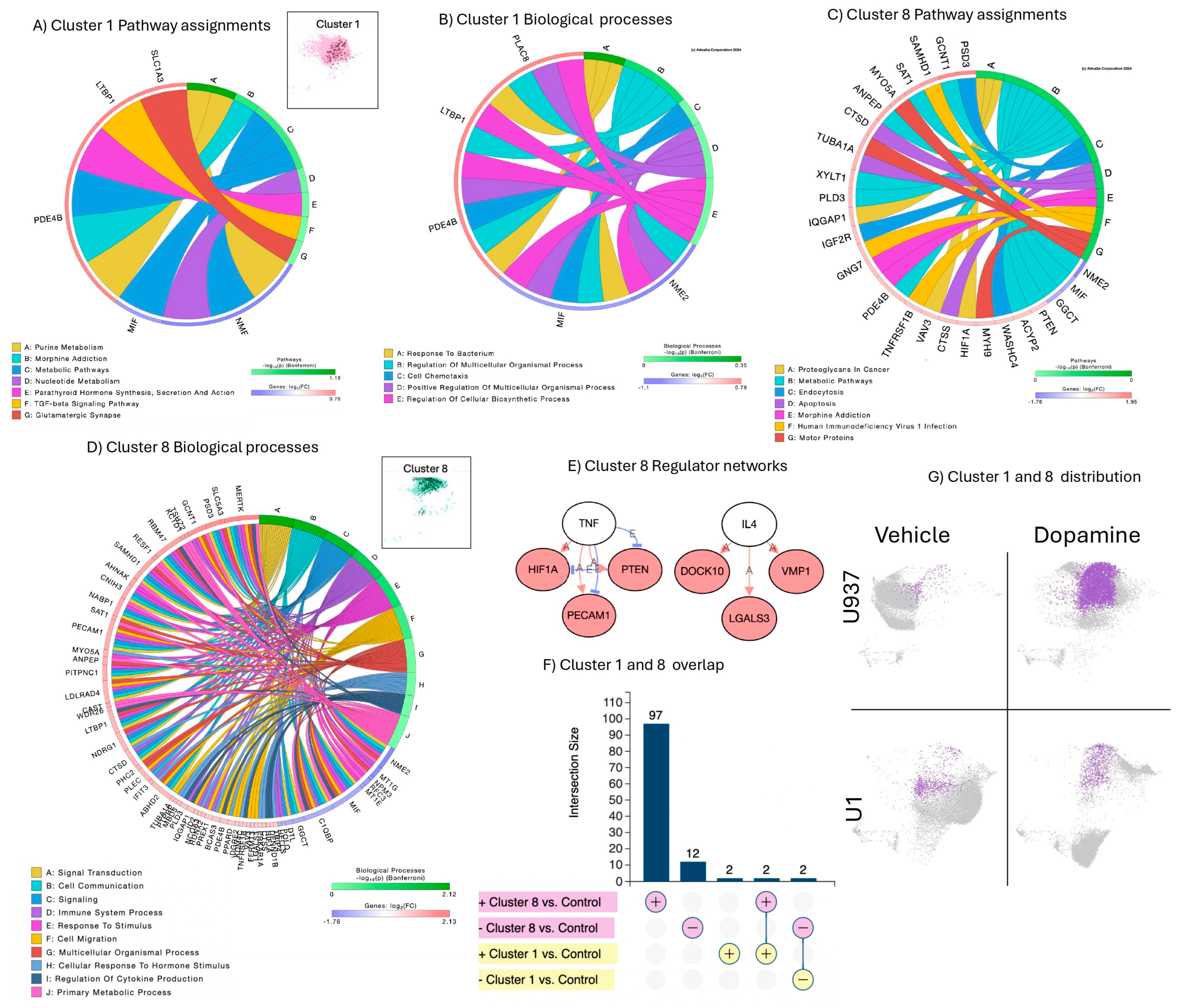
3.1. Uninfected, Unstimulated Control Signatures
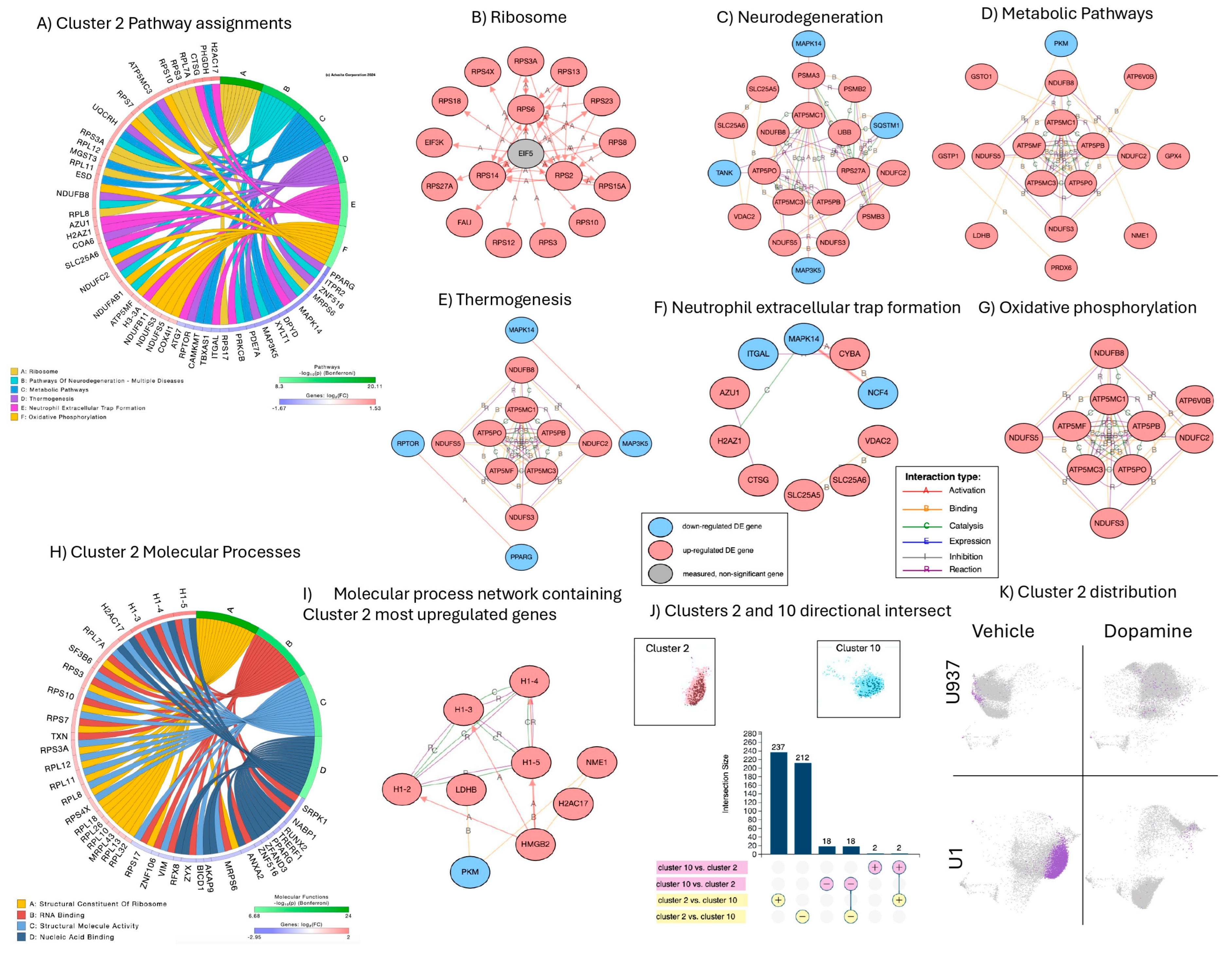
3.2. Signatures of DA Stimulation on Uninfected Control Cells
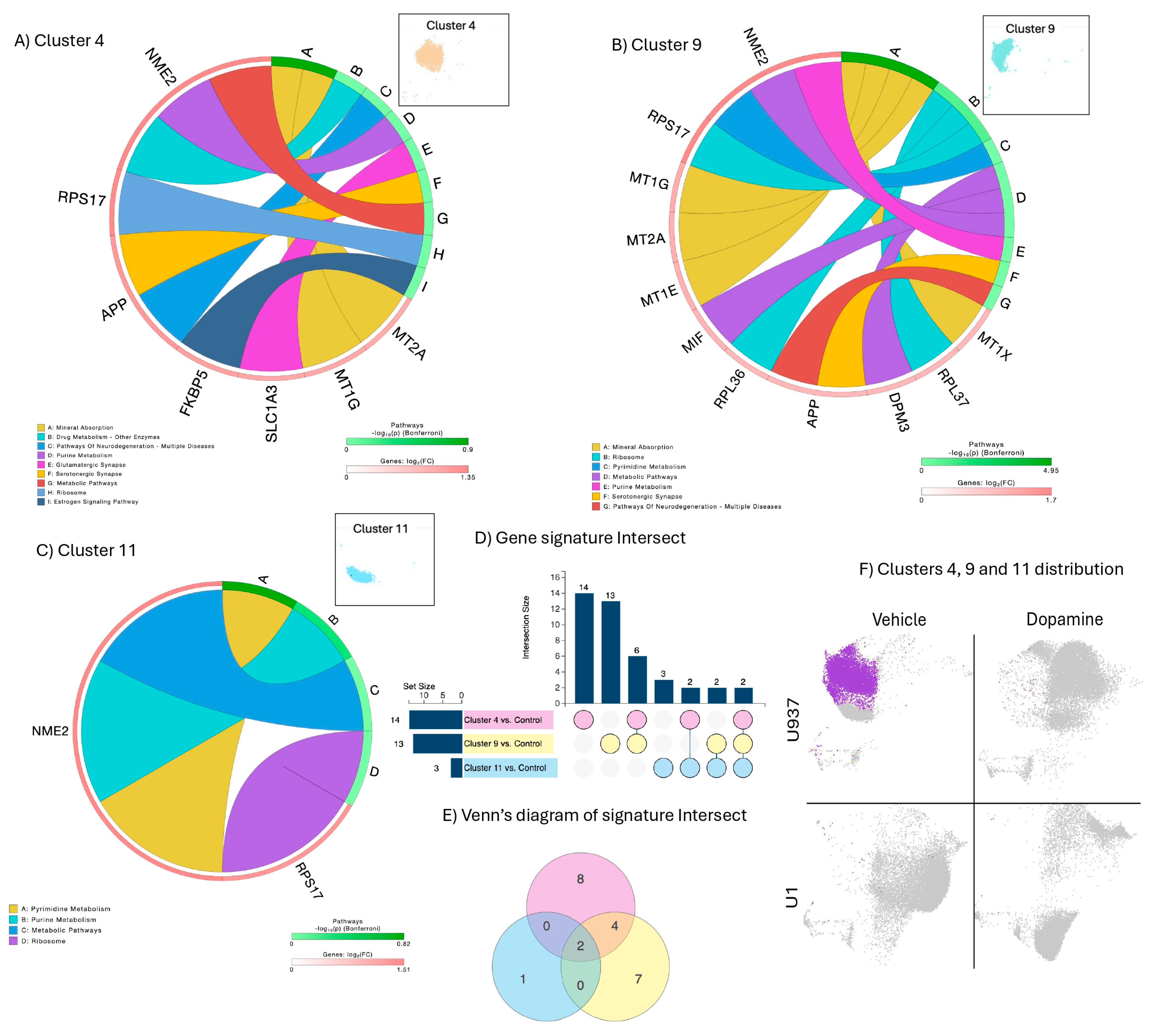
3.3. Signatures Restricted to Unstimulated HIV Latently Infected Cells
3.4. Signatures of DA Stimulation on HIV Latent Cells
3.5. Signatures of Latency Reversal Enhanced by DA Stimulation and Their Regulatory Relationships Defined by Gene Network Analysis
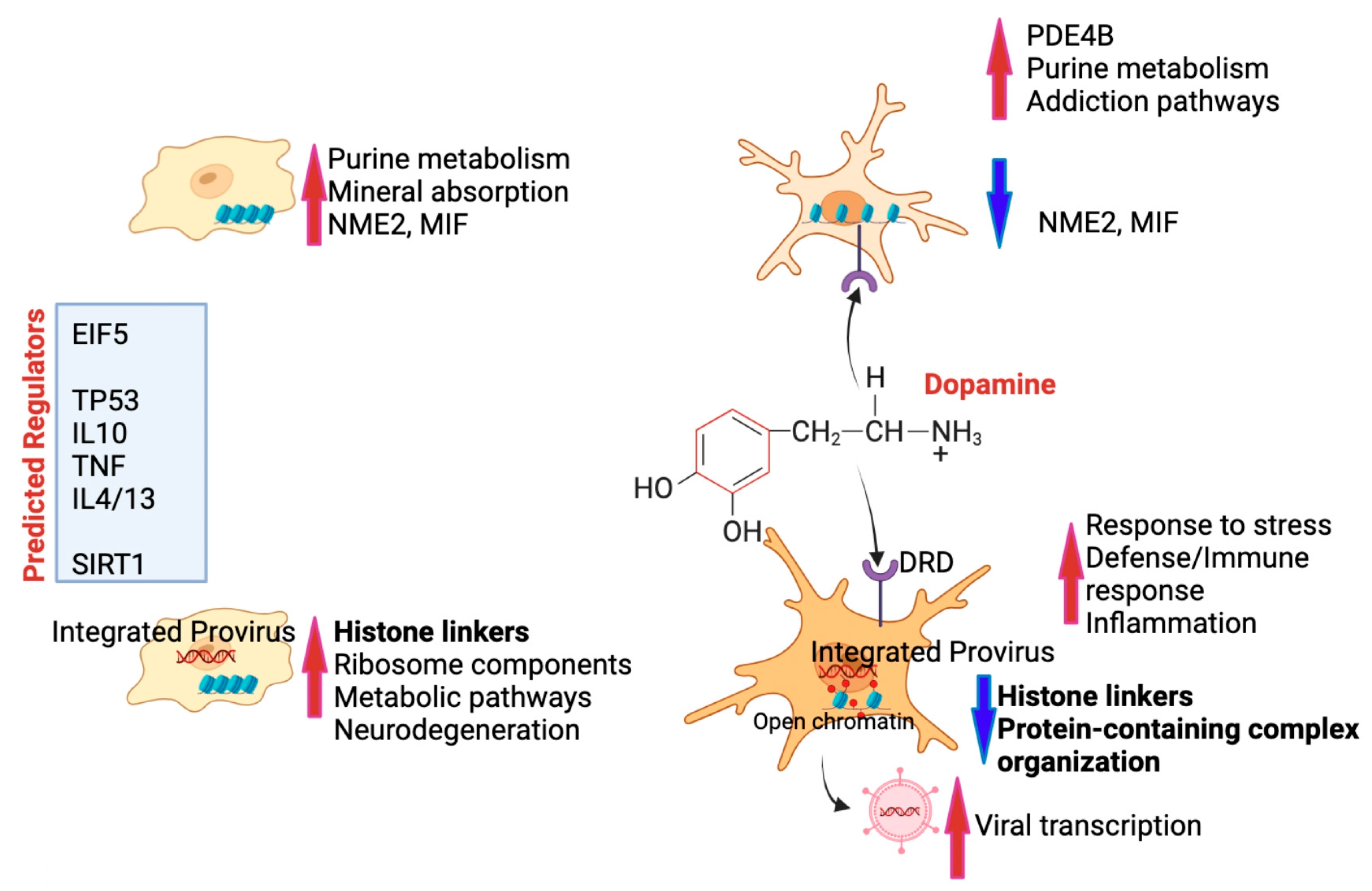
3.6. Summary of Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Meth | Methamphetamine |
| DA | Dopamine |
| HIV | Human Immunodeficiency Virus |
| CNS | Central Nervous System |
| CSF | Cerebrospinal Fluid |
| PWH | People living with HIV |
| MRP8/14 | Myeloid related proteins 8/14 |
| GEM | Gel emulsion |
| UMI | Unique molecular identifiers |
| DGE | Differential gene expression |
| NT | Non-treated |
| UMAP | Uniform manifold approximation and projection |
| NME2 | Nucleoside diphosphate kinase 2 |
| MIF | Macrophage migration inhibitory factor |
| PDE4B | cAMP-specific 3′,5′-cyclic phosphodiesterase 4D |
| LTBP1 | Latent-transforming growth factor beta-binding protein 1 |
| SLC1A3 | Solute carrier family 1 member 3, glutamate transporter |
| PLAC8 | Placenta specific 8 |
| TNF | Tumor necrosis factor |
| IL4 | Interleukin 4 |
| IL13 | Interleukin 13 |
| EIF5 | Eukaryotic initiation factor 5 |
| GSRA | Gene set representation analyses |
| DAVID | Database of Annotation, Visualization and Integrated Discovery |
References
- Basova, L.; Najera, J.A.; Bortell, N.; Wang, D.; Moya, R.; Lindsey, A.; Semenova, S.; Ellis, R.J.; Marcondes, M.C.G. Dopamine and its receptors play a role in the modulation of CCR5 expression in innate immune cells following exposure to Methamphetamine: Implications to HIV infection. PLoS ONE 2018, 13, e0199861. [Google Scholar] [CrossRef] [PubMed]
- Basova, L.V.; Lindsey, A.; McGovern, A.; Rosander, A.; Delorme-Walker, V.; ElShamy, W.M.; Pendyala, V.V.; Gaskill, P.J.; Ellis, R.J.; Cherner, M.; et al. MRP8/14 Is a Molecular Signature Triggered by Dopamine in HIV Latent Myeloid Targets That Increases HIV Transcription and Distinguishes HIV+ Methamphetamine Users with Detectable CSF Viral Load and Brain Pathology. Viruses 2023, 15, 1363. [Google Scholar] [CrossRef] [PubMed]
