The First African Swine Fever Viruses Detected in Wild Boar in Hong Kong, 2021–2023
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Viral DNA Extraction, Real-Time PCR, and Initial ASFV Genotyping
2.3. Virus Isolation and Hemadsorption at the Pirbright Institute
2.4. Whole-Genome Sequencing and Analysis
2.5. Conventional PCR and Sanger Sequencing
2.6. Phylogenetic Analysis
2.7. Recombination Analysis
3. Results
3.1. ASF-Positive Wild Boar Carcasses and Their Distributions
3.2. ASFV p72 Genotyping, Virus Isolation, and HAD
3.3. Characterization of the Complete Genome Sequences of ASFVs
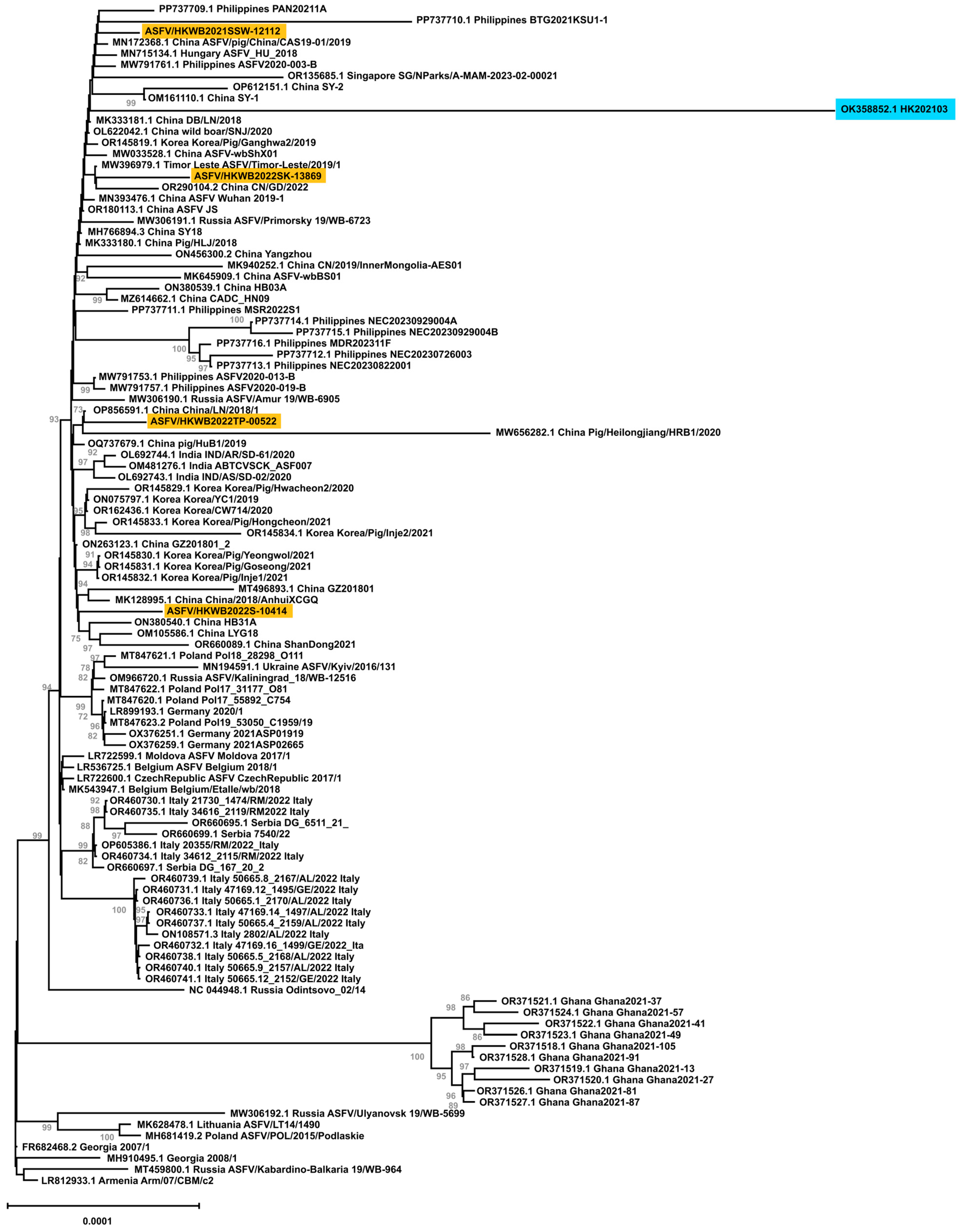
3.4. Whole-Genome Phylogenetic Analysis
3.5. Recombination Analysis of the Four ASFV Strains Identified from Wild Boar
3.6. Data Availability
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yanez, R.J.; Rodriguez, J.M.; Nogal, M.L.; Yuste, L.; Enriquez, C.; Rodriguez, J.F.; Vinuela, E. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 1995, 208, 249–278. [Google Scholar] [CrossRef]
- Le, V.P.; Ahn, M.J.; Kim, J.S.; Jung, M.C.; Yoon, S.W.; Trinh, T.B.N.; Le, T.N.; Kim, H.K.; Kang, J.A.; Lim, J.W.; et al. A Whole-Genome Analysis of the African Swine Fever Virus That Circulated during the First Outbreak in Vietnam in 2019 and Subsequently in 2022. Viruses 2023, 15, 1945. [Google Scholar] [CrossRef]
- de Villiers, E.P.; Gallardo, C.; Arias, M.; da Silva, M.; Upton, C.; Martin, R.; Bishop, R.P. Phylogenomic analysis of 11 complete African swine fever virus genome sequences. Virology 2010, 400, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Goatley, L.C.; Freimanis, G.L.; Tennakoon, C.; Bastos, A.; Heath, L.; Netherton, C.L. African swine fever virus NAM P1/95 is a mixture of genotype I and genotype VIII viruses. Microbiol. Resour. Announc. 2024, 13, e0006724. [Google Scholar] [CrossRef] [PubMed]
- Spinard, E.; Dinhobl, M.; Tesler, N.; Birtley, H.; Signore, A.V.; Ambagala, A.; Masembe, C.; Borca, M.V.; Gladue, D.P. A Re-Evaluation of African Swine Fever Genotypes Based on p72 Sequences Reveals the Existence of Only Six Distinct p72 Groups. Viruses 2023, 15, 2246. [Google Scholar] [CrossRef]
- Dinhobl, M.; Spinard, E.; Tesler, N.; Birtley, H.; Signore, A.; Ambagala, A.; Masembe, C.; Borca, M.V.; Gladue, D.P. Reclassification of ASFV into 7 Biotypes Using Unsupervised Machine Learning. Viruses 2023, 16, 67. [Google Scholar] [CrossRef]
- Penrith, M.-L.; Nyakahuma, D. Recognizing African Swine Fever: A Field Manual; Food and Agriculture Organization of the United Nations: Rome, Italy, 2000. [Google Scholar]
- Njau, E.P.; Machuka, E.M.; Cleaveland, S.; Shirima, G.M.; Kusiluka, L.J.; Okoth, E.A.; Pelle, R. African Swine Fever Virus (ASFV): Biology, Genomics and Genotypes Circulating in Sub-Saharan Africa. Viruses 2021, 13, 2285. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, J.M.; Azevedo, J.R. La peste porcine africaine au Portugal. Bull.-Off. Int. Des. Épizooties 1961, 55, 88–106. [Google Scholar]
- Danzetta, M.L.; Marenzoni, M.L.; Iannetti, S.; Tizzani, P.; Calistri, P.; Feliziani, F. African Swine Fever: Lessons to Learn From Past Eradication Experiences. A Systematic Review. Front. Vet. Sci. 2020, 7, 296. [Google Scholar] [CrossRef]
- Ge, S.; Li, J.; Fan, X.; Liu, F.; Li, L.; Wang, Q.; Ren, W.; Bao, J.; Liu, C.; Wang, H.; et al. Molecular Characterization of African Swine Fever Virus, China, 2018. Emerg. Infect. Dis. 2018, 24, 2131–2133. [Google Scholar] [CrossRef]
- Xin, G.; Kuang, Q.; Le, S.; Wu, W.; Gao, Q.; Gao, H.; Xu, Z.; Zheng, Z.; Lu, G.; Gong, L.; et al. Origin, genomic diversity and evolution of African swine fever virus in East Asia. Virus Evol. 2023, 9, vead060. [Google Scholar] [CrossRef] [PubMed]
