Subtype AD Recombinant HIV-1 Transmitted/Founder Viruses Are Less Sensitive to Type I Interferons than Subtype D
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Ethical Consideration
2.3. Assessment of Mosaic Full-Length Structure of the TF Viruses
2.4. Generation of Virus Stocks
2.5. Virus Replication in PBMCs and Type I Interferon Sensitivity
2.6. Determination of HIV-1 Replicative Capacity
2.7. Determination of Relative Particle Infectivity of TF Viruses
2.8. Statistical Analysis
3. Results
3.1. Mosaic Full-Length Structure of the TF Viruses Tested
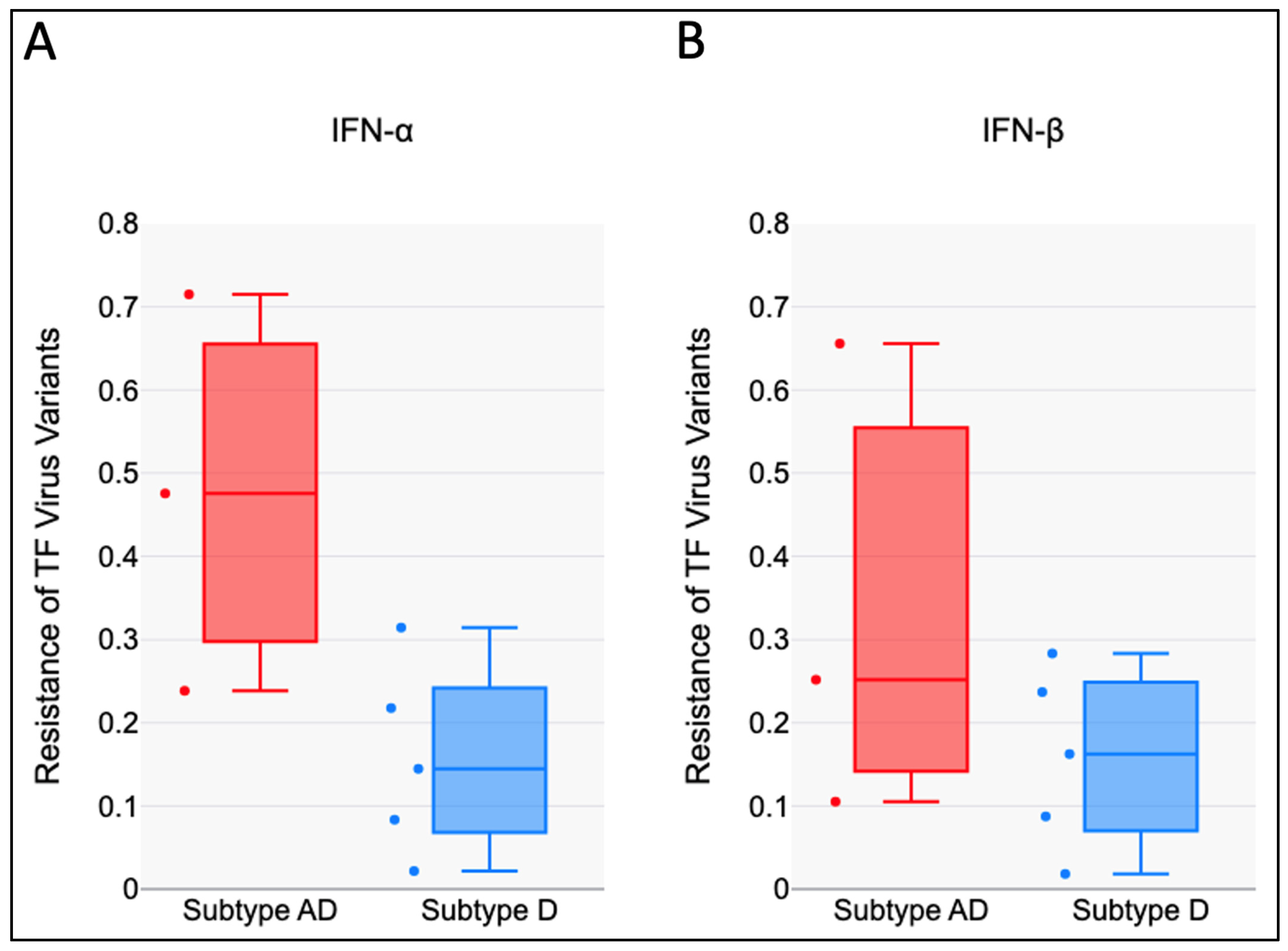
3.2. HIV-1 AD Recombinants Are Less Sensitive to the Antiviral Effects of IFN-I than Subtype D
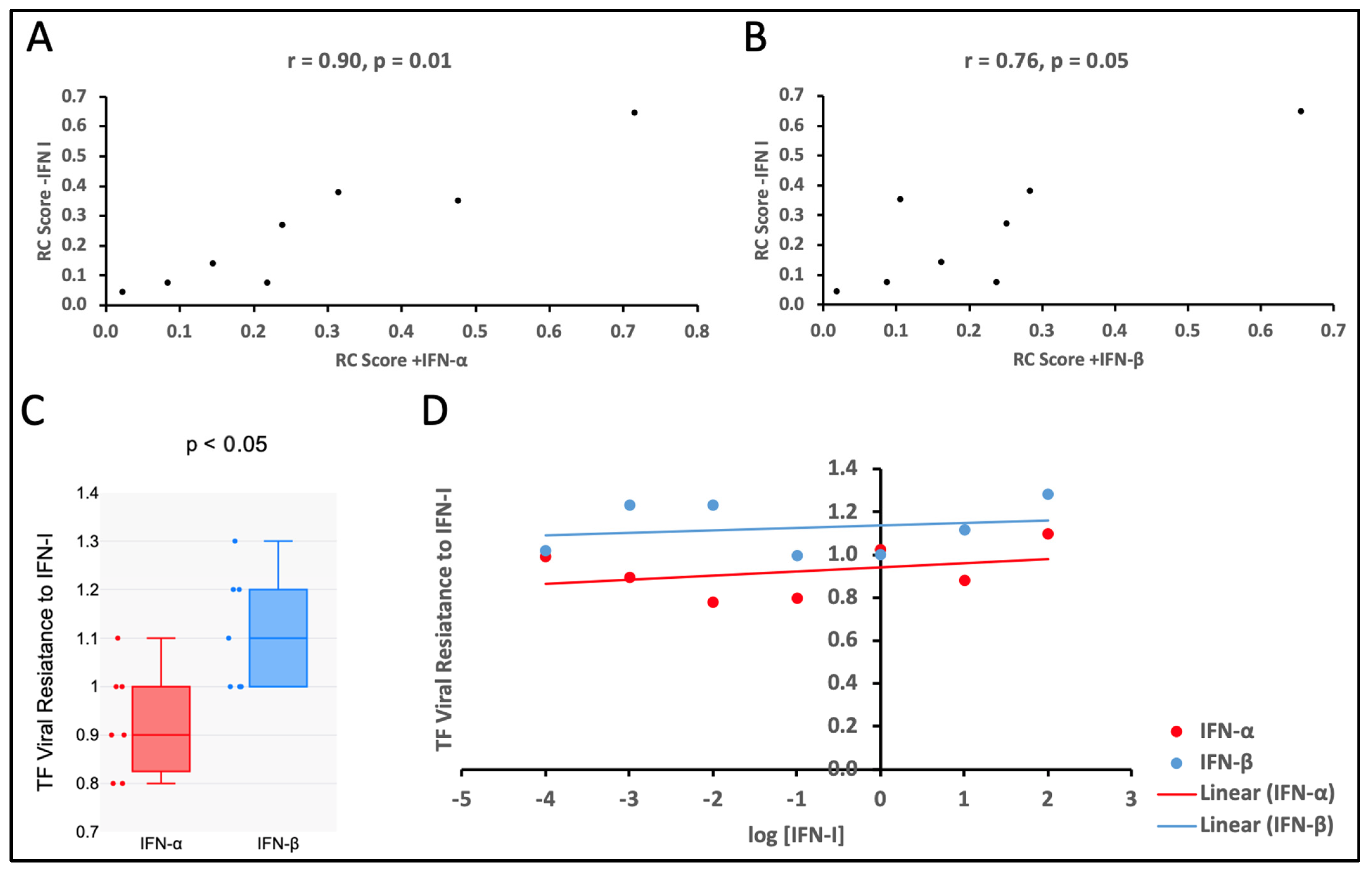
3.3. Viral Replicative Capacities of TF Viruses Increase with Relative Particle Infectivity
3.4. The TF Viruses Tested Were Less Sensitive to the Antiviral Effect of IFN-β than That of IFN-α
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Derdeyn, C.A.; Decker, J.M.; Bibollet-Ruche, F.; Mokili, J.L.; Muldoon, M.; Denham, S.A.; Heil, M.L.; Kasolo, F.; Musonda, R.; Hahn, B.H. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 2004, 303, 2019–2022. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.-T.; Chen, S.S. Emerging roles of interferon-stimulated genes in the innate immune response to hepatitis C virus infection. Cell. Mol. Immunol. 2016, 13, 11–35. [Google Scholar] [CrossRef] [PubMed]
- Stremlau, M.; Owens, C.M.; Perron, M.J.; Kiessling, M.; Autissier, P.; Sodroski, J. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 2004, 427, 848–853. [Google Scholar] [CrossRef]
- Jitobaom, K.; Sirihongthong, T.; Boonarkart, C.; Phakaratsakul, S.; Suptawiwat, O.; Auewarakul, P. Human Schlafen 11 inhibits influenza A virus production. Virus Res. 2023, 334, 199162. [Google Scholar] [CrossRef]
