Investigating Mpox Strain Dynamics Using Computational and Data-Driven Approaches
Abstract
1. Introduction
2. Mathematical Model Formulation
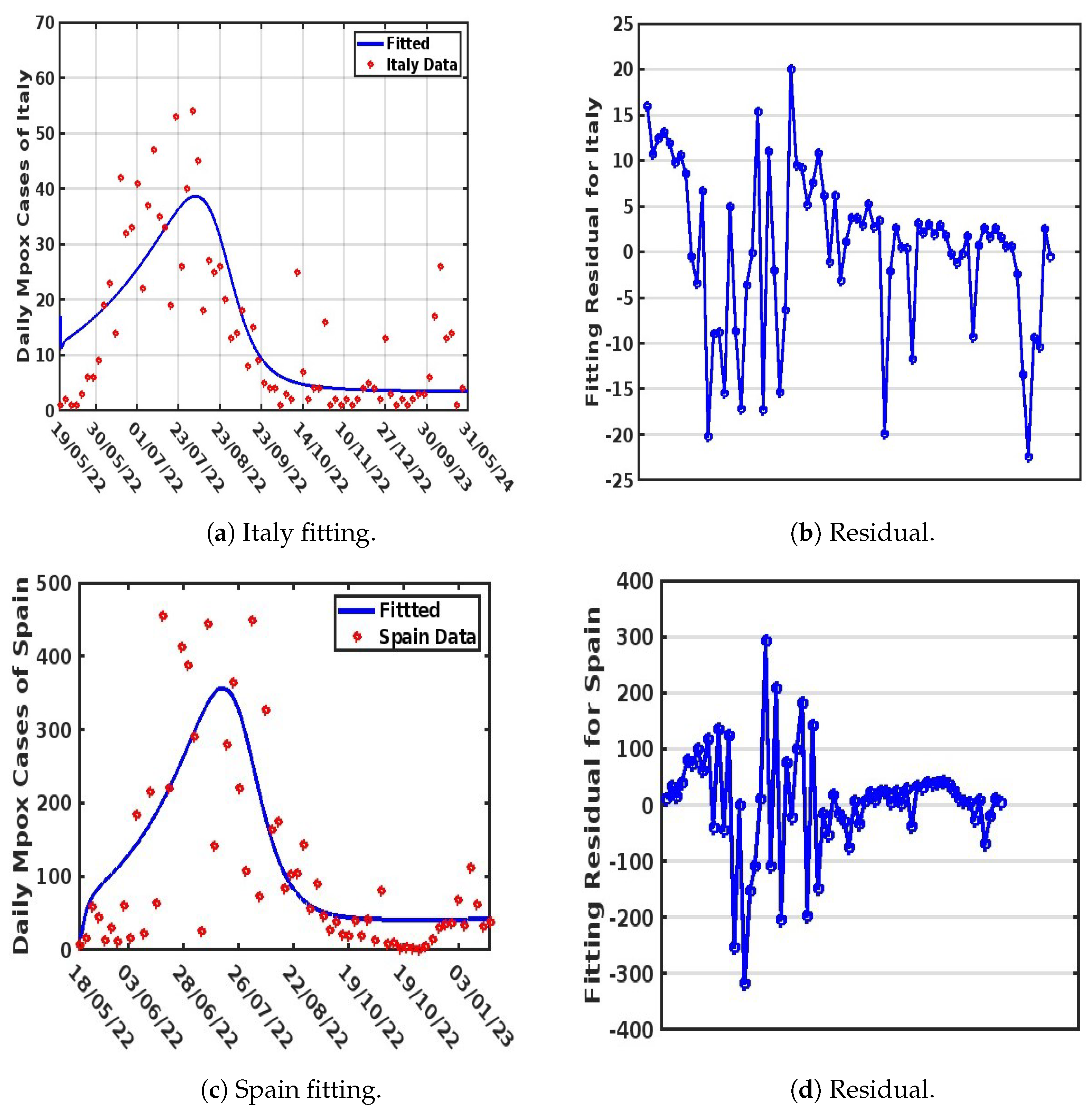
3. Parameter Estimation and Mathematical Model Fitting