- Sun, E.; Zhang, Z.; Wang, Z.; He, X.; Zhang, X.; Wang, L.; Wang, W.; Huang, L.; Xi, F.; Huangfu, H.; et al. Emergence and prevalence of naturally occurring lower virulent African swine fever viruses in domestic pigs in China in 2020. Sci. China Life Sci. 2021, 64, 752–765. [Google Scholar] [CrossRef] [PubMed]
- Sun, E.; Huang, L.; Zhang, X.; Zhang, J.; Shen, D.; Zhang, Z.; Wang, Z.; Huo, H.; Wang, W.; Huangfu, H.; et al. Genotype I African swine fever viruses emerged in domestic pigs in China and caused chronic infection. Emerg. Microbes Infect. 2021, 10, 2183–2193. [Google Scholar] [CrossRef]
- Shi, K.; Liu, H.; Yin, Y.; Si, H.; Long, F.; Feng, S. Molecular Characterization of African Swine Fever Virus From 2019-2020 Outbreaks in Guangxi Province, Southern China. Front. Vet. Sci. 2022, 9, 912224. [Google Scholar] [CrossRef]
- Zhao, D.; Sun, E.; Huang, L.; Ding, L.; Zhu, Y.; Zhang, J.; Shen, D.; Zhang, X.; Zhang, Z.; Ren, T.; et al. Highly lethal genotype I and II recombinant African swine fever viruses detected in pigs. Nat. Commun. 2023, 14, 3096. [Google Scholar] [CrossRef]
- Le, V.P.; Nguyen, V.T.; Le, T.B.; Mai, N.T.A.; Nguyen, V.D.; Than, T.T.; Lai, T.N.H.; Cho, K.H.; Hong, S.K.; Kim, Y.H.; et al. Detection of Recombinant African Swine Fever Virus Strains of p72 Genotypes I and II in Domestic Pigs, Vietnam, 2023. Emerg. Infect. Dis. 2024, 30, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Igolkin, A.; Mazloum, A.; Zinyakov, N.; Chernyshev, R.; Schalkwyk, A.V.; Shotin, A.; Lavrentiev, I.; Gruzdev, K.; Chvala, I. Detection of the first recombinant African swine fever virus (genotypes I and II) in domestic pigs in Russia. Mol. Biol. Rep. 2024, 51, 1011. [Google Scholar] [CrossRef]
- Denstedt, E.; Porco, A.; Hwang, J.; Nga, N.T.T.; Ngoc, P.T.B.; Chea, S.; Khammavong, K.; Milavong, P.; Sours, S.; Osbjer, K.; et al. Detection of African swine fever virus in free-ranging wild boar in Southeast Asia. Transbound. Emerg. Dis. 2021, 68, 2669–2675. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.-S.; Andraud, M.; Kim, E.; Vergne, T. Three Years of African Swine Fever in South Korea (2019–2021): A Scoping Review of Epidemiological Understanding. Transbound. Emerg. Dis. 2023, 2023, 4686980. [Google Scholar] [CrossRef]
- Luskin, M.S.; Moore, J.H.; Mendes, C.P.; Nasardin, M.B.; Onuma, M.; Davies, S.J. The mass mortality of Asia’s native pigs induced by African swine fever. Wildl. Lett. 2023, 1, 8–14. [Google Scholar] [CrossRef]
- Koh, E.Y.; Tan, A.K.S.; Yeo, D.; Lau, C.; Tan, L.Y.; Ng, O.W.; Ong, J.; Chong, S.; Toh, S.; Chen, J.; et al. Detection of African Swine Fever Virus from Wild Boar, Singapore, 2023. Emerg. Infect. Dis. 2023, 29, 2580–2583. [Google Scholar] [CrossRef]
- Ito, S.; Bosch, J.; Jeong, H.; Aguilar-Vega, C.; Park, J.; Martinez-Aviles, M.; Sanchez-Vizcaino, J.M. Spatio-Temporal Epidemiology of the Spread of African Swine Fever in Wild Boar and the Role of Environmental Factors in South Korea. Viruses 2022, 14, 2779. [Google Scholar] [CrossRef]
- Guberti, V.; Khomenko, S.; Masiulis, M.; Kerba, S. African Swine Fever in Wild Boar Ecology and Biosecurity; FAO Animal Production and Health Manual No. 22; FAO: Rome, Italy; OIE: Paris, France; EC: Singapore, 2019. [Google Scholar]
- LCQ14; Nuisances Caused by Wild Pigs. The Government of the Hong Kong Special Adminstrative Region: Hong Kong, China, 2024. Available online: https://www.info.gov.hk/gia/general/202403/13/P2024031300268.htm (accessed on 24 February 2025).