- Kesby, J.P.; Chang, A.; Najera, J.A.; Marcondes, M.C.G.; Semenova, S. Brain Reward Function after Chronic and Binge Methamphetamine Regimens in Mice Expressing the HIV-1 TAT Protein. Curr. HIV Res. 2019, 17, 126–133. [Google Scholar] [CrossRef]
- Mediouni, S.; Marcondes, M.C.; Miller, C.; McLaughlin, J.P.; Valente, S.T. The cross-talk of HIV-1 Tat and methamphetamine in HIV-associated neurocognitive disorders. Front. Microbiol. 2015, 6, 1164. [Google Scholar] [CrossRef] [PubMed]
- Calderon, T.M.; Williams, D.W.; Lopez, L.; Eugenin, E.A.; Cheney, L.; Gaskill, P.J.; Veenstra, M.; Anastos, K.; Morgello, S.; Berman, J.W. Dopamine Increases CD14+CD16+ Monocyte Transmigration across the Blood Brain Barrier: Implications for Substance Abuse and HIV Neuropathogenesis. J. Neuroimmune Pharmacol. 2017, 12, 353–370. [Google Scholar] [CrossRef]
- Coley, J.S.; Calderon, T.M.; Gaskill, P.J.; Eugenin, E.A.; Berman, J.W. Dopamine increases CD14+CD16+ monocyte migration and adhesion in the context of substance abuse and HIV neuropathogenesis. PLoS ONE 2015, 10, e0117450. [Google Scholar] [CrossRef]
- Gaskill, P.J.; Calderon, T.M.; Coley, J.S.; Berman, J.W. Drug induced increases in CNS dopamine alter monocyte, macrophage and T cell functions: Implications for HAND. J. Neuroimmune Pharmacol. 2013, 8, 621–642. [Google Scholar] [CrossRef]
- Gaskill, P.J.; Calderon, T.M.; Luers, A.J.; Eugenin, E.A.; Javitch, J.A.; Berman, J.W. Human immunodeficiency virus (HIV) infection of human macrophages is increased by dopamine: A bridge between HIV-associated neurologic disorders and drug abuse. Am. J. Pathol. 2009, 175, 1148–1159. [Google Scholar] [CrossRef]
- Gaskill, P.J.; Carvallo, L.; Eugenin, E.A.; Berman, J.W. Characterization and function of the human macrophage dopaminergic system: Implications for CNS disease and drug abuse. J. Neuroinflamm. 2012, 9, 203. [Google Scholar] [CrossRef]
- Gaskill, P.J.; Miller, D.R.; Gamble-George, J.; Yano, H.; Khoshbouei, H. HIV, Tat and dopamine transmission. Neurobiol. Dis. 2017, 105, 51–73. [Google Scholar] [CrossRef]
- Gaskill, P.J.; Yano, H.H.; Kalpana, G.V.; Javitch, J.A.; Berman, J.W. Dopamine receptor activation increases HIV entry into primary human macrophages. PLoS ONE 2014, 9, e108232. [Google Scholar] [CrossRef]
