Temporal Trends in HIV-1 Subtypes and Antiretroviral Drug Resistance Mutations in Istanbul, Türkiye (2021–2024): A Next-Generation Sequencing Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Sample Collection
2.2. HIV-1 RNA Detection and Viral Quantification
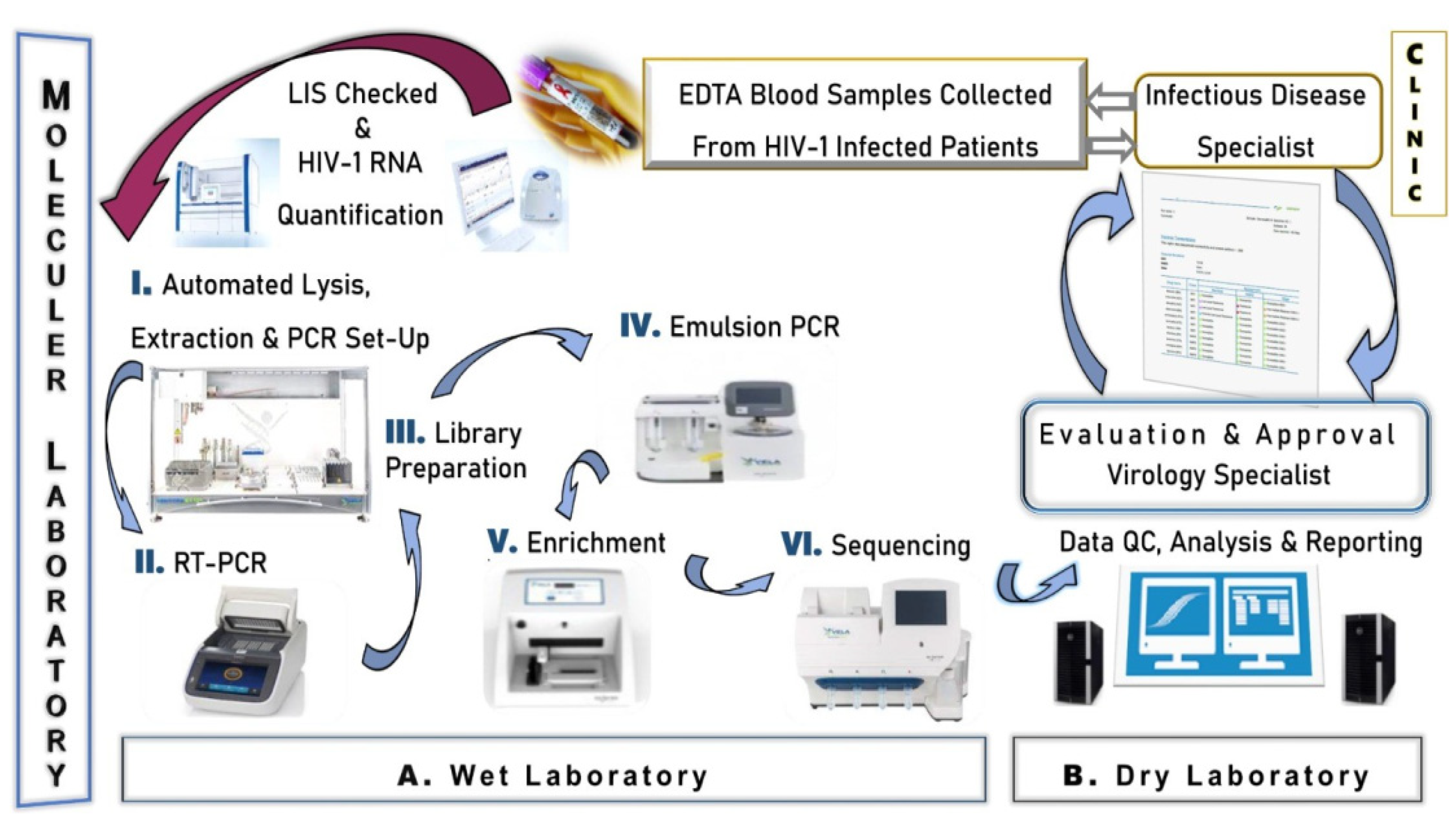
2.3. Next Generation Sequencing
2.3.1. Wet Laboratory Section
2.3.2. Dry Laboratory Section
2.4. Ethics Statement
2.5. Statistical Analysis
3. Results
3.1. Demographic Characteristics of HIV-1 Positive Patients
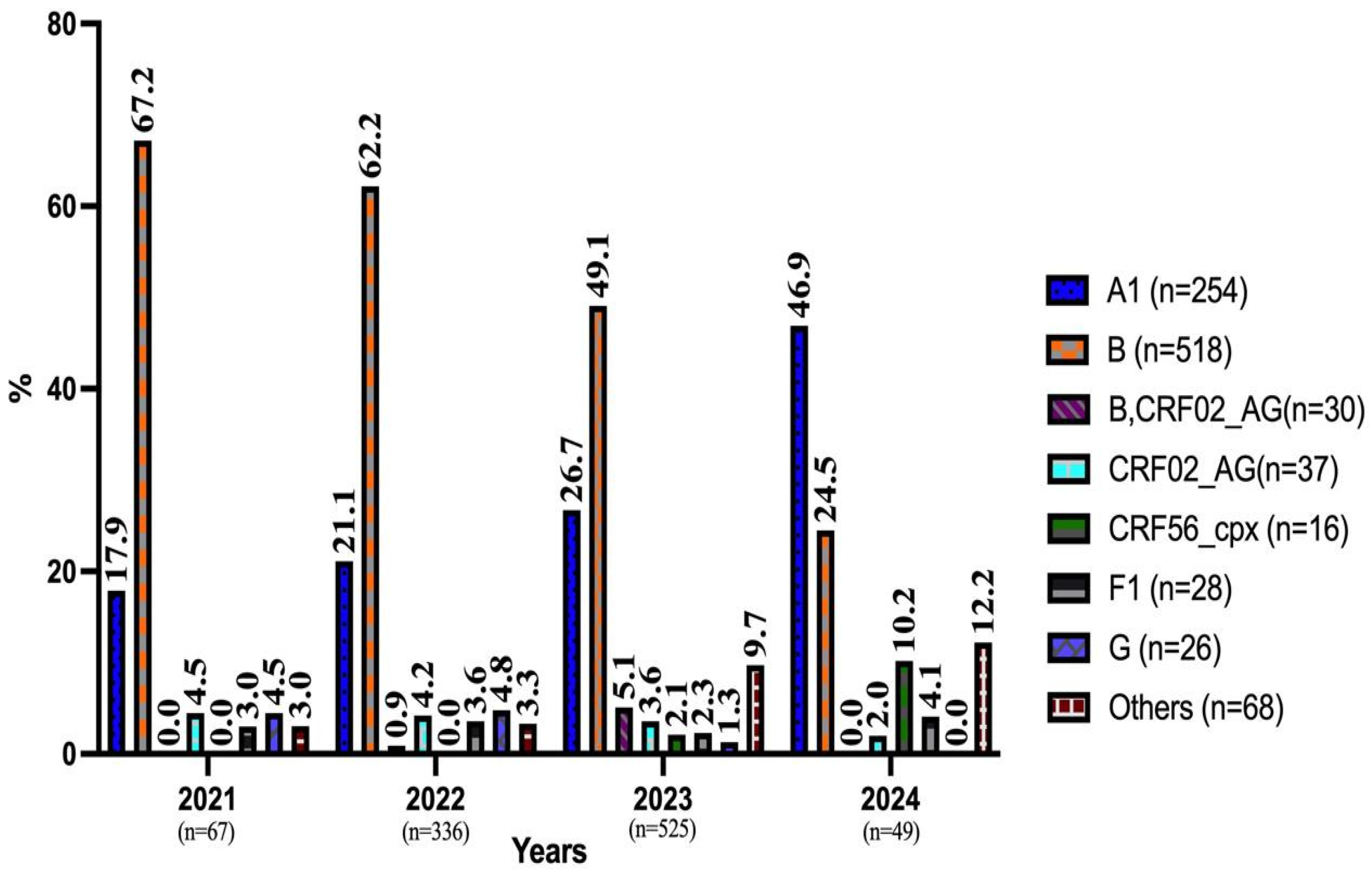
3.2. Prevalence of HIV-1 Genotypes by Year and Demographic Data
3.2.1. Prevalence of RT (NRTI, NNRTI) Resistance and Distribution of Associated Subtype Mutations
3.2.2. Prevalence of PI Drug Resistance and Distribution of Associated Drug Resistance Mutations
3.2.3. Prevalence of INI Drug Resistance and Distribution of Associated Drug Resistance Mutations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mourez, T.; Simon, F.; Plantier, J.-C. Non-M variants of human immunodeficiency virus type 1. Clin. Microbiol. Rev. 2013, 26, 448–461. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.F.; Soares, M.A. HIV Genetic Diversity and Drug Resistance. Viruses 2010, 2, 503–531. [Google Scholar] [CrossRef]
- Konno, Y.; Uriu, K.; Chikata, T.; Takada, T.; Kurita, J.I.; Ueda, M.T.; Islam, S.; Yang Tan, B.J.; Ito, J.; Aso, H.; et al. Two-step evolution of HIV-1 budding system leading to pandemic in the human population. Cell Rep. 2024, 43, 113697. [Google Scholar] [CrossRef]
- Alessandri-Gradt, E.; Moisan, A.; Plantier, J.-C. HIV-1 Non-Group M Strains and ART. Viruses 2023, 15, 780. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, J.; Vallari, A.; McArthur, C.; Sthreshley, L.; Cloherty, G.A.; Berg, M.G.; Rodgers, M.A. Brief Report: Complete Genome Sequence of CG-0018a-01 Establishes HIV-1 Subtype L. J. Acquir. Immune Defic. Syndr. 2020, 83, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Mendes Da Silva, R.K.; Monteiro de Pina Araujo, I.I.; Venegas Maciera, K.; Gonçalves Morgado, M.; Lindenmeyer Guimarães, M. Genetic Characterization of a New HIV-1 Sub-Subtype A in Cabo Verde, Denominated A8. Viruses 2021, 13, 1093. [Google Scholar] [CrossRef]