- Agriculture Fisheries and Conservation Department The Government of the Hong Kong Special Adminstrative Region. Long-Term Control of Wild Pig Population. 2021. Available online: https://www.districtcouncils.gov.hk/central/doc/2020_2023/en/committee_meetings_doc/behwc/20495/20211118_BEHWC_Paper_77_2021_Annex_I_En.pdf (accessed on 8 May 2025).
- Centre for Food Safety The Government of the Hong Kong Special Administrative Region. Government Announces ASF Testing Results. 2019. Available online: https://www.cfs.gov.hk/english/press/20190918_7627.html (accessed on 8 May 2025).
- Food and Health Bureau Agriculture Fisheries and Conservation Department Food and Environmental Hygiene Department The Government of the Hong Kong Special Administrative Region. Work in Response to African Swine Fever Cases in Hong Kong. 2019. Available online: https://www.eeb.gov.hk/food/download/committees/acfeh/doc/2019/paper20190905_05.pdf (accessed on 8 May 2025).
- Go, Y.Y.; Ho, J.H.P.; Tam, K.W.S.; Kamali, M.; Zhang, Y.; Lau, C.C.Y.; Li, S.H.; Wilson, M.T.; Guo, Z.; Li, R.; et al. Investigation of the First African Swine Fever Outbreak in a Domestic Pig Farm in Hong Kong. Transbound. Emerg. Dis. 2023, 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- King, D.P.; Reid, S.M.; Hutchings, G.H.; Grierson, S.S.; Wilkinson, P.J.; Dixon, L.K.; Bastos, A.D.; Drew, T.W. Development of a TaqMan PCR assay with internal amplification control for the detection of African swine fever virus. J. Virol. Methods 2003, 107, 53–61. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health. Terrestrial Manual, Chapter 3.9.1. African Swine Fever (Infection with African Swine Fever Virus). 2024. Available online: https://www.woah.org/fileadmin/Home/fr/Health_standards/tahm/3.09.01_ASF.pdf (accessed on 8 May 2025).
- Bastos, A.D.; Penrith, M.L.; Cruciere, C.; Edrich, J.L.; Hutchings, G.; Roger, F.; Couacy-Hymann, E.; Thomson, G.R. Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Arch. Virol. 2003, 148, 693–706. [Google Scholar] [CrossRef]
- Zhong, H.; Fan, S.; Du, Y.; Zhang, Y.; Zhang, A.; Jiang, D.; Han, S.; Wan, B.; Zhang, G. African Swine Fever Virus MGF110-7L Induces Host Cell Translation Suppression and Stress Granule Formation by Activating the PERK/PKR-eIF2alpha Pathway. Microbiol. Spectr. 2022, 10, e0328222. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. 2013. Available online: http://arxiv.org/abs/1303.3997 (accessed on 8 May 2025).
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Gangavarapu, K.; Quick, J.; Matteson, N.L.; De Jesus, J.G.; Main, B.J.; Tan, A.L.; Paul, L.M.; Brackney, D.E.; Grewal, S.; et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019, 20, 8. [Google Scholar] [CrossRef]
- Tcherepanov, V.; Ehlers, A.; Upton, C. Genome Annotation Transfer Utility (GATU): Rapid annotation of viral genomes using a closely related reference genome. BMC Genom. 2006, 7, 150. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez, R.L.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.K.; Chapman, D.A.; Netherton, C.L.; Upton, C. African swine fever virus replication and genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef]
- Powell, P.P.; Dixon, L.K.; Parkhouse, R.M. An IkappaB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J. Virol. 1996, 70, 8527–8533. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.; Rybicki, E. RDP: Detection of recombination amongst aligned sequences. Bioinformatics 2000, 16, 562–563. [Google Scholar] [CrossRef]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible emergence of new geminiviruses by frequent recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef]
- Martin, D.P.; Posada, D.; Crandall, K.A.; Williamson, C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retroviruses 2005, 21, 98–102. [Google Scholar] [CrossRef]
- Smith, J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-scanning: A Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef]
- Lam, H.M.; Ratmann, O.; Boni, M.F. Improved Algorithmic Complexity for the 3SEQ Recombination Detection Algorithm. Mol. Biol. Evol. 2018, 35, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Nefedeva, M.; Titov, I.; Tsybanov, S.; Malogolovkin, A. Recombination shapes African swine fever virus serotype-specific locus evolution. Sci. Rep. 2020, 10, 18474. [Google Scholar] [CrossRef] [PubMed]
- Reis, A.L.; Abrams, C.C.; Goatley, L.C.; Netherton, C.; Chapman, D.G.; Sanchez-Cordon, P.; Dixon, L.K. Deletion of African swine fever virus interferon inhibitors from the genome of a virulent isolate reduces virulence in domestic pigs and induces a protective response. Vaccine 2016, 34, 4698–4705. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, H.; Yang, C.; Gao, Y.; Yu, X.; Chen, X.; Cui, R.; Zheng, L.; Li, S.; Li, X.; et al. Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues. Nat. Commun. 2019, 10, 387. [Google Scholar] [CrossRef]