4. Statistical Data Modeling
4.1. Generalized Additive Modeling (GAM)
4.2. Generalized Linear Modeling (GLM)
4.3. Comparison of the Strain Dynamics Mathematical Model Developed with Classical Statistical Data Modeling at the Population Level
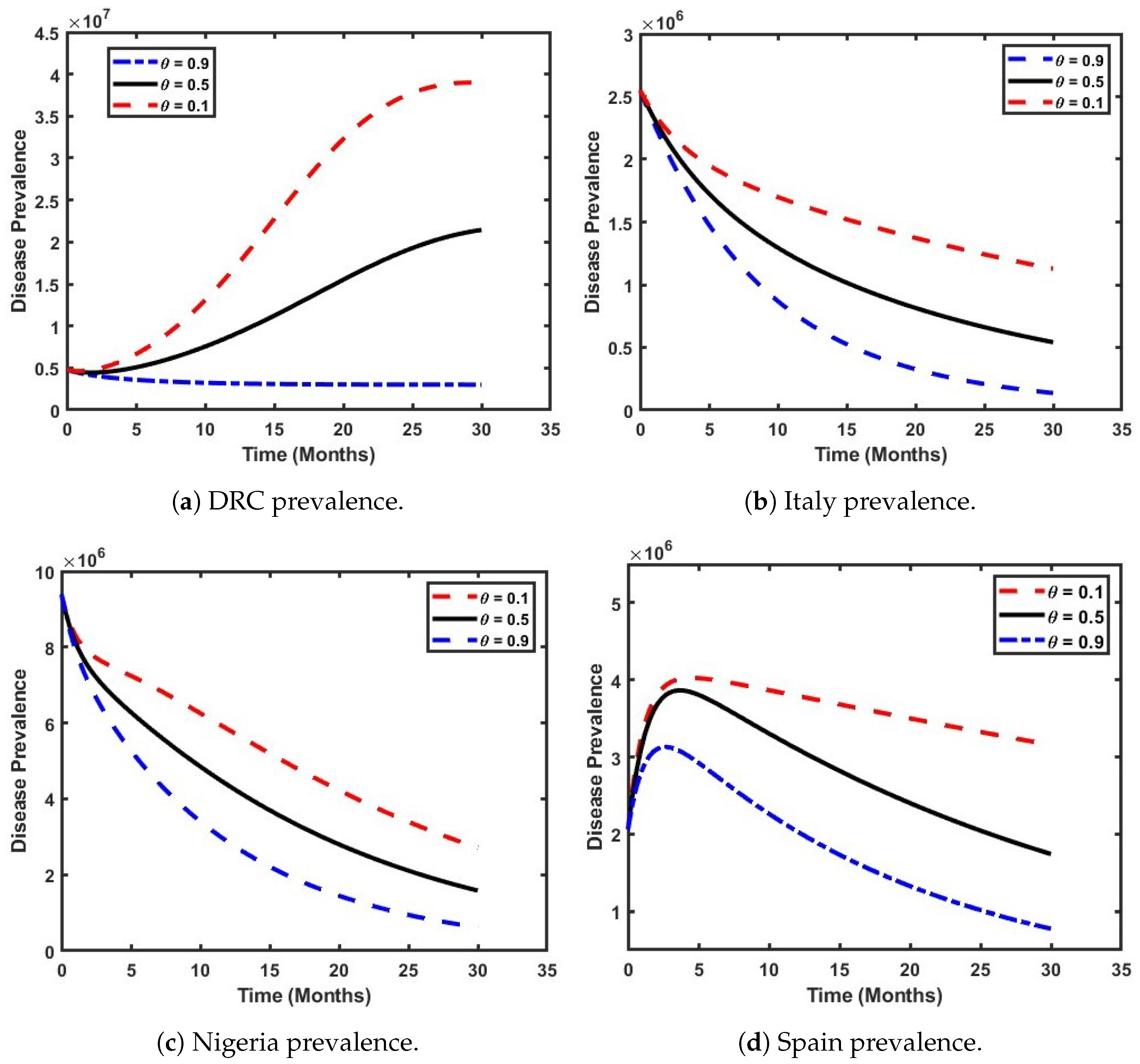
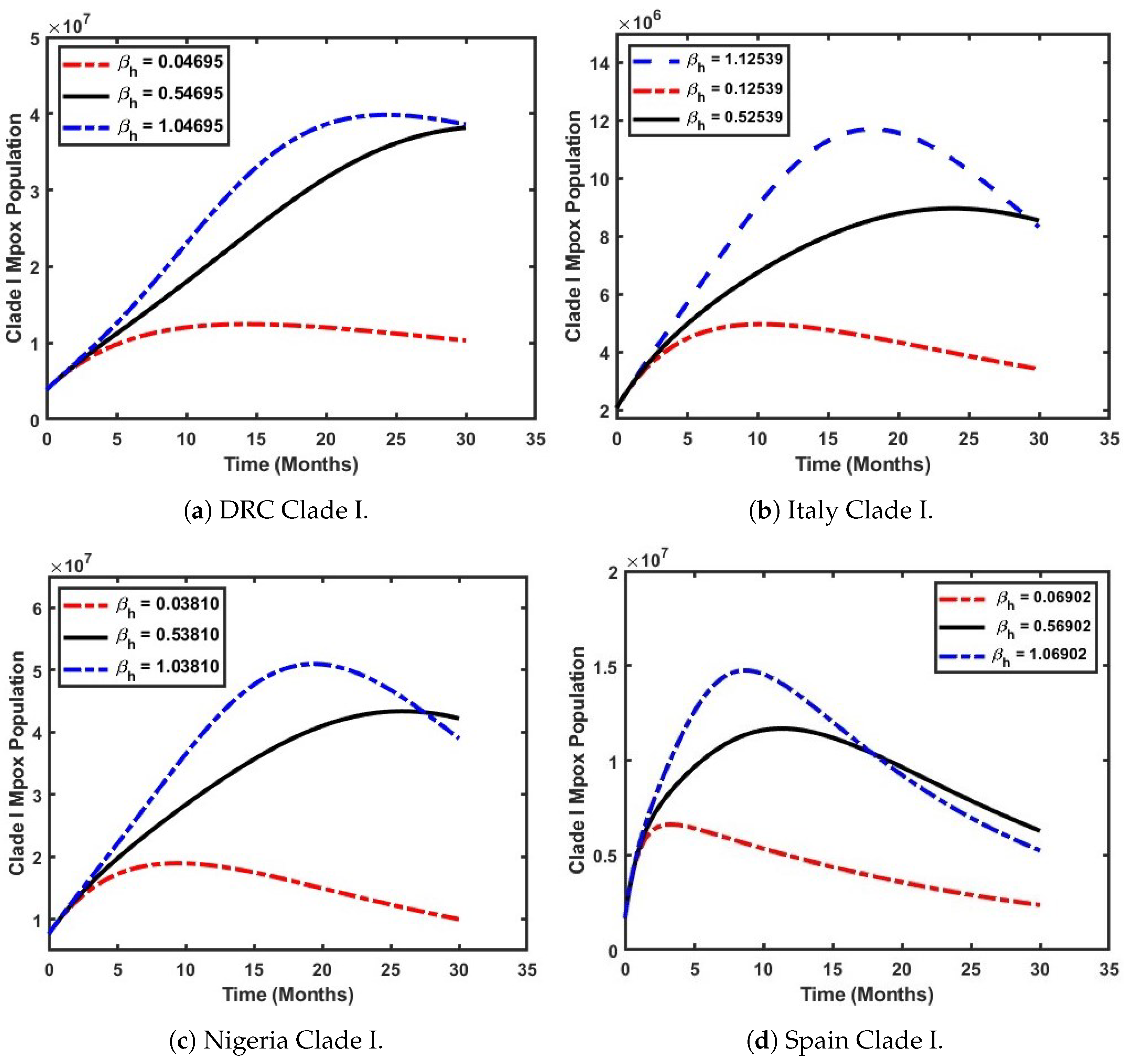
5. Mathematical Model Simulation for Different Scenarios
- i
- The effect of both Clades on the prevalence of Mpox within the population.