- Channer, B.; Matt, S.M.; Nickoloff-Bybel, E.A.; Pappa, V.; Agarwal, Y.; Wickman, J.; Gaskill, P.J. Dopamine, Immunity, and Disease. Pharmacol. Rev. 2023, 75, 62–158. [Google Scholar]
- Gaskill, P.J.; Khoshbouei, H. Dopamine and norepinephrine are embracing their immune side and so should we. Curr. Opin. Neurobiol. 2022, 77, 102626. [Google Scholar] [CrossRef] [PubMed]
- Nickoloff-Bybel, E.A.; Festa, L.; Meucci, O.; Gaskill, P.J. Co-receptor signaling in the pathogenesis of neuroHIV. Retrovirology 2021, 18, 24. [Google Scholar] [CrossRef] [PubMed]
- Heaton, R.K.; Franklin, D.R., Jr.; Deutsch, R.; Letendre, S.; Ellis, R.J.; Casaletto, K.; Marquine, M.J.; Woods, S.P.; Vaida, F.; Atkinson, J.H.; et al. Neurocognitive change in the era of HIV combination antiretroviral therapy: The longitudinal CHARTER study. Clin. Infect. Dis. 2015, 60, 473–480. [Google Scholar] [CrossRef]
- Chang, L.; Ernst, T.; Speck, O.; Grob, C.S. Additive effects of HIV and chronic methamphetamine use on brain metabolite abnormalities. Am. J. Psychiatry 2005, 162, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Basu, B.; Sarkar, C.; Chakroborty, D.; Ganguly, S.; Shome, S.; Dasgupta, P.S.; Basu, S. D1 and D2 dopamine receptor-mediated inhibition of activated normal T cell proliferation is lost in jurkat T leukemic cells. J. Biol. Chem. 2010, 285, 27026–27032. [Google Scholar] [CrossRef]
- Basu, S.; Dasgupta, P.S. Dopamine, a neurotransmitter, influences the immune system. J. Neuroimmunol. 2000, 102, 113–124. [Google Scholar] [CrossRef]
- Cosentino, M.; Ferrari, M.; Kustrimovic, N.; Rasini, E.; Marino, F. Influence of dopamine receptor gene polymorphisms on circulating T lymphocytes: A pilot study in healthy subjects. Hum. Immunol. 2015, 76, 747–752. [Google Scholar] [CrossRef]
- Cosentino, M.; Marino, F.; Bombelli, R.; Ferrari, M.; Lecchini, S.; Frigo, G. Unravelling dopamine (and catecholamine) physiopharmacology in lymphocytes: Open questions. Trends Immunol. 2003, 24, 581–582. [Google Scholar] [CrossRef]