- Jetzt, A.E.; Yu, H.; Klarmann, G.J.; Ron, Y.; Preston, B.D.; Dougherty, J.P. High rate of recombination throughout the human immunodeficiency virus type 1 genome. J. Virol. 2000, 74, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.M.; Perno, C.F. HIV-1 Genetic Variability and Clinical Implications. ISRN Microbiol. 2013, 481314. [Google Scholar] [CrossRef]
- Los Alamos HIV Databases. HIV Circulating Recombinant Forms (CRFs). Available online: https://www.hiv.lanl.gov/components/sequence/HIV/crfdb/crfs.comp (accessed on 24 January 2025).
- Hemelaar, J.; Elangovan, R.; Yun, J.; Dickson-Tetteh, L.; Kirtley, S.; Gouws-Williams, E.; Ghys, P.D. WHO-UNAIDS Network for HIV Isolation and Characterisation Global and regional epidemiology of HIV-1 recombinants in 1990-2015: A systematic review and global survey. Lancet. Infect. Dis. 2020, 7, e772–e781. [Google Scholar] [CrossRef]
- Williams, A.; Menon, S.; Crowe, M.; Agarwal, N.; Biccler, J.; Bbosa, N.; Ssemwanga, D.; Adungo, F.; Moecklinghoff, C.; Macartney, M.; et al. Geographic and Population Distributions of Human Immunodeficiency Virus (HIV)-1 and HIV-2 Circulating Subtypes: A Systematic Literature Review and Meta-analysis (2010–2021). J. Infect. Dis. 2023, 228, 1583–1591. [Google Scholar] [CrossRef]
- Hemelaar, J. Implications of HIV diversity for the HIV-1 pandemic. J. Infect. 2013, 66, 391–400. [Google Scholar] [CrossRef]
- Castley, A.; Sawleshwarkar, S.; Varma, R.; Herring, B.; Thapa, K.; Dwyer, D.; Chibo, D.; Nguyen, N.; Hawke, K.; Ratcliff, R.; et al. Australian Molecular Epidemiology Network-HIV (AMEN-HIV). A national study of the molecular epidemiology of HIV-1 in Australia 2005–2012. PLoS ONE 2017, 12, e0170601. [Google Scholar] [CrossRef]
- ECDC. HIV/AIDS Surveillance in Europe 2023 Stockholm. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/HIV-AIDS_surveillance_in_Europe_2023_(_2022_data_)_0.pdf (accessed on 23 January 2025).
- UNAIDS. UNAIDS Data 2020. Available online: https://www.unaids.org/sites/default/files/media_asset/2020_aids-data-book_en.pdf?utm (accessed on 23 January 2025).