- Bosch-Camos, L.; Lopez, E.; Collado, J.; Navas, M.J.; Blanco-Fuertes, M.; Pina-Pedrero, S.; Accensi, F.; Salas, M.L.; Mundt, E.; Nikolin, V.; et al. M448R and MGF505-7R: Two African Swine Fever Virus Antigens Commonly Recognized by ASFV-Specific T-Cells and with Protective Potential. Vaccines 2021, 9, 508. [Google Scholar] [CrossRef]
- Ran, Y.; Li, D.; Xiong, M.G.; Liu, H.N.; Feng, T.; Shi, Z.W.; Li, Y.H.; Wu, H.N.; Wang, S.Y.; Zheng, H.X.; et al. African swine fever virus I267L acts as an important virulence factor by inhibiting RNA polymerase III-RIG-I-mediated innate immunity. PLoS Pathog. 2022, 18, e1010270. [Google Scholar] [CrossRef]
- Netherton, C.; Rouiller, I.; Wileman, T. The subcellular distribution of multigene family 110 proteins of African swine fever virus is determined by differences in C-terminal KDEL endoplasmic reticulum retention motifs. J. Virol. 2004, 78, 3710–3721. [Google Scholar] [CrossRef] [PubMed]
- Coelho, J.; Martins, C.; Ferreira, F.; Leitao, A. African swine fever virus ORF P1192R codes for a functional type II DNA topoisomerase. Virology 2015, 474, 82–93. [Google Scholar] [CrossRef]
- Galindo, I.; Almazan, F.; Bustos, M.J.; Vinuela, E.; Carrascosa, A.L. African swine fever virus EP153R open reading frame encodes a glycoprotein involved in the hemadsorption of infected cells. Virology 2000, 266, 340–351. [Google Scholar] [CrossRef]
- Dodantenna, N.; Cha, J.W.; Chathuranga, K.; Chathuranga, W.A.G.; Weerawardhana, A.; Ranathunga, L.; Kim, Y.; Jheong, W.; Lee, J.S. The African Swine Fever Virus Virulence Determinant DP96R Suppresses Type I IFN Production Targeting IRF3. Int. J. Mol. Sci. 2024, 25, 2099. [Google Scholar] [CrossRef]
- Shao, Z.; Su, S.; Yang, J.; Zhang, W.; Gao, Y.; Zhao, X.; Zhang, Y.; Shao, Q.; Cao, C.; Li, H.; et al. Structures and implications of the C962R protein of African swine fever virus. Nucleic Acids Res. 2023, 51, 9475–9490. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Mai, Z.; Kong, C.; You, J.; Lin, S.; Gao, C.; Zhang, W.; Chen, X.; Xie, Q.; Wang, H.; et al. African swine fever virus pB475L evades host antiviral innate immunity via targeting STAT2 to inhibit IFN-I signaling. J. Biol. Chem. 2024, 300, 107472. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Yang, J.; Yang, B.; Hao, Y.; Shi, X.; Zhang, D.; Yang, X.; Zhang, T.; Zhao, D.; Yuan, X.; et al. Cyproheptadine hydrochloride inhibits African swine fever viral replication in vitro. Microb. Pathog. 2023, 175, 105957. [Google Scholar] [CrossRef]
- Dolata, K.M.; Pei, G.; Netherton, C.L.; Karger, A. Functional Landscape of African Swine Fever Virus-Host and Virus-Virus Protein Interactions. Viruses 2023, 15, 1634. [Google Scholar] [CrossRef]
- Wen, X.; He, X.; Zhang, X.; Zhang, X.; Liu, L.; Guan, Y.; Zhang, Y.; Bu, Z. Genome sequences derived from pig and dried blood pig feed samples provide important insights into the transmission of African swine fever virus in China in 2018. Emerg. Microbes Infect. 2019, 8, 303–306. [Google Scholar] [CrossRef]
- Vergne, T.; Guinat, C.; Pfeiffer, D.U. Undetected Circulation of African Swine Fever in Wild Boar, Asia. Emerg. Infect. Dis. 2020, 26, 2480–2482. [Google Scholar] [CrossRef]
- Cadenas-Fernandez, E.; Ito, S.; Aguilar-Vega, C.; Sanchez-Vizcaino, J.M.; Bosch, J. The Role of the Wild Boar Spreading African Swine Fever Virus in Asia: Another Underestimated Problem. Front. Vet. Sci. 2022, 9, 844209. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Y.; Gao, H.; Kuang, Q.Y.; Xing, J.B.; Wang, Z.Y.; Cao, X.Y.; Xu, S.J.; Liu, J.; Huang, Z.; Zheng, Z.Z.; et al. Clinical sequencing uncovers the genomic characteristics and mutation spectrum of the 2018 African swine fever virus in Guangdong, China. Front. Vet. Sci. 2022, 9, 978243. [Google Scholar] [CrossRef]
- Wang, X.; Wang, X.; Zhang, X.; He, S.; Chen, Y.; Liu, X.; Guo, C. Genetic Characterization and Variation of African Swine Fever Virus China/GD/2019 Strain in Domestic Pigs. Pathogens 2022, 11, 97. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, C.; Fernandez-Pinero, J.; Pelayo, V.; Gazaev, I.; Markowska-Daniel, I.; Pridotkas, G.; Nieto, R.; Fernandez-Pacheco, P.; Bokhan, S.; Nevolko, O.; et al. Genetic variation among African swine fever genotype II viruses, eastern and central Europe. Emerg. Infect. Dis. 2014, 20, 1544–1547. [Google Scholar] [CrossRef]
- Gallardo, C.; Mwaengo, D.M.; Macharia, J.M.; Arias, M.; Taracha, E.A.; Soler, A.; Okoth, E.; Martin, E.; Kasiti, J.; Bishop, R.P. Enhanced discrimination of African swine fever virus isolates through nucleotide sequencing of the p54, p72, and pB602L (CVR) genes. Virus Genes 2009, 38, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Simulundu, E.; Lubaba, C.H.; van Heerden, J.; Kajihara, M.; Mataa, L.; Chambaro, H.M.; Sinkala, Y.; Munjita, S.M.; Munang’andu, H.M.; Nalubamba, K.S.; et al. The Epidemiology of African Swine Fever in “Nonendemic” Regions of Zambia (1989-2015): Implications for Disease Prevention and Control. Viruses 2017, 9, 236. [Google Scholar] [CrossRef]