- ii
- The influence of the effective contact rate on the prevalence of Mpox Clade I.
- iii
- The impact of the effective contact rate on the prevalence of Mpox Clade II.
5.1. Analyze the Effect of Both Clades on the Prevalence of Mpox Within the Population
5.2. Examine How the Effective Contact Rate Influences the Prevalence of Mpox Clade I
5.3. Assess the Impact of the Effective Contact Rate on the Prevalence of Mpox Clade II
6. Concluding Remarks
6.1. Conclusion and Recommendation
6.2. Limitations and Future Work
- The current formulation of the model (4) does not explicitly incorporate a vaccination compartment. Including a vaccination compartment in the model would significantly improve its ability to capture the effects of vaccination programs on the different Mpox strain dynamics. Vaccination plays a crucial role in mitigating the spread of Mpox, and its absence in the current formulation may lead to an incomplete representation of the real-world epidemiological scenario.
- The exposed compartment in Model (4) is not subdivided to account for the two distinct Mpox Clades: Clade I and Clade II. Subdividing the exposed compartment into these Clades is essential for capturing the heterogeneous nature of Mpox disease dynamics. Clade-specific characteristics, such as differences in transmissibility, severity, and geographic distribution, can have significant implications for understanding the spread of the disease and designing targeted interventions. Incorporating this distinction would make the model more biologically relevant and aligned with observed data.
- For future analyses, the authors propose the adoption of a time-dependent effective contact rate and control reproduction number as part of the modeling framework. Both parameters are critical metrics for assessing the effectiveness of interventions over time. By incorporating this dynamic measure, the model could provide a more detailed understanding of how control measures, such as vaccination and quarantine, impact the spread of the disease during the different stages of an outbreak.
- The authors recommend incorporating forecasting approaches into future analyses of the model. Forecasting techniques, such as machine learning or Bayesian inference, can enhance the predictive capabilities of the model by leveraging historical and real-time data. These methods can provide valuable insights into the potential trajectory of Mpox future outbreaks, allowing policymakers to make proactive and informed decisions regarding resource allocation, vaccination campaigns, and other interventions.
- It was observed that Clade II spread into different countries across the globe from its origin in West Africa due to immigration. We propose that incorporating movement into the model and extending it to a spatial model will account for the spatial spread of the disease, which will help to inform decisions at different spatial scales.
- Disparities in vaccination distribution and access to proper healthcare are still an issue in low-income countries where the disease is predominant; hence, investigating these health inequalities will enhance intervention strategies and the mitigation of Mpox spread in those countries.
- People infected with Mpox with underlying conditions (immunocompromised individuals) face increased risk and mortality from the disease, underscoring the interplay between co-morbid conditions and disease outcomes. Future models need to incorporate this factor into their formulation, perhaps by means of age structure modeling, which will take into account co-morbidity variables in children and young and older populations.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Mathematical Model Analysis
Appendix A.1. Positivity and Boundedness
Appendix A.2. Disease-Free Equilibrium
Appendix A.3. Existence and Uniqueness of Solution
Endemic Equilibrium
Appendix A.4. Local Stability of Mpox-Free Equilibrium
- ;
- ;
- .
References
- WHO. Mpox (Monkeypox). 2024. Available online: https://www.who.int/news-room/fact-sheets/detail/monkeypox (accessed on 2 March 2024).
- NCDC. MonkeyPox. 2022. Available online: https://ncdc.gov.ng/ncdc.gov.ng/diseases/factsheet/55 (accessed on 2 March 2024).
- Idisi, O.I.; Yusuf, T.T.; Adeniyi, E.; Onifade, A.A.; Oyebo, Y.T.; Samuel, A.T.; Kareem, L.A. A new compartmentalized epidemic model to analytically study the impact of awareness on the control and mitigation of the monkeypox disease. Healthc. Anal. 2023, 4, 100267. [Google Scholar] [CrossRef]
- WHO. WHO Recommends New Name for Monkeypox Disease. 2022. Available online: https://www.who.int/news/item/28-11-2022-who-recommends-new-name-for-monkeypox-disease (accessed on 2 March 2024).