- Cosentino, M.; Rasini, E.; Colombo, C.; Marino, F.; Blandini, F.; Ferrari, M.; Samuele, A.; Lecchini, S.; Nappi, G.; Frigo, G. Dopaminergic modulation of oxidative stress and apoptosis in human peripheral blood lymphocytes: Evidence for a D1-like receptor-dependent protective effect. Free Radic. Biol. Med. 2004, 36, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Cosentino, M.; Zaffaroni, M.; Legnaro, M.; Bombelli, R.; Schembri, L.; Baroncini, D.; Bianchi, A.; Clerici, R.; Guidotti, M.; Banfi, P.; et al. Dopaminergic receptors and adrenoceptors in circulating lymphocytes as putative biomarkers for the early onset and progression of multiple sclerosis. J. Neuroimmunol. 2016, 298, 82–89. [Google Scholar] [CrossRef]
- Dominguez-Meijide, A.; Rodriguez-Perez, A.I.; Diaz-Ruiz, C.; Guerra, M.J.; Labandeira-Garcia, J.L. Dopamine modulates astroglial and microglial activity via glial renin-angiotensin system in cultures. Brain Behav. Immun. 2017, 62, 277–290. [Google Scholar] [CrossRef]
- Scheller, C.; Sopper, S.; Jassoy, C.; ter Meulen, V.; Riederer, P.; Koutsilieri, E. Dopamine activates HIV in chronically infected T lymphoblasts. J. Neural Transm. 2000, 107, 1483–1489. [Google Scholar] [CrossRef] [PubMed]
- Marcondes, F.C.; Watry, D.D.; Zandonatti, M.; Fox, H.S. Methamphetamine increases brain viral load and activates NK cells in SIV-infected monkeys. Am. J. Pathol. 2010. in Press. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J.; Childers, M.E.; Cherner, M.; Lazzaretto, D.; Letendre, S.; Grant, I.; HIV Neurobehavioral Research Center Group. Increased human immunodeficiency virus loads in active methamphetamine users are explained by reduced effectiveness of antiretroviral therapy. J. Infect. Dis. 2003, 188, 1820–1826. [Google Scholar] [CrossRef]
- Cohn, L.B.; da Silva, I.T.; Valieris, R.; Huang, A.S.; Lorenzi, J.C.C.; Cohen, Y.Z.; Pai, J.A.; Butler, A.L.; Caskey, M.; Jankovic, M.; et al. Clonal CD4+ T cells in the HIV-1 latent reservoir display a distinct gene profile upon reactivation. Nat. Med. 2018, 24, 604–609. [Google Scholar] [CrossRef]
- Cohn, L.B.; Silva, I.T.; Oliveira, T.Y.; Rosales, R.A.; Parrish, E.H.; Learn, G.H.; Hahn, B.H.; Czartoski, J.L.; McElrath, M.J.; Lehmann, C.; et al. HIV-1 integration landscape during latent and active infection. Cell 2015, 160, 420–432. [Google Scholar] [CrossRef]
- Folks, T.M.; Justement, J.; Kinter, A.; Dinarello, C.A.; Fauci, A.S. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science 1987, 238, 800–802. [Google Scholar] [CrossRef]