- Bosa, N.; Kaleebu, P.; Ssemwanga, D. HIV subtype diversity worldwide. Curr. Opin. HIV AIDS 2019, 14, 153–160. [Google Scholar] [CrossRef]
- Lessells, R.J.; Katzenstein, D.K.; de Oliveira, T. Are subtype differences important in HIV drug resistance? Curr. Opin. Virol. 2012, 2, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Ndashimye, E.; Reyes, P.S.; Arts, E.J. New antiretroviral inhibitors and HIV-1 drug resistance: More focus on %90 HIV-1 isolates? FEMS Microbiol. Rev. 2023, 47, fuac040. [Google Scholar] [CrossRef]
- Johnson, M.M.; Jones, C.E.; Clark, D.N. The Effect of Treatment-Associated Mutations on HIV Replication and Transmission Cycles. Viruses 2023, 15, 107. [Google Scholar] [CrossRef]
- Deeks, S.; Overbaugh, J.; Phillips, A.; Buchbinder, S. HIV infection. Nat. Rev. Dis. Primers 2015, 1, 15035. [Google Scholar] [CrossRef]
- Günthard, H.F.; Calvez, V.; Paredes, R.; Pillay, D.; Shafer, R.W.; Wensing, A.M.; Jacobsen, D.M.; Richman, D.D. Human Immunodeficiency Virus Drug Resistance: 2018 Recommendations of the International Antiviral Society-USA Panel. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2019, 68, 177–187. [Google Scholar] [CrossRef]
- Mbunkah, H.A.; Bertagnolio, S.; Hamers, R.L.; Hunt, G.; Inzaule, S.; Rinke De Wit, T.F.; Paredes, R.; Parkin, N.T.; Jordan, M.R.; Metzner, K.J.; et al. Low-Abundance Drug-Resistant HIV-1 Variants in Antiretroviral Drug-Naive Individuals: A Systematic Review of Detection Methods, Prevalence, and Clinical Impact. J. Infect. Dis. 2020, 221, 1584–1597. [Google Scholar] [CrossRef]
- Metzner, K.J. Technologies for HIV-1 drug resistance testing: Inventory and needs. Curr. Opin. HIV AIDS 2022, 17, 222–228. [Google Scholar] [CrossRef]
- EACS. EACS Guidelines 2024. Available online: https://eacs.sanfordguide.com (accessed on 24 January 2025).
- Stolbova, E.A.; Stolbov, L.A.; Filimonov, D.A.; Poroikov, V.V.; Tarasova, O.A. Quantitative Prediction of Human Immunodeficiency Virus Drug Resistance. Viruses 2024, 16, 1132. [Google Scholar] [CrossRef]
- Abecasis, A.B.; Pingarilho, M.; Vandamme, A.M. Phylogenetic analysis as a forensic tool in HIV transmission investigations. AIDS 2018, 32, 543–554. [Google Scholar] [CrossRef]
- Günthard, H.F.; Saag, M.S.; Benson, C.A.; del Rio, C.; Eron, J.J.; Gallant, J.E.; Hoy, J.F.; Mugavero, M.J.; Sax, P.E.; Thompson, M.A.; et al. Antiretroviral Drugs for Treatment and Prevention of HIV Infection in Adults: 2016 Recommendations of the International Antiviral Society-USA Panel. JAMA 2016, 316, 191–210. [Google Scholar] [CrossRef] [PubMed]
- Saag, M.S.; Gandhi, R.T.; Hoy, J.F.; Landovitz, R.J.; Thompson, M.A.; Sax, P.E.; Smith, D.M.; Benson, C.A.; Buchbinder, S.P.; Del Rio, C.; et al. Antiretroviral Drugs for Treatment and Prevention of HIV Infection in Adults: 2020 Recommendations of the International Antiviral Society-USA Panel. JAMA 2020, 324, 1651–1669. [Google Scholar] [CrossRef] [PubMed]
- Di Giallonardo, F.; Pinto, A.N.; Keen, P.; Shaik, A.; Carrera, A.; Salem, H.; Selvey, C.; Nigro, S.J.; Fraser, N.; Price, K.; et al. Subtype-specific differences in transmission cluster dynamics of HIV-1 B and CRF01_AE in New South Wales, Australia. J. Int. AIDS Soc. 2021, 24, e25655. [Google Scholar] [CrossRef] [PubMed]
- Ávila-Ríos, S.; Parkin, N.; Swanstrom, R.; Paredes, R.; Shafer, R.; Ji, H.; Kantor, R. Next-Generation Sequencing for HIV Drug Resistance Testing: Laboratory, Clinical, and Implementation Considerations. Viruses 2020, 12, 617. [Google Scholar] [CrossRef]
- Tzou, P.L.; Ariyaratne, P.; Varghese, V.; Lee, C.; Rakhmanaliev, E.; Villy, C.; Yee, M.; Tan, K.; Michel, G.; Pinsky, B.A.; et al. Comparison of an In Vitro Diagnostic Next-Generation Sequencing Assay with Sanger Sequencing for HIV-1 Genotypic Resistance Testing. J. Clin. Microbiol. 2018, 56, e00105-18. [Google Scholar] [CrossRef]
- Bonifacio, M.A.; Genchi, C.; Lagioia, A.; Talamo, V.; Volpe, A.; Mariggiò, M.A. Analytical Assessment of the Vela Diagnostics NGS Assay for HIV Genotyping and Resistance Testing: The Apulian Experience. Int. J. Mol. Sci. 2022, 23, 2727. [Google Scholar] [CrossRef]
- FDA. Premarket Notification. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpmn/pmn.cfm?ID=BK050035 (accessed on 23 January 2025).
- May, S.; Adamska, E.; Tang, J. Evaluation of Vela Diagnostics HIV-1 genotyping assay on an automated next generation sequencing platform. J. Clin. Virol. 2020, 127, 104376. [Google Scholar] [CrossRef]
- FDA. BR190330—Sentosa HIV-1 Genotyping Assay. Available online: https://www.fda.gov/vaccines-blood-biologics/substantially-equivalent-510k-device-information/br190330-sentosa-sq-hiv-1-genotyping-assay (accessed on 23 January 2025).
- WHO. HIV Drug Resistance. Available online: https://iris.who.int/bitstream/handle/10665/259731/9789241512879-eng.pdf?sequence=1 (accessed on 23 January 2025).
- Vela Diagnostics. Sentosa® SQ HIV-1 Genotyping Assay v2.0. Available online: https://www.veladx.com./product/virology/sentosa-sq-hiv-genotyping-assay-v2.html (accessed on 23 January 2025).
- WHO. Global Health Sector Strategies on, Respectively, HIV, Viral Hepatitis and Sexually Transmitted Infections for the Period 2022–2030. Available online: https://www.who.int/teams/global-hiv-hepatitis-and-stis-programmes/strategies/global-health-sector-strategies (accessed on 24 January 2025).