- World Health Organization. Weekly Epidemiological Record = Relevé éPidémiologique Hebdomadaire. 2023. Available online: https://iris.who.int/handle/10665/372986 (accessed on 2 March 2024).
- CDC. About Mpox|Mpox|Poxvirus|CDC—cdc.gov. 2024. Available online: https://www.cdc.gov/poxvirus/mpox/about/index.html (accessed on 2 March 2024).
- Simpson, K.; Heymann, D.; Brown, C.S.; Edmunds, W.J.; Elsgaard, J.; Fine, P.; Hochrein, H.; Hoff, N.A.; Green, A.; Ihekweazu, C.; et al. Human monkeypox—After 40 years, an unintended consequence of smallpox eradication. Vaccine 2020, 38, 5077–5081. [Google Scholar] [CrossRef]
- Spirito, F.; Guida, A.; Caponio, V.C.A.; Lo Muzio, L. Monkeypox: A New Challenge for Global Health System? Life 2023, 13, 1250. [Google Scholar] [CrossRef]
- Chatterjee, A.; Mukherjee, S.; Sengupta, A.; Das, A. The Psychological Influence of Online Stigmatization in Social Life due to Monkeypox. In Proceedings of the International Conference on Innovations in Computational Intelligence and Computer Vision, Online, 24–25 November 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 661–674. [Google Scholar]
- Khattak, S.; Rauf, M.A.; Ali, Y.; Yousaf, M.T.; Liu, Z.; Wu, D.D.; Ji, X.Y. The monkeypox diagnosis, treatments and prevention: A review. Front. Cell. Infect. Microbiol. 2023, 12, 1088471. [Google Scholar] [CrossRef] [PubMed]
- Banuet-Martinez, M.; Yang, Y.; Jafari, B.; Kaur, A.; Butt, Z.A.; Chen, H.H.; Yanushkevich, S.; Moyles, I.R.; Heffernan, J.M.; Korosec, C.S. Monkeypox: A review of epidemiological modelling studies and how modelling has led to mechanistic insight. Epidemiol. Infect. 2023, 151, e121. [Google Scholar] [CrossRef] [PubMed]
- Silenou, B.C.; Tom-Aba, D.; Adeoye, O.; Arinze, C.C.; Oyiri, F.; Suleman, A.K.; Yinka-Ogunleye, A.; Dörrbecker, J.; Ihekweazu, C.; Krause, G. Use of surveillance outbreak response management and analysis system for human monkeypox outbreak, Nigeria, 2017–2019. Emerg. Infect. Dis. 2020, 26, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Yinka-Ogunleye, A.; Dalhat, M.; Akinpelu, A.; Aruna, O.; Garba, F.; Ahmad, A.; Adeleye, A.; Botson, I.; Oluwafemi, B.; Ogunbode, O.; et al. Mpox (monkeypox) risk and mortality associated with HIV infection: A national case-control study in Nigeria. BMJ Glob. Health 2023, 8, 13126. [Google Scholar] [CrossRef] [PubMed]
- Beeson, A.M.; Haston, J.; McCormick, D.W.; Reynolds, M.; Chatham-Stephens, K.; McCollum, A.M.; Godfred-Cato, S. Mpox in Children and Adolescents: Epidemiology, Clinical Features, Diagnosis, and Management. Pediatrics 2023, 151, e2022060179. [Google Scholar] [CrossRef]
- Lu, J.; Xing, H.; Wang, C.; Tang, M.; Wu, C.; Ye, F.; Yin, L.; Yang, Y.; Tan, W.; Shen, L. Mpox (formerly monkeypox): Pathogenesis, prevention, and treatment. Signal Transduct. Target. Ther. 2023, 8, 458. [Google Scholar] [CrossRef] [PubMed]