- Matt, S.M.; Gaskill, P.J. Where Is Dopamine and how do Immune Cells See it?: Dopamine-Mediated Immune Cell Function in Health and Disease. J. Neuroimmune Pharmacol. 2020, 15, 114–164. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, S.; Draghici, S. Identifying Significantly Impacted Pathways and Putative Mechanisms with iPathwayGuide. Curr. Protoc. Bioinform. 2017, 57, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Cline, M.S.; Smoot, M.; Cerami, E.; Kuchinsky, A.; Landys, N.; Workman, C.; Christmas, R.; Avila-Campilo, I.; Creech, M.; Gross, B.; et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007, 2, 2366–2382. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014, 15, 550. [Google Scholar] [CrossRef]
- Montojo, J.; Zuberi, K.; Rodriguez, H.; Bader, G.D.; Morris, Q. GeneMANIA: Fast gene network construction and function prediction for Cytoscape. F1000Res 2014, 3, 153. [Google Scholar] [CrossRef]
- Montojo, J.; Zuberi, K.; Rodriguez, H.; Kazi, F.; Wright, G.; Donaldson, S.L.; Morris, Q.; Bader, G.D. GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics 2010, 26, 2927–2928. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef]
- Oughtred, R.; Stark, C.; Breitkreutz, B.J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef] [PubMed]
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Zak, D.E.; Andersen-Nissen, E.; Peterson, E.R.; Sato, A.; Hamilton, M.K.; Borgerding, J.; Krishnamurty, A.T.; Chang, J.T.; Adams, D.J.; Hensley, T.R.; et al. Merck Ad5/HIV induces broad innate immune activation that predicts CD8+ T-cell responses but is attenuated by preexisting Ad5 immunity. Proc. Natl. Acad. Sci. USA 2012, 109, E3503–E3512. [Google Scholar] [CrossRef]
- Smith, D.B.; Simmonds, P.; Bell, J.E. Brain viral burden, neuroinflammation and neurodegeneration in HAART-treated HIV positive injecting drug users. J. Neurovirol 2014, 20, 28–38. [Google Scholar] [CrossRef]
- Clouse, K.A.; Powell, D.; Washington, I.; Poli, G.; Strebel, K.; Farrar, W.; Barstad, P.; Kovacs, J.; Fauci, A.S.; Folks, T.M. Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. J. Immunol. 1989, 142, 431–438. [Google Scholar] [CrossRef]
- Fujinaga, K.; Cary, D.C. Experimental Systems for Measuring HIV Latency and Reactivation. Viruses 2020, 12, 1279. [Google Scholar] [CrossRef]