- Cui, W.; Braun, E.; Wang, W.; Tang, J.; Zheng, Y.; Slater, B.; Li, N.; Chen, C.; Liu, Q.; Wang, B. Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins. Proc. Natl. Acad. Sci. USA 2021, 118, e2022269118. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, K.; Wang, S.; Du, J. Multi-functional BST2/tetherin against HIV-1, other viruses and LINE-1. Front. Cell. Infect. Microbiol. 2022, 12, 979091. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.B.; Swanstrom, R.; Kashuba, A.D.; Cohen, M.S. Bottlenecks in HIV-1 transmission: Insights from the study of founder viruses. Nat. Rev. Microbiol. 2015, 13, 414–425. [Google Scholar] [CrossRef]
- Deymier, M.J.; Ende, Z.; Fenton-May, A.E.; Dilernia, D.A.; Kilembe, W.; Allen, S.A.; Borrow, P.; Hunter, E. Heterosexual transmission of subtype C HIV-1 selects consensus-like variants without increased replicative capacity or interferon-α resistance. PLoS Pathog. 2015, 11, e1005154. [Google Scholar] [CrossRef]
- Esparza, J.; Osmanov, S. The development and evaluation of HIV vaccines. Curr. Opin. Infect. Dis. 1993, 6, 218–229. [Google Scholar] [CrossRef]
- Price, M.A.; Kilembe, W.; Ruzagira, E.; Karita, E.; Inambao, M.; Sanders, E.J.; Anzala, O.; Allen, S.; Edward, V.A.; Kaleebu, P. Cohort Profile: IAVI’s HIV epidemiology and early infection cohort studies in Africa to support vaccine discovery. Int. J. Epidemiol. 2021, 50, 29–30. [Google Scholar] [CrossRef]
- Balinda, S.N.; Kapaata, A.; Xu, R.; Salazar, M.G.; Mezzell, A.T.; Qin, Q.; Herard, K.; Dilernia, D.; Kamali, A.; Ruzagira, E. Characterization of Near Full-Length Transmitted/Founder HIV-1 Subtype D and A/D Recombinant Genomes in a Heterosexual Ugandan Population (2006–2011). Viruses 2022, 14, 334. [Google Scholar] [CrossRef]
- Ochsenbauer, C.; Edmonds, T.G.; Ding, H.; Keele, B.F.; Decker, J.; Salazar, M.G.; Salazar-Gonzalez, J.F.; Shattock, R.; Haynes, B.F.; Shaw, G.M. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J. Virol. 2012, 86, 2715–2728. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Decker, J.M.; Liu, H.; Zhang, Z.; Arani, R.B.; Kilby, J.M.; Saag, M.S.; Wu, X.; Shaw, G.M.; Kappes, J.C. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 2002, 46, 1896–1905. [Google Scholar] [CrossRef]