- Karagoz, A.; Tombuloglu, H.; Alsaeed, M.; Tombuloglu, G.; AlRubaish, A.A.; Mahmoud, A.; Smajlović, S.; Ćordić, S.; Rabaan, A.A.; Alsuhaimi, E. Monkeypox (mpox) virus: Classification, origin, transmission, genome organization, antiviral drugs, and molecular diagnosis. J. Infect. Public Health 2023, 16, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Hazra, A.; Zucker, J.; Bell, E.; Flores, J.; Gordon, L.; Mitjà, O.; Suñer, C.; Lemaignen, A.; Jamard, S.; Nozza, S.; et al. Mpox in people with past infection or a complete vaccination course: A global case series. Lancet Infect. Dis. 2024, 24, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Akazawa, D.; Ohashi, H.; Hishiki, T.; Morita, T.; Iwanami, S.; Kim, K.S.; Jeong, Y.D.; Park, E.S.; Kataoka, M.; Shionoya, K.; et al. Potential Anti-Mpox Virus Activity of Atovaquone, Mefloquine, and Molnupiravir, and Their Potential Use as Treatments. J. Infect. Dis. 2023, 228, 591–603. [Google Scholar] [CrossRef]
- Patel, M.; Adnan, M.; Aldarhami, A.; Bazaid, A.S.; Saeedi, N.H.; Alkayyal, A.A.; Saleh, F.M.; Awadh, I.B.; Saeed, A.; Alshaghdali, K. Current Insights into Diagnosis, Prevention Strategies, Treatment, Therapeutic Targets, and Challenges of Monkeypox (Mpox) Infections in Human Populations. Life 2023, 13, 249. [Google Scholar] [CrossRef] [PubMed]
- Alshehri, A.; Ullah, S. Optimal control analysis of Monkeypox disease with the impact of environmental transmission. AIMS Math. 2023, 8, 16926–16960. [Google Scholar] [CrossRef]
- Oyebanji, O.; Ofonagoro, U.; Akande, O.; Nsofor, I.; Ukenedo, C.; Mohammed, T.B.; Anueyiagu, C.; Agenyi, J.; Yinka-Ogunleye, A.; Ihekweazu, C. Lay media reporting of monkeypox in Nigeria. BMJ Glob. Health 2019, 4, e002019. [Google Scholar] [CrossRef]
- Leandry, L.; Mureithi, E.W. An investigation on the monkeypox virus dynamics in human and rodent populations for a deterministic mathematical model. Inform. Med. Unlocked 2023, 41, 101325. [Google Scholar] [CrossRef]
- Rabiu, M.; Dansu, E.J.; Mogbojuri, O.A.; Idisi, I.O.; Yahaya, M.M.; Chiwira, P.; Abah, R.T.; Adeniji, A.A. Modeling the sexual transmission dynamics of mpox in the United States of America. Eur. Phys. J. Plus 2024, 139, 250. [Google Scholar] [CrossRef]
- Bhunu, C.; Mushayabasa, S. Modelling the transmission dynamics of pox-like infections. IAENG Int. J. Appl. Math. 2011, 41, 2. [Google Scholar]
- Akinyemi, S.T.; Idisi, I.O.; Rabiu, M.; Okeowo, V.I.; Iheonu, N.; Dansu, E.J.; Abah, R.T.; Mogbojuri, O.A.; Audu, A.M.; Yahaya, M.M.; et al. A tale of two countries: Optimal control and cost-effectiveness analysis of monkeypox disease in Germany and Nigeria. Healthc. Anal. 2023, 4, 100258. [Google Scholar] [CrossRef]
- Batiha, I.M.; Abubaker, A.A.; Jebril, I.H.; Al-Shaikh, S.B.; Matarneh, K.; Almuzini, M. A Mathematical Study on a Fractional-Order SEIR Mpox Model: Analysis and Vaccination Influence. Algorithms 2023, 16, 418. [Google Scholar] [CrossRef]
- Iftikhar, H.; Daniyal, M.; Qureshi, M.; Tawaiah, K.; Ansah, R.K.; Afriyie, J.K. A hybrid forecasting technique for infection and death from the mpox virus. Digit. Health 2023, 9, 20552076231204748. [Google Scholar] [CrossRef] [PubMed]