- Leibbrandt, M.E.; Koropatnick, J. Activation of human monocytes with lipopolysaccharide induces metallothionein expression and is diminished by zinc. Toxicol. Appl. Pharmacol. 1994, 124, 72–81. [Google Scholar] [CrossRef]
- Puts, G.S.; Leonard, M.K.; Pamidimukkala, N.V.; Snyder, D.E.; Kaetzel, D.M. Nuclear functions of NME proteins. Lab. Investig. 2018, 98, 211–218. [Google Scholar] [CrossRef]
- Massanella, M.; Fromentin, R.; Chomont, N. Residual inflammation and viral reservoirs: Alliance against an HIV cure. Curr. Opin. HIV AIDS 2016, 11, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Nolan, R.A.; Reeb, K.L.; Rong, Y.; Matt, S.M.; Johnson, H.S.; Runner, K.; Gaskill, P.J. Dopamine activates NF-kappaB and primes the NLRP3 inflammasome in primary human macrophages. Brain Behav. Immun. Health 2020, 2, 100030. [Google Scholar] [CrossRef] [PubMed]
- Herbein, G.; Varin, A. The macrophage in HIV-1 infection: From activation to deactivation? Retrovirology 2010, 7, 33. [Google Scholar] [CrossRef]
- Wu, Y.; Hu, Y.; Wang, B.; Li, S.; Ma, C.; Liu, X.; Moynagh, P.N.; Zhou, J.; Yang, S. Dopamine Uses the DRD5-ARRB2-PP2A Signaling Axis to Block the TRAF6-Mediated NF-κB Pathway and Suppress Systemic Inflammation. Mol. Cell 2020, 78, 42–56.e6. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Sugino, Y.; Shibagaki, F.; Yamamuro, A.; Ishimaru, Y.; Maeda, S. Dopamine attenuates lipopolysaccharide-induced expression of proinflammatory cytokines by inhibiting the nuclear translocation of NF-kappaB p65 through the formation of dopamine quinone in microglia. Eur. J. Pharmacol. 2020, 866, 172826. [Google Scholar] [CrossRef]
- Mogensen, T.H.; Melchjorsen, J.; Larsen, C.S.; Paludan, S.R. Innate immune recognition and activation during HIV infection. Retrovirology 2010, 7, 54. [Google Scholar] [CrossRef]
- Bortell, N.; Basova, L.; Najera, J.A.; Morsey, B.; Fox, H.S.; Marcondes, M.C.G. Sirtuin 1-Chromatin-Binding Dynamics Points to a Common Mechanism Regulating Inflammatory Targets in SIV Infection and in the Aging Brain. J. Neuroimmune Pharmacol. 2018, 13, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Emiliani, S.; Fischle, W.; Ott, M.; Van Lint, C.; Amella, C.A.; Verdin, E. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol. 1998, 72, 1666–1670. [Google Scholar] [CrossRef]