- Gallardo, C.; Casado, N.; Soler, A.; Djadjovski, I.; Krivko, L.; Madueno, E.; Nieto, R.; Perez, C.; Simon, A.; Ivanova, E.; et al. A multi gene-approach genotyping method identifies 24 genetic clusters within the genotype II-European African swine fever viruses circulating from 2007 to 2022. Front. Vet. Sci. 2023, 10, 1112850. [Google Scholar] [CrossRef]
- Vilem, A.; Nurmoja, I.; Niine, T.; Riit, T.; Nieto, R.; Viltrop, A.; Gallardo, C. Molecular Characterization of African Swine Fever Virus Isolates in Estonia in 2014–2019. Pathogens 2020, 9, 582. [Google Scholar] [CrossRef]
- Mazloum, A.; van Schalkwyk, A.; Chernyshev, R.; Igolkin, A.; Heath, L.; Sprygin, A. A Guide to Molecular Characterization of Genotype II African Swine Fever Virus: Essential and Alternative Genome Markers. Microorganisms 2023, 11, 642. [Google Scholar] [CrossRef]
- Cho, K.H.; Yoo, D.S.; Hong, S.K.; Kim, D.Y.; Jang, M.K.; Kang, H.E.; Kim, Y.H. Genetic Profile of African Swine Fever Viruses Circulating at Pig Farms in South Korea during the Outbreaks between 2022 and April 2023. Viruses 2023, 15, 1552. [Google Scholar] [CrossRef]
- Kim, G.; Park, J.E.; Kim, S.J.; Kim, Y.; Kim, W.; Kim, Y.K.; Jheong, W. Complete genome analysis of the African swine fever virus isolated from a wild boar responsible for the first viral outbreak in Korea, 2019. Front. Vet. Sci. 2022, 9, 1080397. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, S.I.; Jeong, H.G.; Yoo, J.; Jeong, H.; Choi, Y.; Son, K.; Jheong, W.H. Rapid emergence of African swine fever virus variants with different numbers of a tandem repeat sequence in South Korea. Transbound. Emerg. Dis. 2021, 68, 1726–1730. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.T.; Truong, A.D.; Dang, A.K.; Ly, D.V.; Nguyen, C.T.; Chu, N.T.; Hoang, T.V.; Nguyen, H.T.; Dang, H.V. Circulation of two different variants of intergenic region (IGR) located between the I73R and I329L genes of African swine fever virus strains in Vietnam. Transbound. Emerg. Dis. 2021, 68, 2693–2695. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ren, Z.; Wang, Q.; Ge, S.; Liu, Y.; Liu, C.; Liu, F.; Hu, Y.; Li, J.; Bao, J.; et al. Infection of African swine fever in wild boar, China, 2018. Transbound. Emerg. Dis. 2019, 66, 1395–1398. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.T.; Cho, K.H.; Mai, N.T.A.; Park, J.Y.; Trinh, T.B.N.; Jang, M.K.; Nguyen, T.T.H.; Vu, X.D.; Nguyen, T.L.; Nguyen, V.D.; et al. Multiple variants of African swine fever virus circulating in Vietnam. Arch. Virol. 2022, 167, 1137–1140. [Google Scholar] [CrossRef]
- Mazur-Panasiuk, N.; Walczak, M.; Juszkiewicz, M.; Wozniakowski, G. The Spillover of African Swine Fever in Western Poland Revealed Its Estimated Origin on the Basis of O174L, K145R, MGF 505-5R and IGR I73R/I329L Genomic Sequences. Viruses 2020, 12, 1094. [Google Scholar] [CrossRef]
- Mai, N.T.A.; Dam, V.P.; Cho, K.H.; Nguyen, V.T.; Van Tuyen, N.; Nguyen, T.L.; Ambagala, A.; Park, J.Y.; Le, V.P. Emergence of a novel intergenic region (IGR) IV variant of african swine fever virus genotype II in domestic pigs in Vietnam. Vet. Res. Commun. 2023, 47, 1773–1776. [Google Scholar] [CrossRef]
- Dixon, L.K.; Abrams, C.C.; Bowick, G.; Goatley, L.C.; Kay-Jackson, P.C.; Chapman, D.; Liverani, E.; Nix, R.; Silk, R.; Zhang, F. African swine fever virus proteins involved in evading host defence systems. Vet. Immunol. Immunopathol. 2004, 100, 117–134. [Google Scholar] [CrossRef]
- Sanna, G.; Dei Giudici, S.; Bacciu, D.; Angioi, P.P.; Giammarioli, M.; De Mia, G.M.; Oggiano, A. Improved Strategy for Molecular Characterization of African Swine Fever Viruses from Sardinia, Based on Analysis of p30, CD2V and I73R/I329L Variable Regions. Transbound. Emerg. Dis. 2017, 64, 1280–1286. [Google Scholar] [CrossRef]
- Sun, Y.; Xu, Z.; Gao, H.; Xu, S.; Liu, J.; Xing, J.; Kuang, Q.; Chen, Y.; Wang, H.; Zhang, G. Detection of a Novel African Swine Fever Virus with Three Large-Fragment Deletions in Genome, China. Microbiol. Spectr. 2022, 10, e0215522. [Google Scholar] [CrossRef]
- Ankhanbaatar, U.; Auer, A.; Ulziibat, G.; Settypalli, T.B.K.; Gombo-Ochir, D.; Basan, G.; Takemura, T.; Tseren-Ochir, E.O.; Ouled Ahmed, H.; Meki, I.K.; et al. Comparison of the Whole-Genome Sequence of the African Swine Fever Virus from a Mongolian Wild Boar with Genotype II Viruses from Asia and Europe. Pathogens 2023, 12, 1143. [Google Scholar] [CrossRef]
- Philippe, H.; Brinkmann, H.; Lavrov, D.V.; Littlewood, D.T.; Manuel, M.; Worheide, G.; Baurain, D. Resolving difficult phylogenetic questions: Why more sequences are not enough. PLoS Biol. 2011, 9, e1000602. [Google Scholar] [CrossRef]
- Hakizimana, J.N.; Yona, C.; Makange, M.R.; Kasisi, E.A.; Netherton, C.L.; Nauwynck, H.; Misinzo, G. Complete genome analysis of African swine fever virus genotypes II, IX and XV from domestic pigs in Tanzania. Sci. Rep. 2023, 13, 5318. [Google Scholar] [CrossRef]
- LCQ12; Measures to Control Wild Pigs. The Government of the Hong Kong Special Adminstrative Region: Hong Kong, China, 2024. Available online: https://www.info.gov.hk/gia/general/202412/18/P2024121800200.htm (accessed on 8 May 2025).