- Carlson, J.M.; Schaefer, M.; Monaco, D.C.; Batorsky, R.; Claiborne, D.T.; Prince, J.; Deymier, M.J.; Ende, Z.S.; Klatt, N.R.; DeZiel, C.E. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science 2014, 345, 1254031. [Google Scholar] [CrossRef] [PubMed]
- Umviligihozo, G.; Muok, E.; Nyirimihigo Gisa, E.; Xu, R.; Dilernia, D.; Herard, K.; Song, H.; Qin, Q.; Bizimana, J.; Farmer, P. Increased frequency of inter-subtype HIV-1 recombinants identified by near full-length virus sequencing in Rwandan acute transmission cohorts. Front. Microbiol. 2021, 12, 734929. [Google Scholar] [CrossRef] [PubMed]
- Klingler, J.; Anton, H.; Réal, E.; Zeiger, M.; Moog, C.; Mély, Y.; Boutant, E. How HIV-1 gag manipulates its host cell proteins: A focus on interactors of the nucleocapsid domain. Viruses 2020, 12, 888. [Google Scholar] [CrossRef]
- Chintala, K.; Mohareer, K.; Banerjee, S. Dodging the host interferon-stimulated gene mediated innate immunity by HIV-1: A brief update on intrinsic mechanisms and counter-mechanisms. Front. Immunol. 2021, 12, 716927. [Google Scholar] [CrossRef]
- Duchon, A.; Santos, S.; Chen, J.; Brown, M.; Nikolaitchik, O.A.; Tai, S.; Chao, J.A.; Freed, E.O.; Pathak, V.K.; Hu, W.-S. Plasma membrane anchoring and Gag: Gag multimerization on viral RNA are critical properties of HIV-1 Gag required to mediate efficient genome packaging. MBio 2021, 12, e0325421. [Google Scholar] [CrossRef]
- Ohainle, M.; Kim, K.; Komurlu Keceli, S.; Felton, A.; Campbell, E.; Luban, J.; Emerman, M. TRIM34 restricts HIV-1 and SIV capsids in a TRIM5α-dependent manner. PLoS Pathog. 2020, 16, e1008507. [Google Scholar] [CrossRef]