- Mude, W.; Mwenyango, H.; Preston, R.; O’Mullan, C.; Vaughan, G.; Jones, G. HIV Testing Disruptions and Service Adaptations During the COVID-19 Pandemic: A Systematic Literature Review. AIDS Behav. 2024, 28, 186–200. [Google Scholar] [CrossRef]
- Çerçi, P.; İnkaya, A.Ç.; Alp, Ş.; Tümer, A.; Ünal, S. Evaluation of 255 HIV/AIDS cases: Hacettepe cohort, Ankara, Turkey. Mikrobiyoloji Bul. 2016, 50, 94–103. [Google Scholar] [CrossRef]
- Republic of Turkıye Ministry of Health General Directorate of Public Health. Statistical Data: HIV/AIDS. Available online: https://hsgm.saglik.gov.tr/depo/birimler/bulasici-hastaliklar-ve-erken-uyari-db/Dokumanlar/Istatistikler/Ek_HIV-AIDS_Istatistikleri.pdf (accessed on 24 January 2025).
- Qiagen. Qiasymphony Virus Kit Handbook. Available online: https://www.qiagen.com/us/resources/resourcedetail?id=7621a8dd-9144-4a4b-af31-5c229a3177cc&lang=en (accessed on 24 January 2025).
- Fleiss, J.L. Statistical Methods for Rates and Proportions, 1st ed; John Wiley & Son: London, UK, 1981; 218p. [Google Scholar]
- Perez, S.M.; Panneer, N.; France, A.M.; Carnes, N.; Curran, K.G.; Denson, D.J.; Oster, A.M. Clusters of Rapid HIV Transmission Among Gay, Bisexual, and Other Men Who Have Sex with Men—United States, 2018–2021. MMWR Morb. Mortal. Wkly. Rep. 2022, 71, 1201–1206. [Google Scholar] [CrossRef]
- Magiorkinis, G.; Angelis, K.; Mamais, I.; Katzourakis, A.; Hatzakis, A.; Albert, J.; Lawyer, G.; Hamouda, O.; Struck, D.; Vercauteren, J.; et al. The global spread of HIV-1 subtype B epidemic. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2016, 46, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Tekin, D.; Gokengin, D.; Onay, H.; Erensoy, S.; Sertoz, R. Investigation of drug resistance against protease, reverse transcriptase, and integrase inhibitors by next-generation sequencing in HIV-positive patients. J. Med. Virol. 2021, 93, 3627–3633. [Google Scholar] [CrossRef] [PubMed]
- Köksal, M.O.; Beka, H.; Lübke, N.; Verheyen, J.; Eraksoy, H.; Cagatay, A.; Kaiser, R.; Akgül, B.; Agacfidan, A. HIV-1 subtypes and drug resistance profiles in a cohort of heterosexual patients in Istanbul, Turkey. Med. Microbiol. Immunol. 2015, 204, 551–555. [Google Scholar] [CrossRef]
- Sayan, M.; Gündüz, A.; Ersöz, G.; İnan, A.; Deveci, A.; Özgür, G.; Sargın, F.; Karagöz, G.; İnci, A.; İnan, D.; et al. Integrase Strand Transfer Inhibitors (INSTIs) Resistance Mutations in HIV-1 Infected Turkish Patients. HIV Clin. Trials 2016, 17, 109–113. [Google Scholar]
- Jovanovic, L.; Siljic, M.; Cirkovic, V.; Salemovic, D.; Jevtovic, D.; Alexiev, I.; Zidovec-Lepej, S.; Oroz, M.; Begovac, J.; Paraskevis, D.; et al. HIV-1 subtype B spread through cross-border clusters in the Balkans: A molecular analysis in view of incidence trends. AIDS 2023, 37, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Kantzanou, M.; Karalexi, M.A.; Papachristou, H.; Vasilakis, A.; Rokka, C.; Katsoulidou, A. Transmitted drug resistance among HIV-1 drug-naïve patients in Greece. Int. J. Infect. Dis. 2021, 105, 42–48. [Google Scholar] [CrossRef]
- Áy, É.; Müller, V.; Mezei, M.; Pocskay, Á.; Koroknai, A.; Müller, D.; Győri, Z.; Marschalkó, M.; Tóth, B.; Kárpáti, S.; et al. Transmitted drug resistance in newly diagnosed and treatment-naïve HIV type 1-infected patients in Hungary. J. Glob. Antimicrob. Resist. 2020, 20, 124–130. [Google Scholar] [CrossRef]
- Gartner, M.J.; Roche, M.; Churchill, M.J.; Gorry, P.R.; Flynn, J.K. Understanding the mechanisms driving the spread of subtype C HIV-1. EBioMedicine 2020, 53, 102682. [Google Scholar] [CrossRef]
- Kiros, M.; Biset, S.; Gebremariam, B.; Yalew, G.T.; Abegaz, W.E.; Geteneh, A. Trends in HIV-1 pretreatment drug resistance and HIV-1 variant dynamics among antiretroviral therapy-naive Ethiopians from 2003 to 2018: A pooled sequence analysis. Virol. J. 2023, 20, 243. [Google Scholar] [CrossRef] [PubMed]
- Miranda, M.N.S.; Pingarilho, M.; Pimentel, V.; Martins, M.D.R.O.; Kaiser, R.; Seguin-Devaux, C.; Paredes, R.; Zazzi, M.; Incardona, F.; Abecasis, A.B. Trends of Transmitted and Acquired Drug Resistance in Europe from 1981 to 2019: A Comparison Between the Populations of Late Presenters and Non-late Presenters. Front. Microbiol. 2022, 13, 846943. [Google Scholar] [CrossRef] [PubMed]
- Mimtsoudis, I.; Tsachouridou, O.; Akinosoglou, K.; Metallidis, S. Treatment Management Challenges in Naïve and Experienced HIV-1-Infected Individuals Carrying the M184V Mutation. Viruses 2024, 16, 1392. [Google Scholar] [CrossRef]
- Etta, E.M.; Mavhandu, L.; Manhaeve, C.; McGonigle, K.; Jackson, P.; Rekosh, D.; Hammarskjold, M.L.; Bessong, P.O.; Tebit, D.M. High level of HIV-1 drug resistance mutations in patients with unsuppressed viral loads in rural northern South Africa. AIDS Res. Ther. 2017, 14, 36. [Google Scholar] [CrossRef]