- Mebus, C.A.; House, C.; Gonzalvo, F.R.; Pineda, J.M.; Tapiador, J.; Pire, J.J.; Bergada, J.; Yedloutschnig, R.J.; Sahu, S.; Becerra, V.; et al. Survival of foot-and-mouth disease, African swine fever, and hog cholera viruses in Spanish serrano cured hams and Iberian cured hams, shoulders and loins. Food Microbiol. 1993, 10, 133–143. [Google Scholar] [CrossRef]
- McKercher, P.D.; Hess, W.R.; Hamdy, F. Residual viruses in pork products. Appl. Environ. Microbiol. 1978, 35, 142–145. [Google Scholar] [CrossRef]
- Farez, S.; Morley, R.S. Potential animal health hazards of pork and pork products. Rev. Sci. Tech. Off. Int. Des. épizooties 1997, 16, 65–78. [Google Scholar] [CrossRef]
- Wang, W.H.; Lin, C.Y.; Chang Ishcol, M.R.; Urbina, A.N.; Assavalapsakul, W.; Thitithanyanont, A.; Lu, P.L.; Chen, Y.H.; Wang, S.F. Detection of African swine fever virus in pork products brought to Taiwan by travellers. Emerg. Microbes Infect. 2019, 8, 1000–1002. [Google Scholar] [CrossRef]
- Kim, H.J.; Lee, M.J.; Lee, S.K.; Kim, D.Y.; Seo, S.J.; Kang, H.E.; Nam, H.M. African Swine Fever Virus in Pork Brought into South Korea by Travelers from China, August 2018. Emerg. Infect. Dis. 2019, 25, 1231–1233. [Google Scholar] [CrossRef]
- Ratnawati, A.; Hartawan, R.; Sendow, I.; Saepulloh, M.; Sumarningsih, S.; Hewajuli, D.A.; Zainuddin, N.; Dharmayanti, N.; Wibawan, I.W.T.; Mayasari, N. Transboundary risk of African swine fever (ASF): Detection of ASF virus genotype II in pork products carried by international travelers to Indonesia. Vet. World 2025, 18, 280–286. [Google Scholar] [CrossRef]
- Serdena, A.P.R.; Bernardo, J.M.G.; Pangga, G.M.V.; Salamat, S.E.A.; Agulto, T.N.; Desamero, M.J.M.; Atienza, C.P.G.; Calumpang, G.J.A.; Canlas, R.M.P.; Castillo, M.S.M.; et al. Molecular detection of African swine fever virus in pork and pork products and associated risk factors in the Philippines. J. Vet. Med. Sci. 2025, 87, 13–27. [Google Scholar] [CrossRef]

| Case | Date | Location Found | Weight (kg) | Sample Type | RT-PCR (Ct) | Gel PCR | VI | HAD |
|---|---|---|---|---|---|---|---|---|
| ASFV/HKWB2021SSW-12112 | 2 September 2021 | Siu Sai Wan | 55 | Spleen | 18.48 | Positive | Not Isolated | - |
| Tonsil # | 17.89 | Positive | Not Isolated | - | ||||
| Lymph node | 18.31 | Positive | Not Isolated | - | ||||
| Kidney | 19.11 | Positive | Not Isolated | - | ||||
| Oronasal swab | 25.86 | - | Not Isolated | - | ||||
| ASFV/HKWB2022TP-00522 | 12 January 2022 | Tai Po | 29 | Spleen # | 19.33 | Positive | Not Isolated | - |
| Tonsil | 25.97 | Positive | - | - | ||||
| Lymph node | 20.85 | Positive | Not Isolated | - | ||||
| Kidney | 21.02 | Positive | - | - | ||||
| Oronasal swab | 26.69 | Positive | Not Isolated | - | ||||
| ASFV/HKWB2022S-10414 | 24 February 2022 | Stanley | 74 | Spleen # | 17.14 | Positive | Not Isolated | - |
| Tonsil | 23.96 | Positive | - | - | ||||
| Lymph node | 17.07 | Positive | - | - | ||||
| Kidney | 21.94 | Positive | - | - | ||||
| Oronasal swab | 20.85 | Positive | Positive | Positive | ||||
| ASFV/HKWB2022SK-13869 | 27 May 2022 | Sai Kung | 43 | Spleen | 19.73 | Positive | - | - |
| Tonsil | 18.80 | Positive | - | - | ||||
| Lymph node | 20.02 | Positive | - | - | ||||
| Kidney # | 17.95 | Positive | Positive | Positive | ||||
| Oronasal swab | 22.02 | - | Positive | Positive |
| Strain | Genome Length (bp) | GC Content (%) | Number of ORFs | Number of MGF Members | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LVR | RVR | ||||||||||
| MGF 360 | MGF 110 | MGF 505 | MGF 300 | MGF 100 | MGF 360 | MGF 100 | MGF 505 | ||||
| ASFV/HKWB2021SSW-12112 | 190,165 | 38.40 | 194 | 1 | 12 * | 3 | 14 | 9 | 2 | 5 | 1 |
| ASFV/HKWB2022TP-00522 | 190,181 | 38.40 | 194 | 1 | 12 * | 3 | 14 | 9 | 2 | 5 | 1 |
| ASFV/HKWB2022S-10414 | 189,730 | 38.39 | 192 | 1 | 11 * | 3 | 14 | 9 | 2 | 5 | 1 |
| ASFV/HKWB2022SK-13869 | 191,076 | 38.36 | 193 | 1 | 11 * | 3 | 14 | 9 | 2 | 5 | 1 |
| Strain | Gene | Function | CDS Position * | Polymorphism Type | Change | Effect | Reference |
|---|---|---|---|---|---|---|---|
| ASFV/HKWB2021SSW-12112 | MGF 110-1L CDS | Unknown | 590 | Substitution | G>A | Nonsense mutation | |
| MGF 110-7L CDS | Induces host cell translation suppression and stress granule formation | 170–183 | Deletion | TGTGAAGATGGGAT | Frame shift and early truncation | [33] | |
| MGF 110-10L—MGF110-14L fusion | Unknown | 346–347 | Deletion | GG | Frame shift and early truncation. | ||
| MGF 360-10L CDS | Modulates type I interferon response and is associated with viral virulence | 986 | Substitution | A>G | Nonsynonymous substitution (N329S) | [54] | |