- Giandhari, J. Genetic Changes in HIV-1 Gag and Pol genes Associated with Protease Inhibitor-Based Therapy Failure in Paediatric Patients; University of the Witwatersrand: Johannesburg, South Africa, 2015. [Google Scholar]
- Galli, A.; Lai, A.; Corvasce, S.; Saladini, F.; Riva, C.; Deho, L.; Caramma, I.; Franzetti, M.; Romano, L.; Galli, M. Recombination analysis and structure prediction show correlation between breakpoint clusters and RNA hairpins in the pol gene of human immunodeficiency virus type 1 unique recombinant forms. J. Gen. Virol. 2008, 89, 3119–3125. [Google Scholar] [CrossRef]
- Robinson, C.A.; Lyddon, T.D.; Gil, H.M.; Evans, D.T.; Kuzmichev, Y.V.; Richard, J.; Finzi, A.; Welbourn, S.; Rasmussen, L.; Nebane, N.M. Novel compound inhibitors of HIV-1NL4-3 Vpu. Viruses 2022, 14, 817. [Google Scholar] [CrossRef]
- Zhang, M.-D.; Chen, F.; He, W.-Q.; Lu, Y.; Liu, F.-L.; Zhang, H.-G.; Yang, L.-M.; Dong, C.-S.; Xiong, S.-D.; Zheng, Y.-T. The Deubiquitinase OTUD1 Influences HIV-1 Release by Regulating the Host Restriction Factor BST-2. Viruses 2025, 17, 260. [Google Scholar] [CrossRef] [PubMed]
- Neil, S.J.; Zang, T.; Bieniasz, P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 2008, 451, 425–430. [Google Scholar] [CrossRef]
- Elalouf, A.; Maoz, H.; Rosenfeld, A.Y. Comprehensive Insights into the Molecular Basis of HIV Glycoproteins. Appl. Sci. 2024, 14, 8271. [Google Scholar] [CrossRef]
- Marceau, T.; Braibant, M. Role of viral envelope proteins in determining susceptibility of viruses to IFITM proteins. Viruses 2024, 16, 254. [Google Scholar] [CrossRef] [PubMed]
- Kwong, P.D.; Wyatt, R.; Robinson, J.; Sweet, R.W.; Sodroski, J.; Hendrickson, W.A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 1998, 393, 648–659. [Google Scholar] [CrossRef]
- Pang, W.; Song, J.-H.; Lu, Y.; Zhang, X.-L.; Zheng, H.-Y.; Jiang, J.; Zheng, Y.-T. Host restriction factors APOBEC3G/3F and other interferon-related gene expressions affect early HIV-1 infection in northern pig-tailed macaque (Macaca leonina). Front. Immunol. 2018, 9, 1965. [Google Scholar] [CrossRef]