- Maldonado, J.O.; Mansky, L.M. The HIV-1 Reverse Transcriptase A62V Mutation Influences Replication Fidelity and Viral Fitness in the Context of Multi-Drug-Resistant Mutations. Viruses 2018, 10, 376. [Google Scholar] [CrossRef]
- Iyidogan, P.; Anderson, K.S. Current perspectives on HIV-1 antiretroviral drug resistance. Viruses 2014, 6, 4095–4139. [Google Scholar] [CrossRef] [PubMed]
- Hofstra, L.M.; Sauvageot, N.; Albert, J.; Alexiev, I.; Garcia, F.; Struck, D.; Van de Vijver, D.A.M.C.; Åsjö, B.; Beshkov, D.; Coughlan, S.; et al. Transmission of HIV Drug Resistance and the Predicted Effect on Current First-line Regimens in Europe. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2016, 62, 655–663. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, M.; Wang, J.; Liu, L.; Chen, J.; Zhang, R.; Tang, Y.; Shen, Y.; Qi, T.; Song, W.; et al. Efficacy of Efavirenz-Based Regimen in Antiretroviral-Naïve Patients with HIV-1 V179D/E Mutations in Shanghai, China. Infect. Dis. Ther. 2023, 12, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, V.; Pingarilho, M.; Sebastião, C.S.; Miranda, M.; Gonçalves, F.; Cabanas, J.; Costa, I.; Diogo, I.; Fernandes, S.; Costa, O.; et al. Applying Next-Generation Sequencing to Track HIV-1 Drug Resistance Mutations Circulating in Portugal. Viruses 2024, 16, 622. [Google Scholar] [CrossRef]
- Ye, J.; Dong, Y.; Lan, Y.; Chen, J.; Zhou, Y.; Liu, J.; Yuan, D.; Lu, X.; Guo, W.; Zheng, M.; et al. Trends and Patterns of HIV Transmitted Drug Resistance in China From 2018 to 2023. J. Infect. Dis. 2024, 230, 1410–1421. [Google Scholar] [CrossRef]
- Varghese, V.; Mitsuya, Y.; Fessel, W.J.; Liu, T.F.; Melikian, G.L.; Katzenstein, D.A.; Schiffer, C.A.; Holmes, S.P.; Shafer, R.W. Prototypical Recombinant Multi-Protease-Inhibitor-Resistant Infectious Molecular Clones of Human Immunodeficiency Virus Type 1. Antimicrob. Agents Chemother. 2013, 57, 4290–4299. [Google Scholar] [CrossRef] [PubMed]
- Schapiro, J.M.; Scherer, J.; Boucher, C.A.; Baxter, J.D.; Tilke, C.; Perno, C.F.; Maggiolo, F.; Santoro, M.M.; Hall, D.B. Improving the prediction of virological response to tipranavir: The development and validation of a tipranavir-weighted mutation score. Antivir. Ther. 2010, 15, 1011–1019. [Google Scholar] [CrossRef]
- Charpentier, C.; Malet, I.; Andre-Garnier, E.; Storto, A.; Bocket, L.; Amiel, C.; Morand-Joubert, L.; Tumiotto, C.; Nguyen, T.; Maillard, A.; et al. Phenotypic analysis of HIV-1 E157Q integrase polymorphism and impact on virological outcome in patients initiating an integrase inhibitor-based regimen. J. Antimicrob. Chemother. 2018, 73, 1039–1044. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, R.T.; Bedimo, R.; Hoy, J.F.; Landovitz, R.J.; Smith, D.M.; Eaton, E.F.; Lehmann, C.; Springer, S.A.; Sax, P.E.; Thompson, M.A.; et al. Antiretroviral Drugs for Treatment and Prevention of HIV Infection in Adults: 2022 Recommendations of the International Antiviral Society-USA Panel. JAMA 2023, 329, 63–84. [Google Scholar] [CrossRef]
- Wagner, S.; Kurz, M.; Klimkait, T.; Swiss HIV Cohort Study. Algorithm evolution for drug resistance prediction: Comparison of systems for HIV-1 genotyping. Antivir. Ther. 2015, 20, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; May, S.; Richman, D.D.; Hecht, F.M.; Markowitz, M.; Daar, E.S.; Routy, J.P.; Margolick, J.B.; Collier, A.C.; Woelk, C.H.; et al. Comparison of algorithms that interpret genotypic HIV-1 drug resistance to determine the prevalence of transmitted drug resistance. AIDS 2008, 22, 835–839. [Google Scholar] [CrossRef]
- Ventosa-Cubillo, J.; Pinzón, R.; González-Alba, J.M.; Estripeaut, D.; Navarro, M.L.; Holguín, Á. Drug resistance in children and adolescents with HIV in Panama. J. Antimicrob. Chemother. 2023, 78, 423–435. [Google Scholar] [CrossRef]
- Rubio-Garrido, M.; Reina, G.; Ndarabu, A.; Rodriguez-Galet, A.; Valadés-Alcaraz, A.; Barquín, D.; Carlos, S.; Holguín, Á. High drug resistance levels could compromise the control of HIV infection in pediatric and adolescent population in Kinshasa, the Democratic Republic of Congo. PLoS ONE 2021, 16, e0248835. [Google Scholar] [CrossRef]
- Sohn, A.H.; Nuttall, J.J.; Zhang, F. Sequencing of antiretroviral therapy in children in low- and middle-income countries. Curr. Opin. HIV AIDS 2010, 5, 54–60. [Google Scholar] [CrossRef]
- WHO. HIV Drug Resistance Report 2019. Geneva, Switzerland: (WHO/CDS/HIV/19.21). Licence: CC BY-NC-SA 3.0 IGO. Available online: https://iris.who.int/handle/10665/325891 (accessed on 8 March 2025).