| MGF 360-14L CDS | Involved in virus cell tropism; may be required for efficient virus replication in macrophages | 845–846 | Insertion | G | Frame shift and early truncation | [55] | |
| MGF 505-9R CDS | Unknown | 967 | Substitution | A>G | Nonsynonymous substitution (K323E) | ||
| ASFV G ACD 00190 CDS | Unknown | 13 | Deletion | T | Frame shift and early truncation | ||
| ASFV G ACD 00350 CDS | Unknown | 37–42 | Deletion | GGGGGG | In-frame deletion | ||
| EP1242L CDS | Involved in viral gene transcription | 2767 | Substitution | G>A | Nonsynonymous substitution (V923I) | [41] | |
| NP419L CDS | DNA ligase; potentially involved in repair mechanisms | 1241 | Substitution | A>G | Nonsynonymous substitution (N414S) | [56] | |
| DP60R CDS | Unknown | 53–54 | Insertion | A | Frame shift and extension | ||
| M448R CDS | T-cell antigen with protective potential | 163 | Substitution | G>A | Nonsynonymous substitution (E55K) | [57] | |
| I267L CDS | Acts as a virulence factor by inhibiting RNA polymerase III-RIG-I-mediated immunity | 583 | Substitution | A>T | Nonsynonymous substitution (I195F) | [58] | |
| ASFV/HKWB2022TP-00522 | MGF 110-1L CDS | Unknown | 590 | Substitution | G>A | Nonsense mutation | |
| MGF 110-4L CDS | Involved in virion assembly | 260 | Substitution | A>G | Nonsynonymous substitution (Q87R) | [59] | |
| MGF 110-7L CDS | Induces host cell translation suppression and stress granule formation | 110 | Deletion | C | Frame shift and early truncation | [33] | |
| MGF 110-10L—MGF110-14L fusion | Unknown | 343–347 | Deletion | GGGGG | Frame shift and early truncation. | ||
| MGF 360-1La CDS | Unknown | 363 | Substitution | T>G | Nonsynonymous substitution (S121R) | ||
| MGF 360-10L CDS | Modulates type I interferon response and is associated with viral virulence | 986 | Substitution | A>G | Nonsynonymous substitution (N329S) | [54] | |
| MGF 505-9R CDS | Unknown | 967 | Substitution | A>G | Nonsynonymous substitution (K323E) | ||
| ASFV G ACD 00190 CDS | Unknown | 13 | Deletion | T | Frame shift and early truncation | ||
| ASFV G ACD 00350 CDS | Unknown | 37–42 | Deletion | GGGGGG | In-frame deletion | ||
| NP419L CDS | DNA ligase; potentially involved in repair mechanisms | 1241 | Substitution | A>G | Nonsynonymous substitution (N414S) | [56] | |
| DP60R CDS | Unknown | 53–54 | Insertion | A | Frame shift and extension | ||
| I267L CDS | Acts as a virulence factor by inhibiting RNA polymerase III-RIG-I-mediated immunity | 583 | Substitution | A>T | Nonsynonymous substitution (I195F) | [58] | |
| P1192R CDS | Topoisomerase; involved in virus transcription and replication | 645–648 | Substitution | GGCG>AACA | Nonsynonymous substitution (A216T) | [60] | |
| I243L CDS | Transcription factor | 506 | Substitution | G>C | Nonsynonymous substitution (G169A) | [41] | |
| EP153R CDS | Involved in hemadsorption | 421 | Substitution | A>G | Nonsynonymous substitution (S141G) | [61] | |
| ASFV/HKWB2022S-10414 | MGF 110-1L CDS | Unknown | 590 | Substitution | G>A | Nonsense mutation | |
| MGF 110-2L CDS | Involved in virus cell tropism; may be required for efficient virus replication in macrophages | 5 | Substitution | G>A | R2K | [55] | |
| MGF 110-3L CDS | Unknown | Entire CDS | Deletion | Deletion | Gene deletion | ||
| MGF 110-7L CDS | Induces host cell translation suppression and stress granule formation | 110–111 | Insertion | C | Frame shift and early truncation | [33] | |
| MGF 110-10L—MGF110-14L fusion | Unknown | 346–347 | Deletion | GG | Frame shift and early truncation. | ||
| MGF 110-13Lb CDS | Unknown | 359–366 | Deletion | GGGGGGGG | Frame shift and extension | ||
| MGF 360-1La CDS | Unknown | 94 | Substitution | G>A | Nonsynonymous substitution (E32K) | ||
| 463 | Substitution | G>A | Nonsynonymous substitution (E155K) | ||||
| MGF 360-4L CDS | Involved in virus cell tropism; may be required for efficient virus replication in macrophages | 574 | Substitution | G>A | Nonsynonymous substitution (A192T) | [55] | |
| MGF 360-19Ra CDS | Involved in virus cell tropism; may be required for efficient virus replication in macrophages | 355 | Substitution | C>T | Nonsynonymous substitution (L119F) | [55] | |
| 452 | Substitution | C>T | Nonsynonymous substitution (P151L) | ||||
| 691 | Substitution | C>T | Nonsynonymous substitution (H231Y) | ||||
| MGF 360-10L CDS | Modulates type I interferon response and is associated with viral virulence | 986 | Substitution | A>G | Nonsynonymous substitution (N329S) | [54] | |