- Okada, A.; Iwatani, Y. APOBEC3G-mediated G-to-A hypermutation of the HIV-1 genome: The missing link in antiviral molecular mechanisms. Front. Microbiol. 2016, 7, 2027. [Google Scholar] [CrossRef]
- Claiborne, D.T.; Prince, J.L.; Scully, E.; Macharia, G.; Micci, L.; Lawson, B.; Kopycinski, J.; Deymier, M.J.; Vanderford, T.H.; Nganou-Makamdop, K. Replicative fitness of transmitted HIV-1 drives acute immune activation, proviral load in memory CD4+ T cells, and disease progression. Proc. Natl. Acad. Sci. USA 2015, 112, E1480–E1489. [Google Scholar] [CrossRef]
- Rouzine, I.M.; Weinberger, A.D.; Weinberger, L.S. An evolutionary role for HIV latency in enhancing viral transmission. Cell 2015, 160, 1002–1012. [Google Scholar] [CrossRef]
- Abel, K.; Rocke, D.M.; Chohan, B.; Fritts, L.; Miller, C.J. Temporal and anatomic relationship between virus replication and cytokine gene expression after vaginal simian immunodeficiency virus infection. J. Virol. 2005, 79, 12164–12172. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Estes, J.D.; Schlievert, P.M.; Duan, L.; Brosnahan, A.J.; Southern, P.J.; Reilly, C.S.; Peterson, M.L.; Schultz-Darken, N.; Brunner, K.G. Glycerol monolaurate prevents mucosal SIV transmission. Nature 2009, 458, 1034–1038. [Google Scholar] [CrossRef] [PubMed]
- Etienne, L.; Hahn, B.H.; Sharp, P.M.; Matsen, F.A.; Emerman, M. Gene loss and adaptation to hominids underlie the ancient origin of HIV-1. Cell Host Microbe 2013, 14, 85–92. [Google Scholar] [CrossRef]
- Sandler, N.G.; Bosinger, S.E.; Estes, J.D.; Zhu, R.T.; Tharp, G.K.; Boritz, E.; Levin, D.; Wijeyesinghe, S.; Makamdop, K.N.; Del Prete, G.Q. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature 2014, 511, 601–605. [Google Scholar] [CrossRef]
- Parrish, N.F.; Gao, F.; Li, H.; Giorgi, E.E.; Barbian, H.J.; Parrish, E.H.; Zajic, L.; Iyer, S.S.; Decker, J.M.; Kumar, A. Phenotypic properties of transmitted founder HIV-1. Proc. Natl. Acad. Sci. USA 2013, 110, 6626–6633. [Google Scholar] [CrossRef] [PubMed]