| All | Turkish | Others | |||||||
|---|---|---|---|---|---|---|---|---|---|
| All | M | F | All | M | F | All | M | F | |
| n (%) | 1029 (100) | 922 (89.6) | 107 (10.4) | 967 (100) | 871 (90.1) | 96 (9.9) | 62 (100) | 51 (82.3) | 11 (17.7) |
| Age Groups | |||||||||
| <25 n | 118 | 109 | 9 | 114 | 105 | 9 | 4 | 4 | - |
| 25–34 n | 366 | 341 | 25 | 339 | 317 | 22 | 27 | 24 | 3 |
| 35−44 n | 291 | 257 | 34 | 273 | 243 | 30 | 18 | 14 | 4 |
| 45−54 n | 148 | 128 | 20 | 137 | 119 | 18 | 11 | 9 | 2 |
| >54 n | 106 | 87 | 19 | 104 | 87 | 17 | 2 | - | 2 |
| Age Median | 36 | 35 | 41 | 36 | 35 | 40 | 34 | 34 | 41 |
| IQR | 28−44 | 28−44 | 33−49 | 28−44 | 28−44 | 32−48 | 29–43 | 29−42 | 33−53 |
| Viral Load (log10) | 1.78 | 1.84 | 1.03 | 1.72 | 1.81 | 0.98 | 2.29 | 2.39 | 1.50 |
| IQR | 3.99 × 104– 6.74 × 105 | 4.31 × 104– 6.81 × 105 | 1.10 × 104– 5.72 × 105 | 4.03 × 104– 6.81 × 105 | 4.36 × 104– 6.84 × 105 | 1.15 × 104– 5.26 × 105 | 2.56 × 104– 5.78 × 105 | 2.81 × 104– 5.27 × 105 | 4.61 × 103– 1.12 × 106 |
| A1 | B | B & CRF02_AG | CRF02_AG | CRF56_cpx | F1 | G | Others | |
|---|---|---|---|---|---|---|---|---|
| Gender | ||||||||
| Male n (%) | 230 (26.3) | 459 (52.5) | 28 (3.2) | 31 (3.5) | 14 (1.6) | 26 (3.0) | 26 (3.0) | 60 (6.9) |
| Female n (%) | 24 (23.3) | 59 (57.3) | 2 (1.9) | 6 (5.8) | 2 (1.9) | 2 (1.9) | 0 | 8 (7.8) |
| Etnicity | ||||||||
| Turkish n (%) | 227 (24.8) | 498 (54.4) | 28 (3.1) | 33 (3.6) | 16 (1.7) | 28 (3.1) | 23 (2.5) | 62 (6.8) |
| Others n (%) | 27 (43.5) | 20 (32.3) | 2 (3.2) | 4 (6.5) | 0 | 0 | 3 (4.8) | 6 (9.7) |
| NRTI (n = 116) | NNRTI (n = 207) | ||||||
|---|---|---|---|---|---|---|---|
| AA | Mutation | Prevalence n (%) | Only Mutation n (%) | AA | Mutation | Prevalence n (%) | Only Mutation n (%) |
| M41 | L | 21 (18.1) | 4 (19.0) | A98 | G | 3 (1.4) | 2 (66.6) |
| E44 | D | 6 (5.2) | 6 (100) | L100 | V | 2 (0.9) | 1 (50) |
| A62 | V | 45 (38.7) | 41 (91.1) | K101 | E | 8 (3.8) | 2 (25) |
| K65 | R | 5 (4.3) | 0 (0) | Q | 2 (0.9) | 0 (0) | |
| D67 | R | 3 (2.6) | 1 (33.3) | R | 8 (3.8) | 7 (87.5) | |
| G | 1 (0.8) | 0 (0) | K103 | E | 6 (2.8) | 2 (33.3) | |
| N | 2 (1.7) | 0 (0) | N | 7 (3.3) | 6 (85.7) | ||
| T69 | D | 1 (0.8) | 0 (0) | R | 2 (0.9) | 1 (50) | |
| insertion | 1 (0.8) | 0 (0) | V108 | I | 11 (5.3) | 10 (90.9) | |
| K70 | E | 2 (1.7) | 1 (50.0) | E138 | A | 115 (55.5) | 102 (88.6) |
| N | 5 (4.3) | 3 (60.0) | G | 24 (11.5) | 24 (100) | ||
| Q | 1 (0.8) | 0 (0) | K | 1 (0.4) | 0 (0) | ||
| R | 1 (0.8) | 0 (0) | Q | 2 (0.9) | 0 (0) | ||
| T | 2 (1.7) | 0 (0) | V179 | T | 9 (4.3) | 8 (88.8) | |
| L74 | I | 1 (0.8) | 0 (0) | D | 14 (6.7) | 10 (71.4) | |
| V75 | A | 1 (0.8) | 1 (100) | E | 5 (2.4) | 5 (100) | |
| M | 1 (0.8) | 0 (0) | Y188 | C | 1 (0.4) | 1 (100) | |
| Y115 | F | 1 (0.8) | 0 (0) | L | 2 (0.9) | 1 (50) | |
| M184 | I | 3 (2.6) | 1 (33.3) | G190 | S | 3 (1.4) | 2 (66.6) |
| V | 26 (22.4) | 8 (30.7) | M230 | I | 1 (0.4) | 1 (100) | |
| L210 | W | 1 (0.8) | 1 (100) | Y318 | F | 0 (0) | 0 (0) |
| T215 | C | 12 (10.3) | 3 (25) | N348 | I | 4 (1.9) | 3 (75) |
| D | 5 (4.3) | 1 (20.0) | |||||
| E | 8 (6.8) | 4 (50) | |||||
| I | 1 (0.8) | 0 (0) | |||||
| S | 5 (4.3) | 1 (20.0) | |||||
| Y | 2 (1.7) | 0 (0) | |||||
| K219 | R | 2 (1.7) | 2 (100) | ||||
| Q | 3 (2.6) | 2 (66.6) | |||||
| PI Major (n = 10) | PI Accessory (n = 16) | ||||||
|---|---|---|---|---|---|---|---|
| AA | Mutation | Prevalence n (%) | Only Mutation n (%) | AA | Mutation | Prevalence n (%) | Only Mutation n (%) |