| MGF 505-9R CDS | Unknown | 967 | Substitution | A>G | Nonsynonymous substitution (K323E) | ||
| ASFV G ACD 00120 CDS | Unknown | 1–155 | Deletion | Deletion | Gene deletion | ||
| ASFV G ACD 00190 CDS | Unknown | 13 | Deletion | T | Frame shift and early truncation | ||
| NP419L CDS | DNA ligase; potentially involved in repair mechanisms | 1241 | Substitution | A>G | Nonsynonymous substitution (N414S) | [56] | |
| DP60R CDS | Unknown | 53–54 | Insertion | A | Frame shift and extension | ||
| DP96R CDS | Uridine kinase; inhibits interferons production and associated with virus virulence | 196 | Substitution | C>T | Nonsynonymous substitution (P66S) | [62] | |
| I267L CDS | Acts as a virulence factor by inhibiting RNA polymerase III-RIG-I-mediated immunity | 583 | Substitution | A>T | Nonsynonymous substitution (I195F) | [58] | |
| C962R | Potentially involved in repair mechanisms | 1538 | Substitution | G>A | Nonsynonymous substitution (R513H) | [63] | |
| A238L CDS | Downregulates host inflammatory responses | 394 | Substitution | G>A | Nonsynonymous substitution (A132T) | [42] | |
| ASFV/HKWB2022SK-13869 | MGF 110-1L CDS | Unknown | 590 | Substitution | G>A | Nonsense mutation | |
| MGF 110-3L CDS | Unknown | 272 | Substitution | G>A | Nonsynonymous substitution (G91D) | ||
| MGF 110-7L CDS | Induces host cell translation suppression and stress granule formation | 110–111 | Insertion | C | Frame shift and early truncation | [33] | |
| MGF 110-13Lb CDS | Unknown | 366 | Deletion | G | Frame shift and fusion with MGF 110-13 La | ||
| MGF 110-4L CDS | Involved in virion assembly | 325 | Substitution | A>G | Nonsynonymous substitution (N109D) | [59] | |
| MGF 110-10L—MGF110-14L fusion | Unknown | 344–347 | Deletion | GGGG | Frame shift and early truncation. | ||
| MGF 360-10L CDS | Modulates type I interferon response and is associated with viral virulence | 986 | Substitution | A>G | Nonsynonymous substitution (N329S) | [54] | |
| MGF 360-14L CDS | Involved in virus cell tropism; may be required for efficient virus replication in macrophages | 845–846 | Insertion | G | Frame shift and early truncation | [55] | |
| MGF 360-15R CDS | Involved in virus cell tropism; may be required for efficient virus replication in macrophages | 697 | Substitution | G>A | Nonsynonymous substitution (A233T) | [55] | |
| MGF 505-9R CDS | Unknown | 967 | Substitution | A>G | Nonsynonymous substitution (K323E) | ||
| ASFV G ACD 00190 CDS | Unknown | 13 | Deletion | T | Frame shift and early truncation | ||
| ASFV G ACD 00350 CDS | Unknown | 39–42 | Deletion | GGGG | In-frame deletion | ||
| NP419L CDS | DNA ligase; potentially involved in repair mechanisms | 1241 | Substitution | A>G | Nonsynonymous substitution (N414S) | [56] | |
| DP60R CDS | Unknown | 53–54 | Insertion | A | Frame shift and extension | ||
| I267L CDS | Acts as a virulence factor by inhibiting RNA polymerase III-RIG-I-mediated immunity | 583 | Substitution | A>T | Nonsynonymous substitution (I195F) | [58] | |
| B646L CDS | Viral capsid | 1175 | Substitution | G>A | Nonsynonymous substitution (R392H) | [55] | |
| B475L CDS | Potentially involved in the host antiviral innate immunity evasion | 853 | Substitution | G>A | Nonsynonymous substitution (E285K) | [64] | |
| D1133L CDS | Helicase; potentially involved in viral replication | 13 | Substitution | G>A | Nonsynonymous substitution (E5K) | [65] | |
| E199L CDS | Involved in virus entry and induce autophagy | 374 | Substitution | A>T | Nonsynonymous substitution (E125V) | [66] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tam, K.W.S.; Lau, C.C.Y.; Ng, T.T.L.; Ip, S.M.; Pun, S.F.; Corla, A.; Batten, C.; Brackman, C.J. The First African Swine Fever Viruses Detected in Wild Boar in Hong Kong, 2021–2023. Viruses 2025, 17, 896. https://doi.org/10.3390/v17070896
Tam KWS, Lau CCY, Ng TTL, Ip SM, Pun SF, Corla A, Batten C, Brackman CJ. The First African Swine Fever Viruses Detected in Wild Boar in Hong Kong, 2021–2023. Viruses. 2025; 17(7):896. https://doi.org/10.3390/v17070896
Chicago/Turabian StyleTam, Karina W. S., Candy C. Y. Lau, Timothy T. L. Ng, Sin Ming Ip, Sin Fat Pun, Amanda Corla, Carrie Batten, and Christopher J. Brackman. 2025. "The First African Swine Fever Viruses Detected in Wild Boar in Hong Kong, 2021–2023" Viruses 17, no. 7: 896. https://doi.org/10.3390/v17070896
APA StyleTam, K. W. S., Lau, C. C. Y., Ng, T. T. L., Ip, S. M., Pun, S. F., Corla, A., Batten, C., & Brackman, C. J. (2025). The First African Swine Fever Viruses Detected in Wild Boar in Hong Kong, 2021–2023. Viruses, 17(7), 896. https://doi.org/10.3390/v17070896