- Fenton-May, A.E.; Dibben, O.; Emmerich, T.; Ding, H.; Pfafferott, K.; Aasa-Chapman, M.M.; Pellegrino, P.; Williams, I.; Cohen, M.S.; Gao, F. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology 2013, 10, 146. [Google Scholar] [CrossRef]
- Etemad, B.; Gonzalez, O.A.; White, L.; Laeyendecker, O.; Kirk, G.D.; Mehta, S.; Sagar, M. Characterization of HIV-1 envelopes in acutely and chronically infected injection drug users. Retrovirology 2014, 11, 106. [Google Scholar] [CrossRef][Green Version]
- Oberle, C.S.; Joos, B.; Rusert, P.; Campbell, N.K.; Beauparlant, D.; Kuster, H.; Weber, J.; Schenkel, C.D.; Scherrer, A.U.; Magnus, C. Tracing HIV-1 transmission: Envelope traits of HIV-1 transmitter and recipient pairs. Retrovirology 2016, 13, 62. [Google Scholar] [CrossRef]
- Luthuli, B.; Gounder, K.; Deymier, M.J.; Dong, K.L.; Balazs, A.B.; Mann, J.K.; Ndung’u, T. Generation and characterization of infectious molecular clones of transmitted/founder HIV-1 subtype C viruses. Virology 2023, 583, 14–26. [Google Scholar] [CrossRef]
- Iyer, S.S.; Bibollet-Ruche, F.; Sherrill-Mix, S.; Learn, G.H.; Plenderleith, L.; Smith, A.G.; Barbian, H.J.; Russell, R.M.; Gondim, M.V.; Bahari, C.Y. Resistance to type 1 interferons is a major determinant of HIV-1 transmission fitness. Proc. Natl. Acad. Sci. USA 2017, 114, E590–E599. [Google Scholar] [CrossRef]
- Shirazi, Y.; Pitha, P.M. Alpha interferon inhibits early stages of the human immunodeficiency virus type 1 replication cycle. J. Virol. 1992, 66, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
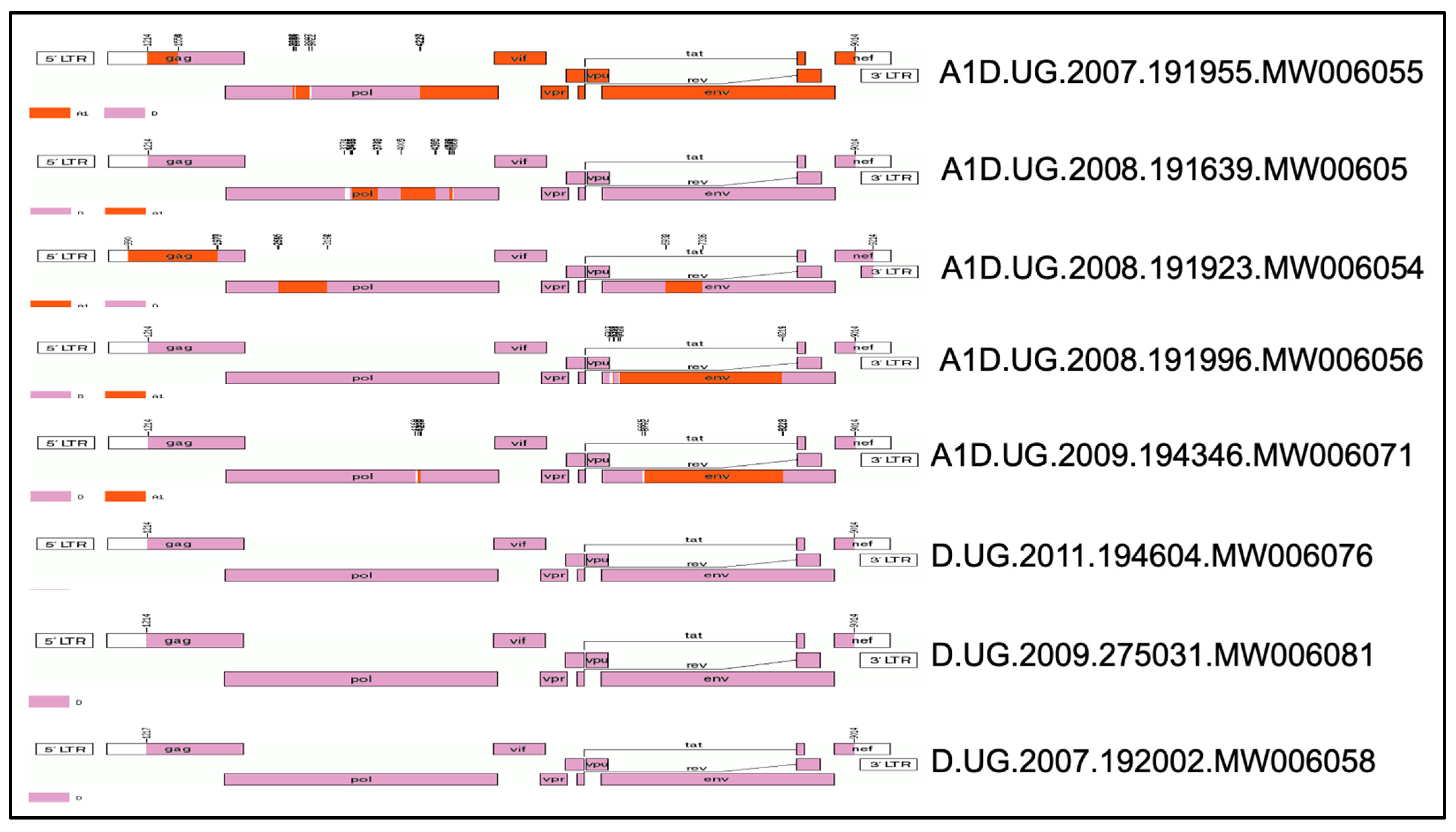

| Sample ID | Sample Date | Estimated Date of Infection | Days Post Infection | Initial Viral Load (Copies/mL) | Gender | Age | Subtype (Full Genome) | GenBank No |
|---|---|---|---|---|---|---|---|---|