| D30 | N | 1 (8.3) | 1 (100) | K20 | T | 2 (11.7) | 2 (100) |
| M46 | I | 4 (33.3) | 3 (75) | L33 | F | 2 (11.7) | 1 (50) |
| L | 3 (25) | 3 (100) | K43 | T | 9 (52.9) | 8 (88.8) | |
| I50 | L | 1 (8.3) | 1 (100) | F53 | L | 1 (5.8) | 1 (100) |
| V | 1 (8.3) | 1 (100) | Q58 | E | 3 (17.6) | 3 (100) | |
| 154 | L | 1 (8.3) | 0 (0) | N63 | D | 1 (5.8) | 1 (100) |
| V | 1 (8.3) | 0 (0) | |||||
| L76 | V | 1 (8.3) | 0 (0) | ||||
| V82 | A | 1 (8.3) | 0 (0) | ||||
| I84 | V | 2 (16.6) | 1 (50) | ||||
| Integrase Inhibitor Mutation Major (n = 11) | Integrase Inhibitor Mutation Accessory (n = 44) | ||||||
|---|---|---|---|---|---|---|---|
| AA | Mutation | Prevalence n (%) | Only Mutation n (%) | AA | Mutation | Prevalence n (%) | Only Mutation n (%) |
| T66 | A | 2 (11.7) | 2 (100) | Q95 | K | 2 (4.1) | 1 (50) |
| E92 | G | 3 (17.6) | 2 (66.6) | T97 | A | 14 (29.1) | 12 (85.7) |
| Q | 3 (17.6) | 3 (100) | A128 | T | 6 (12.5) | 5 (83.3) | |
| E138 | K A | 2 (11.7) | 1 (50.0) | E157 | Q | 16 (33.3) | 15 (93.7) |
| G140 | S | 2 (11.7) | 0 (0) | G163 | K | 3 (6.2) | 3 (100) |
| Y143 | C | 3 (17.6) | 1 (33.3) | R | 10 (20.8) | 7 (70.0) | |
| R | 1 (5.8) | 0 (0) | S230 | R | 1(2.0) | 1 (100) | |
| S147 | G | 1 (5.8) | 0 (0) | ||||
| Q148 | H | 2 (11.7) | 0 (0) | ||||
| R | 1 (5.8) | 0 (0) | |||||
| N155 | H | 1 (5.8) | 0 (0) | ||||
| T | 1 (5.8) | 1 (100) | |||||
| R263 | K | 1 (5.8) | 1 (100) | ||||
| Methods | Statistics | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Standford | ANRS | Rega | ||||||||
| S | R (I + H) | S | R (I + H) | S | R (I + H) | |||||
| Drug Groups | Drugs | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | p-Value | Fleiss’s Kappa (κ) | |
| RT (n = 931) | N R T I | ABC | 890 (95.6) | 41 (4.4) | 889 (95.5) | 42 (4.5) | 919 (99) | 12 (1.3) | <0.001 | 0.608 |
| AZT | 886 (95.2) | 45 (4.8) | 898 (96.5) | 33 (3.5) | 899 (96.6) | 32 (3.4) | <0.001 | 0.858 | ||
| D4T | 876 (94.1) | 55 (5.9) | 900 (96.7) | 31 (3.3) | 898 (96.5) | 33 (3.5) | <0.001 | 0.719 | ||
| DDI | 874 (93.9) | 57 (6.1) | 924 (99.2) | 7 (0.8) | 903 (97) | 28 (3.0) | <0.001 | 0.236 | ||
| N N R T I | FTC | 900 (96.7) | 31 (3.3) | 905 (97.2) | 26 (2.8) | 904 (97.1) | 27 (2.9) | <0.001 | 0.939 | |
| 3TC | 899 (96.6) | 32 (3.4) | 904 (97.1) | 27 (2.9) | 903 (97) | 28 (3.0) | <0.001 | 0.941 | ||
| TDF | 904 (97.1) | 27 (2.9) | 922 (99) | 9 (1.0) | 923 (99.1) | 8 (0.9) | <0.001 | 0.561 | ||
| EFV | 850 (91.3) | 81 (8.7) | 907 (97.4) | 24 (2.6) | 909 (97.6) | 22 (2.4) | <0.001 | 0.439 | ||
| ETR | 768 (82.5) | 163 (17.5) | 772 (82.9) | 159 (17.1) | 926 (99.5) | 5 (0.5) | <0.001 | 0.363 | ||
| NVP | 844 (90.7) | 87 (9.3) | 906 (97.3) | 25 (2.7) | 909 (97.6) | 22 (2.4) | <0.001 | 0.443 | ||
| RPV | 763 (82) | 168 (18) | 778 (83.6) | 153 (16.4) | 789 (84.7) | 142 (15.3) | <0.001 | 0.933 | ||
| PI (n = 912) | ATV/r | 897 (98.4) | 15 (1.6) | 730 (80) | 182 (20) | 905 (99.2) | 7 (0.8) | 0.294 | 0.020 | |
| DRV/r | 908 (99.6) | 4 (0.4) | 911 (99.9) | 1 (0.1) | 911 (99.9) | 1 (0.1) | <0.001 | 0.499 | ||
| FPV/r | 893 (97.9) | 19 (2.1) | 905 (99.2) | 7 (0.8) | 907(99.4) | 5 (0.6) | <0.001 | 0.511 | ||
| IDV/r | 897 (98.4) | 15 (1.6) | 901 (98.8) | 11 (1.2) | 905 (99.2) | 7 (0.8) | <0.001 | 0.724 | ||
| LPV/r | 897 (98.4) | 15 (1.6) | 897 (98.4) | 15 (1.6) | 909 (99.7) | 3 (0.3) | <0.001 | 0.356 | ||
| NFV | 879 (96.4) | 33 (3.6) | 906 (99.3) | 6 (0.7) | 904 (99.1) | 8 (0.9) | <0.001 | 0.329 | ||
| SQV/r | 898 (98.5) | 14 (1.5) | 653 (71.6) | 259 (28.4) | 908 (99.6) | 4 (0.4) | 0.002 | −0.060 | ||