- Wang, X.; Sun, B.; Mbondji, C.; Biswas, S.; Zhao, J.; Hewlett, I. Differences in Activation of HIV-1 Replication by Superinfection With HIV-1 and HIV-2 in U1 Cells. J. Cell Physiol. 2017, 232, 1746–1753. [Google Scholar] [CrossRef]
- Biswas, P.; Poli, G.; Kinter, A.L.; Justement, J.S.; Stanley, S.K.; Maury, W.J.; Bressler, P.; Orenstein, J.M.; Fauci, A.S. Interferon gamma induces the expression of human immunodeficiency virus in persistently infected promonocytic cells (U1) and redirects the production of virions to intracytoplasmic vacuoles in phorbol myristate acetate-differentiated U1 cells. J. Exp. Med. 1992, 176, 739–750. [Google Scholar] [CrossRef]
- Devadas, K.; Hardegen, N.J.; Wahl, L.M.; Hewlett, I.K.; Clouse, K.A.; Yamada, K.M.; Dhawan, S. Mechanisms for macrophage-mediated HIV-1 induction. J. Immunol. 2004, 173, 6735–6744. [Google Scholar] [CrossRef] [PubMed]
- Symons, J.; Chopra, A.; Malatinkova, E.; De Spiegelaere, W.; Leary, S.; Cooper, D.; Abana, C.O.; Rhodes, A.; Rezaei, S.D.; Vandekerckhove, L.; et al. HIV integration sites in latently infected cell lines: Evidence of ongoing replication. Retrovirology 2017, 14, 2, Erratum in Retrovirology 2017, 14, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Zhao, X.; Zhang, Y.; Li, Y.; Liu, S.; Han, J.; Sun, Z.; Wang, C.; Deng, D.; Wang, S.; et al. Single cell multi-omics reveal intra-cell-line heterogeneity across human cancer cell lines. Nat. Commun. 2023, 14, 8170. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, L.; Nazlie Mohebiany, A.; Premereur, J.; Polanco Miquel, P.; Bijnens, B.; Van de Walle, P.; Fattorelli, N.; Mancuso, R. Microglia heterogeneity, modeling and cell-state annotation in development and neurodegeneration. Nat. Neurosci. 2025, 1–12. [Google Scholar] [CrossRef]
- Healy, L.M.; Zia, S.; Plemel, J.R. Towards a definition of microglia heterogeneity. Commun. Biol. 2022, 5, 1114. [Google Scholar] [CrossRef]
- Schlachetzki, J.C.; Gianella, S.; Ouyang, Z.; Lana, A.J.; Yang, X.; O’Brien, S.; Challacombe, J.F.; Gaskill, P.J.; Jordan-Sciutto, K.L.; Chaillon, A.; et al. Gene expression and chromatin conformation of microglia in virally suppressed people with HIV. Life Sci. Alliance 2024, 7, e202403085. [Google Scholar] [CrossRef] [PubMed]