- Bragazzi, N.L.; Iyaniwura, S.A.; Han, Q.; Woldegerima, W.A.; Kong, J.D.; Woldegerima, W.A.; Kong, J.D.; Han, Q. Quantifying the basic reproduction number and the under-estimated fraction of mpox cases around the world at the onset of the outbreak: A mathematical modeling and machine learning-based study. Available at SSRN 4533567. Lancet 2023, preprint. [Google Scholar]
- Singh, V.; Khan, S.A.; Yadav, S.K.; Akhter, Y. Modeling Global Monkeypox Infection Spread Data: A Comparative Study of Time Series Regression and Machine Learning Models. Curr. Microbiol. 2024, 81, 15. [Google Scholar] [CrossRef]
- Soto-Ferrari, M.; Carrasco-Pena, A.; Prieto, D. Deep Learning Architectures Framework for Emerging Outbreak Forecasting of Mpox: A Bagged Ensemble Scheme to Model Accurate Prediction Intervals; Research Square: Durham, NC, USA, 2023; pp. 237–2276. [Google Scholar] [CrossRef]
- Jaradat, A.S.; Mamlook, R.E.A.; Almakayeel, N.; Alharbe, N.; Almuflih, A.S.; Nasayreh, A.; Gharaibeh, H.; Gharaibeh, M.; Gharaibeh, A.; Bzizi, H. Automated Monkeypox Skin Lesion Detection Using Deep Learning and Transfer Learning Techniques. Int. J. Environ. Res. Public Health 2023, 20, 4422. [Google Scholar] [CrossRef] [PubMed]
- Chadaga, K.; Prabhu, S.; Sampathila, N.; Nireshwalya, S.; Katta, S.S.; Tan, R.S.; Acharya, U.R. Application of Artificial Intelligence Techniques for Monkeypox: A Systematic Review. Diagnostics 2023, 13, 824. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.J.; Patel, D.S.; Tanguy, T.A.; Rychtář, J.; Taylor, D. Mathematical Models of Lyme Disease: An Overview. UMAP J. 2024, 45, 11. [Google Scholar]
- Lee, W.; Kim, Y.J.; Lee, S.J.; Ahn, D.G.; Kim, S.J. Current Status of Epidemiology, Diagnosis, Therapeutics, and Vaccines for the Re-Emerging Human Monkeypox Virus. J. Microbiol. Biotechnol. 2023, 33, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Besombes, C.; Breban, R. The peculiar emergence of monkeypox/mpox: A modeling perspective. HAL 2023, preprint. [Google Scholar]
- Ngungu, M.; Addai, E.; Adeniji, A.; Adam, U.M.; Oshinubi, K. Mathematical epidemiological modeling and analysis of monkeypox dynamism with non-pharmaceutical intervention using real data from United Kingdom. Front. Public Health 2023, 11, 1101436. [Google Scholar] [CrossRef] [PubMed]
- Peter, O.J.; Kumar, S.; Kumari, N.; Oguntolu, F.A.; Oshinubi, K.; Musa, R. Transmission dynamics of Monkeypox virus: A mathematical modelling approach. Model. Earth Syst. Environ. 2022, 8, 3423–3434. [Google Scholar] [CrossRef]
- Mathieu, E.; Spooner, F.; Dattani, S.; Ritchie, H.; Roser, M. Mpox. 2022. Available online: https://ourworldindata.org/mpox (accessed on 2 March 2024).
- OWID. Population. 2024. Available online: https://www.worldometers.info/population/europe/southern-europe/x (accessed on 2 March 2024).
- Wood, S.N. Generalized Additive Models; Chapman and Hall: London, UK; CRC: Boca Raton, FL, USA, 2017. [Google Scholar] [CrossRef]
- Haman, J.; Avery, M.; Institute for Defense Analyses. Package ‘ciTools’, version 0.6.1; R Package Version; CRAN: Vienna, Austria, 2022; Volume 3.