| TPV/r | 889 (97.5) | 23 (2.5) | 282 (30.9) | 630 (69.1) | 911 (99.9) | 1 (0.1) | <0.001 | −0.277 | ||
| INI (n= 977) | DTG | 964 (98.7) | 13 (1.3) | 959 (98.1) | 18 (1.9) | 972 (99.5) | 5 (0.5) | <0.001 | 0.297 | |
| EVG | 921 (94.3) | 56 (5.7) | 936 (95.8) | 41 (4.2) | 961 (98.4) | 16 (1.6) | <0.001 | 0.595 | ||
| RAL | 921 (94.3) | 56 (5.7) | 951 (97.3) | 26 (2.7) | 946 (96.8) | 31 (3.2) | <0.001 | 0.383 | ||
| 2021 | 2022 | 2023 | 2024 a | p-Value | |||||
|---|---|---|---|---|---|---|---|---|---|
| S | R (I + H) | S | R (I + H) | S | R (I + H) | S | R (I + H) | ||
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | ||
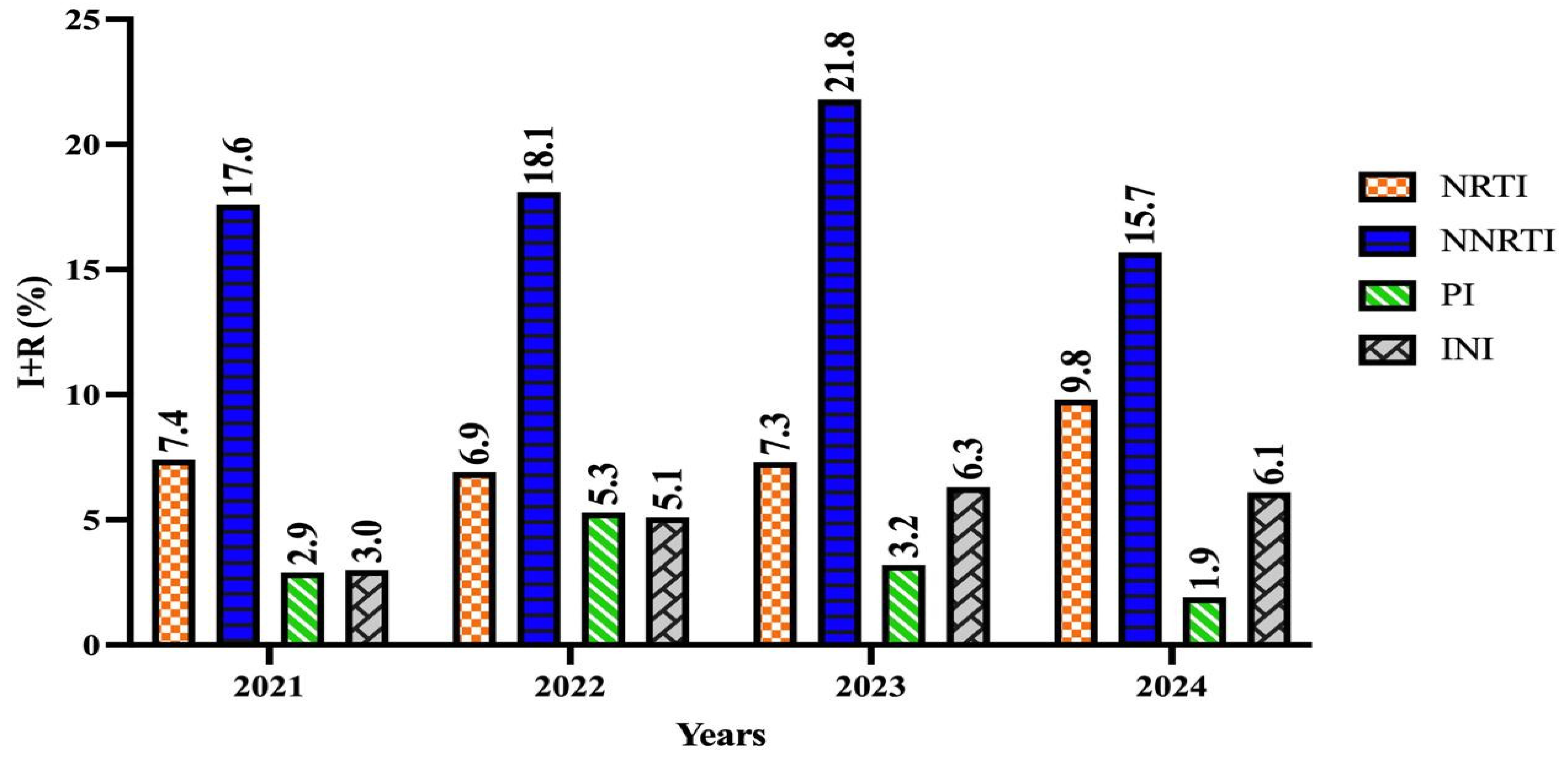
| NRTI | 63 (92.6) | 5 (7.4) | 299 (93.1) | 22 (6.9) | 455 (92.7) | 36 (7.3) | 46 (90.2) | 5 (9.8) | 0.904 |
| NNRTI | 56 (82.4) | 12 (17.6) | 263 (81.9) | 58 (18.1) | 384 (78.2) | 107 (21.8) | 43 (84.3) | 8 (15.7) | 0.464 |
| PI | 66 (97.1) | 2 (2.9) | 303 (94.7) | 17 (5.3) | 457 (96.8) | 15 (3.2) | 50 (98) | 1 (1.9) | 0.380 |
| INI | 65 (97) | 2 (3) | 319 (94.9) | 17 (5.1) | 492 (93.7) | 33 (6.3) | 46 (93.9) | 3 (6.1) | 0.674 |
| N | Gender (M/F) | Etnicity | Age | Viral Load Copy/mL | Subtype | NRTI | NNRTI | PI | INI | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | IR | HR | S | IR | HR | S | IR | HR | S | IR | HR | ||||||
| 1 | M | TR | 13 | 51,220 | CRF02_AG | TDF | ABC AZT D4T DDI | FTC 3TC | ETR RVP | EFV NVP | - | DRV/r | ATV/r FPV/r IDV/r LPV/r SQV/r TPV/r | NFV | DTG | EVG RAL | - |
| 2 | M | TR | 15 | 54,814,294 | B | All | - | - | All | - | - | All | - | - | All | - | - |
| 3 | M | TR | 1 | 16,263 | A1 | All | - | - | All | - | - | All | - | - | All | - | - |
| 4 | M | TR | 17 | 105,000 | B + CRF02_AG + G + J | All | - | - | All | - | - | All | - | - | All | - | - |
| 5 | F | TR | 2 | 124,854 | B | AZT D4T TDF | ABC DDI | FTC 3TC | - | NVP RPV DOR EFV ETR | - | All | - | - | All | - | - |
| 6 | M | Other | 17 | 1,040,000 | B | All | - | - | All | - | - | All | - | - | All | - | - |
| 7 | M | TR | 16 | 1,580,000 | CRF02_AG | All | - | - | All | - | - | All | - | - | All | - | - |
| 8 | M | TR | 16 | 1,100,120 | CRF02_AG + G | All | - | - | All | - | - | All | - | - | All | - | - |
| 9 | M | TR | 1 | 794,000 | D + G | All | - | - | All | - | - | All | - | - | All | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yaman, M.; Gülcen, B.S.; Özgüler, K.; Köksal, M.O.; Tekol, S.D.; İlki, A. Temporal Trends in HIV-1 Subtypes and Antiretroviral Drug Resistance Mutations in Istanbul, Türkiye (2021–2024): A Next-Generation Sequencing Study. Viruses 2025, 17, 478. https://doi.org/10.3390/v17040478
Yaman M, Gülcen BS, Özgüler K, Köksal MO, Tekol SD, İlki A. Temporal Trends in HIV-1 Subtypes and Antiretroviral Drug Resistance Mutations in Istanbul, Türkiye (2021–2024): A Next-Generation Sequencing Study. Viruses. 2025; 17(4):478. https://doi.org/10.3390/v17040478
Chicago/Turabian StyleYaman, Murat, Begüm Saran Gülcen, Kübra Özgüler, Muammer Osman Köksal, Serap Demir Tekol, and Arzu İlki. 2025. "Temporal Trends in HIV-1 Subtypes and Antiretroviral Drug Resistance Mutations in Istanbul, Türkiye (2021–2024): A Next-Generation Sequencing Study" Viruses 17, no. 4: 478. https://doi.org/10.3390/v17040478
APA StyleYaman, M., Gülcen, B. S., Özgüler, K., Köksal, M. O., Tekol, S. D., & İlki, A. (2025). Temporal Trends in HIV-1 Subtypes and Antiretroviral Drug Resistance Mutations in Istanbul, Türkiye (2021–2024): A Next-Generation Sequencing Study. Viruses, 17(4), 478. https://doi.org/10.3390/v17040478