- Basova, L.V.; Bortell, N.; Conti, B.; Fox, H.S.; Milner, R.; Marcondes, M.C.G. Age-associated changes in microglia activation and Sirtuin-1- chromatin binding patterns. Aging 2022, 14, 8205–8220. [Google Scholar] [CrossRef]
- Basova, L.; Lindsey, A.; McGovern, A.M.; Ellis, R.J.; Marcondes, M.C.G. Detection of H3K4me3 Identifies NeuroHIV Signatures, Genomic Effects of Methamphetamine and Addiction Pathways in Postmortem HIV+ Brain Specimens that Are Not Amenable to Transcriptome Analysis. Viruses 2021, 13, 544. [Google Scholar] [CrossRef]

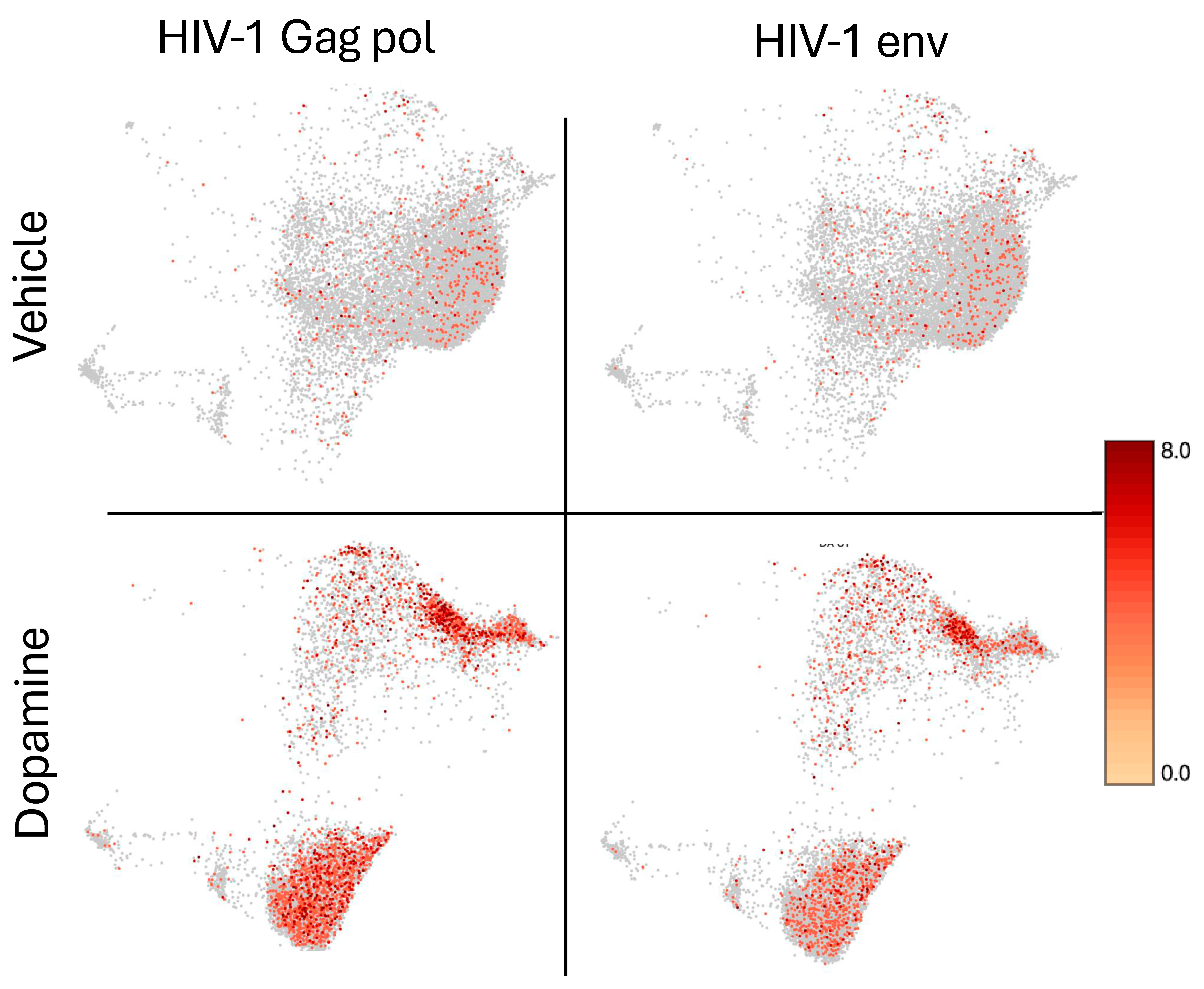
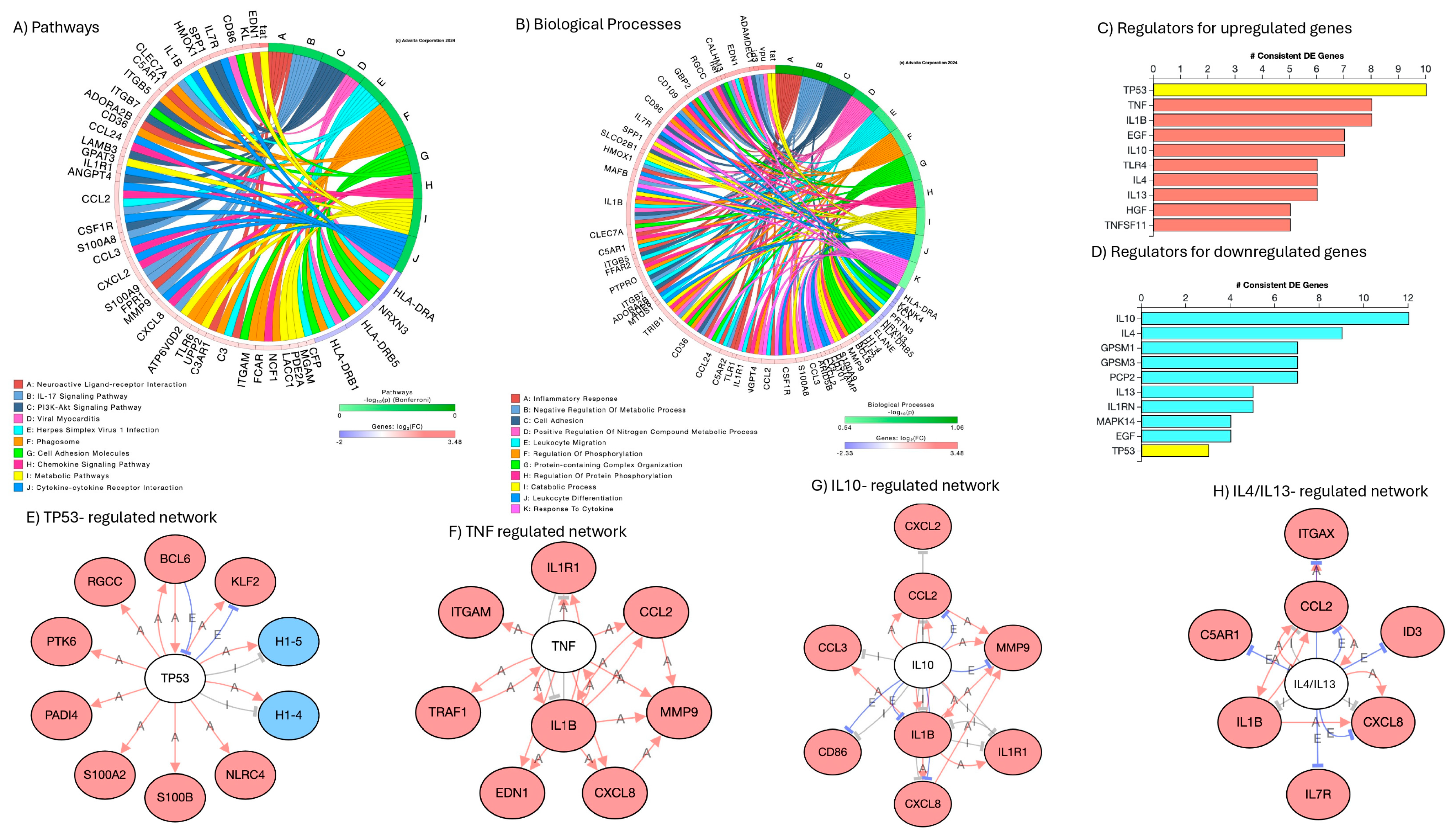
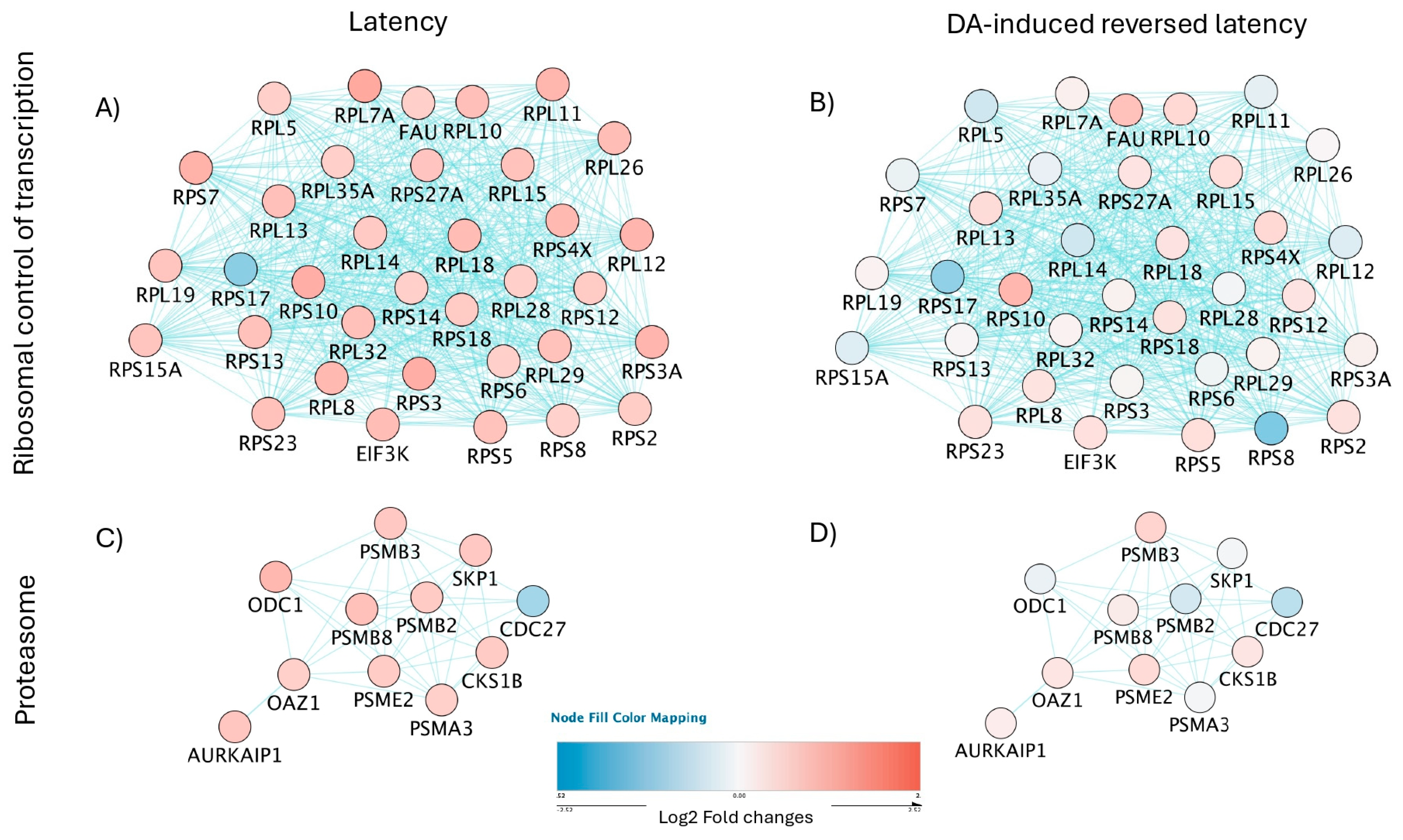
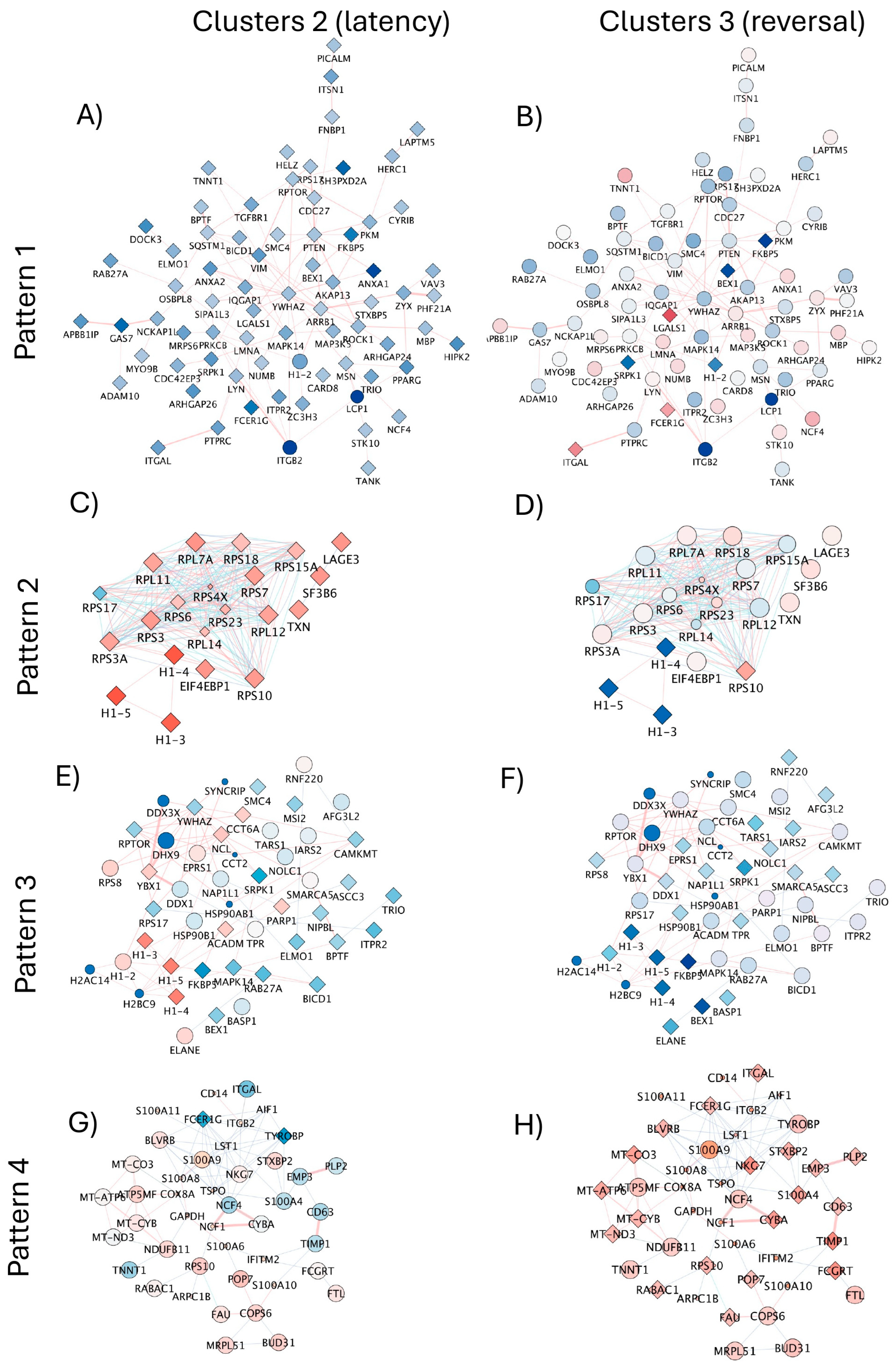
| Estimated Number of Cells | Total Genes Detected | Median UMI Count per Cell | Median Genes per Cell | |
|---|---|---|---|---|
| U1 NT | 11,322 | 15,430 | 12,664 | 3349 |
| U1 DA | 8706 | 15,222 | 4702 | 1875 |
| U937 NT | 8721 | 24,571 | 8852 | 3106 |
| U937 DA | 11,933 | 24,866 | 7518 | 2858 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basova, L.V.; Lim, W.L.; Delorme-Walker, V.; Riley, T.; Au, K.; Lima, D.S.; Lusic, M.; Ellis, R.J.; Fox, H.S.; Marcondes, M.C.G. A Single-Cell Perspective on the Effects of Dopamine in the Regulation of HIV Latency Phenotypes in a Myeloid Cell Model. Viruses 2025, 17, 895. https://doi.org/10.3390/v17070895
Basova LV, Lim WL, Delorme-Walker V, Riley T, Au K, Lima DS, Lusic M, Ellis RJ, Fox HS, Marcondes MCG. A Single-Cell Perspective on the Effects of Dopamine in the Regulation of HIV Latency Phenotypes in a Myeloid Cell Model. Viruses. 2025; 17(7):895. https://doi.org/10.3390/v17070895
Chicago/Turabian StyleBasova, Liana V., Wei Ling Lim, Violaine Delorme-Walker, Tera Riley, Kaylin Au, Daniel Siqueira Lima, Marina Lusic, Ronald J. Ellis, Howard S. Fox, and Maria Cecilia Garibaldi Marcondes. 2025. "A Single-Cell Perspective on the Effects of Dopamine in the Regulation of HIV Latency Phenotypes in a Myeloid Cell Model" Viruses 17, no. 7: 895. https://doi.org/10.3390/v17070895
APA StyleBasova, L. V., Lim, W. L., Delorme-Walker, V., Riley, T., Au, K., Lima, D. S., Lusic, M., Ellis, R. J., Fox, H. S., & Marcondes, M. C. G. (2025). A Single-Cell Perspective on the Effects of Dopamine in the Regulation of HIV Latency Phenotypes in a Myeloid Cell Model. Viruses, 17(7), 895. https://doi.org/10.3390/v17070895