- Idisi, O.I.; Yusuf, T.T. A Mathematical Model For Lassa Fever Transmission Dynamics With Impacts of Control Measures: Analysis And Simulation. Eur. J. Math. Stat. 2021, 2, 19–28. [Google Scholar] [CrossRef]






| Variable | Interpretation |
|---|---|
| S | Population of susceptible individuals |
| E | Population of individuals exposed to both Clades |
| Population of Clade I infectious individuals | |
| Population of Clade II infectious individuals | |
| H | Population of hospitalized individuals |
| R | Population of recovered individuals |
| Population of susceptible mammals | |
| Population of infectious mammals | |
| Parameter | Interpretation |
| Recruitment rates into the human population | |
| Transmission rate of individuals to the exposed class from the susceptible class | |
| Re-infection rate or loss of immunity of recovered individuals for both Clades | |
| Transmission rate of individuals from exposed class to infectious compartment for Clade I and Clade II | |
| The proportion of individuals infected | |
| Hospitalized rate of individuals in Clade I infectious population | |
| Hospitalized rate of individuals in Clade II infectious population | |
| Disease-induced death rate in Clade I infectious population | |
| Disease-induced death rate in Clade II infectious population | |
| Disease-induced death rate in infectious population in the hospitalized class | |
| Recovery rate of individuals from hospitalized class | |
| Recovery rate of individuals from Clade II infectious population | |
| Natural mortality rate of human population | |
| Recruitment rates into the vector population | |
| Transmission rate of susceptible vector to infectious vector | |
| Natural mortality rate of the vector population | |
| Modification parameters that reduce the infection transmission rate between humans and mammals |
| Country | |||||||
|---|---|---|---|---|---|---|---|
| Italy | 687.30 | 0.0121 | 0.0714 | 1393.5 | 0.0180 | 0.1039 | 0.0004 |
| Spain | 2330.3 | 0.0118 | 0.0714 | 9559.5 | 0.0009 | 0.4551 | 0.0003 |
| Nigeria | 5329.8 | 0.0185 | 0.0714 | 1083.9 | 0.0320 | 0.5761 | 0.0076 |
| DRC | 8259.8 | 0.0163 | 0.0714 | 956.8 | 0.0001 | 0.5739 | 0.0102 |
| Parameter | Spain | Nigeria | DRC | Italy |
|---|---|---|---|---|
| 2.300 | 8.800 | 1.870 | 4.3 | |
| 4.466 | 5.419 | 6.728 | 2.5404 | |
| 5.000 | 3.300 | 2.000 | 5.0 | |
| 4.10 | 2.300 | 2.151 | 5.043 | |
| 3.090 | 1.160 | 5.300 | 2.212 | |
| 0.789 | 1.038 | 8.023 | 1.3080 | |
| 0.4645 | 0.4954 | 0.4610 | 0.96082 | |
| 3.853 | 3.922 | 0.1553 | 0.27450 | |
| 0.2677 | 0.1810 | 0.2254 | 1.25396 | |
| 0.31741 | 0.4117 | 0.9819 | 0.18953 | |
| 0.6902 | 0.3810 | 0.4695 | 1.25396 | |
| 0.1072 | 6.284 | 8.703 | 0.14317 | |
| 0.2311 | 3.3360 | 3.888 | 0.75697 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Idisi, I.O.; Oshinubi, K.; Sewanu, V.B.; Yahaya, M.M.; Olagbami, O.S.; Edogbanya, H.O. Investigating Mpox Strain Dynamics Using Computational and Data-Driven Approaches. Viruses 2025, 17, 154. https://doi.org/10.3390/v17020154
Idisi IO, Oshinubi K, Sewanu VB, Yahaya MM, Olagbami OS, Edogbanya HO. Investigating Mpox Strain Dynamics Using Computational and Data-Driven Approaches. Viruses. 2025; 17(2):154. https://doi.org/10.3390/v17020154
Chicago/Turabian StyleIdisi, Isaiah Oke, Kayode Oshinubi, Vigbe Benson Sewanu, Mukhtar Muhammed Yahaya, Oluwafemi Samson Olagbami, and Helen Olaronke Edogbanya. 2025. "Investigating Mpox Strain Dynamics Using Computational and Data-Driven Approaches" Viruses 17, no. 2: 154. https://doi.org/10.3390/v17020154
APA StyleIdisi, I. O., Oshinubi, K., Sewanu, V. B., Yahaya, M. M., Olagbami, O. S., & Edogbanya, H. O. (2025). Investigating Mpox Strain Dynamics Using Computational and Data-Driven Approaches. Viruses, 17(2), 154. https://doi.org/10.3390/v17020154