| 191955 | 26-Mar-07 | 03-Mar-07 | 23 | 4190 | F | 39 | AD | MW006055 |
| 192002 | 24-Jul-07 | 04-Jun-07 | 50 | 2420 | F | 27 | D | MW006058 |
| 275031 | 05-Jun-09 | 11-May-09 | 25 | 3750 | M | 31 | D | MW006081 |
| 191923 | 24-Jan-08 | 30-Nov-07 | 55 | 198,000 | F | 31 | AD | MW006054 |
| 191639 | 03-Apr-08 | 13-Feb-08 | 50 | 102,000 | M | 50 | AD | MW006052 |
| 191996 | 19-Sep-08 | 26-Jul-08 | 55 | 201 | F | 37 | AD | MW006056 |
| 194346 | 31-Mar-09 | 28-Feb-09 | 31 | 259,000 | M | 29 | AD | MW006071 |
| 194604 | 28-Mar-11 | Feb-11 | 44 | 254,516 | F | 35 | D | MW006076 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omara, D.; Natwijuka, F.; Kapaata, A.; Kato, F.; Kato, L.; Ndekezi, C.; Nakyanzi, A.; Ayebale, M.L.; Yue, L.; Hunter, E.; et al. Subtype AD Recombinant HIV-1 Transmitted/Founder Viruses Are Less Sensitive to Type I Interferons than Subtype D. Viruses 2025, 17, 486. https://doi.org/10.3390/v17040486
Omara D, Natwijuka F, Kapaata A, Kato F, Kato L, Ndekezi C, Nakyanzi A, Ayebale ML, Yue L, Hunter E, et al. Subtype AD Recombinant HIV-1 Transmitted/Founder Viruses Are Less Sensitive to Type I Interferons than Subtype D. Viruses. 2025; 17(4):486. https://doi.org/10.3390/v17040486
Chicago/Turabian StyleOmara, Denis, Fortunate Natwijuka, Anne Kapaata, Frank Kato, Laban Kato, Christian Ndekezi, Angella Nakyanzi, Mercy L. Ayebale, Ling Yue, Eric Hunter, and et al. 2025. "Subtype AD Recombinant HIV-1 Transmitted/Founder Viruses Are Less Sensitive to Type I Interferons than Subtype D" Viruses 17, no. 4: 486. https://doi.org/10.3390/v17040486
APA StyleOmara, D., Natwijuka, F., Kapaata, A., Kato, F., Kato, L., Ndekezi, C., Nakyanzi, A., Ayebale, M. L., Yue, L., Hunter, E., Sande, O. J., Ochsenbauer, C., Kaleebu, P., & Balinda, S. N. (2025). Subtype AD Recombinant HIV-1 Transmitted/Founder Viruses Are Less Sensitive to Type I Interferons than Subtype D. Viruses, 17(4), 486. https://doi.org/10.3390/v17040486








