Chronic Chikungunya Arthritis in Northeastern Brazil: An Association with Very Severe Joint Pain and Lack of Correlation with IL-6 and TNFα Gene Polymorphisms
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Study Population, Samples, and Clinical Data
2.3. Enzyme-Linked Immunosorbent Assay
2.4. DNA Extraction
2.5. Genotyping
2.6. Data Analysis
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amdekar, S.; Parashar, D.; Alagarasu, K. Chikungunya virus-induced arthritis: Role of host and viral factors in the pathogenesis. Viral Immunol. 2017, 30, 691–702. [Google Scholar] [CrossRef]
- Avila-Trejo, A.M.; Rodríguez-Páez, L.I.; Alcántara-Farfán, V.; Aguilar-Faisal, J.L. Multiple factors involved in bone damage caused by chikungunya virus infection. Int. J. Mol. Sci. 2023, 24, 13087. [Google Scholar] [CrossRef]
- Alla, S.A.O.; Combe, B. Arthritis after infection with Chikungunya virus. Best Pract. Res. Clin. Rheumatol. 2012, 25, 337–346. [Google Scholar] [CrossRef]
- De Jesus, P.B.; Brasil, M.Q.A.; Silva, J.D.J.; Cristal, J.R.; Carvalho, I.D.P.; Miranda, M.C.P.; Paixão, P.; Khouri, R.; Cerqueira-Silva, T.; Boulos, F.C.; et al. Chikungunya in a pediatric cohort: Asymptomatic infection, seroconversion, and chronicity rates. PLoS Neglected Trop. Dis. 2025, 19, e0013254. [Google Scholar] [CrossRef]
- Silva, J.V.J., Jr.; Ludwig-Begall, L.F.; Oliveira-Filho, E.F.; Oliveira, R.A.S.; Durães-Carvalho, R.; Lopes, T.R.R.; Silva, D.E.A.; Gil, L.H.V.G. A scoping review of Chikungunya virus infection: Epidemiology, clinical characteristics, viral co-circulation complications, and control. Acta Trop. 2018, 188, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Couzigou, B.; Criquet-Hayot, A.; Javelle, E.; Tignac, S.; Mota, E.; Rigaud, F.; Alain, A.; Troisgros, O.; Pierre-Francois, S.; Abel, S.; et al. Occurrence of chronic stage chikungunya in the general population of Martinique during the first 2014 epidemic: A prospective epidemiological study. Am. J. Trop. Med. Hyg. 2018, 99, 182–190. [Google Scholar] [CrossRef]
- Amaral, J.K.; Taylor, P.C.; Teixeira, M.M.; Morrison, T.E.T.; Schoen, R.T. The clinical features, pathogenesis and methotrexate therapy of chronic chikungunya arthritis. Viruses 2019, 11, 289. [Google Scholar] [CrossRef]
- Ng, W.H.; Amaral, K.; Javelle, E.; Mahalingam, S. Chronic chikungunya disease (CCD): Clinical insights, immunopathogenesis and therapeutic perspectives. QJM Int. J. Med. 2024, 117, 489–494. [Google Scholar] [CrossRef]
- Silva, L.A.; Dermody, T.S. Chikungunya virus: Epidemiology, replication, disease mechanisms, and prospective intervention strategies. J. Clin. Investig. 2017, 127, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, A.F.L.; Okano, A.H.; Micussi, M.T.; Souza, C.G.D.; Passos, J.O.S.; Morya, E.; Freitas, R.P.D.A. Chronic Chikungunya arthralgia reduces functionality, quality of life and occupational performance: Descriptive cross-sectional study. BrJP 2022, 5, 233–238. [Google Scholar] [CrossRef]
- Dos Santos, G.R.; Jawed, F.; Mukandavire, C.; Deol, A.; Scarponi, D.; Mboera, L.E.; Seruyange, E.; Poirier, M.J.P.; Bosomprah, S.; Udeze, A.O.; et al. Global burden of chikungunya virus infections and the potential benefit of vaccination campaigns. Nat. Med. 2025, 31, 2343–2349. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Geographical Expansion of Cases of Dengue and Chikungunya Beyond the Historical Areas of Transmission in the Region of the Americas. 2023. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON448 (accessed on 10 October 2025).
- De Brito, C.A.A. Alert: Severe cases and deaths associated with Chikungunya in Brazil. Rev. Soc. Bras. Med. Trop. 2017, 50, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Simião, A.R.; Barreto, F.K.D.A.; Oliveira, R.D.M.A.B.; Cavalcante, J.W.; Lima Neto, A.S.; Barbosa, R.B.; Lins, C.S.; Meira, A.G.; Araújo, F.M.C.; Lemos, D.R.Q.; et al. A major chikungunya epidemic with high mortality in northeastern Brazil. Rev. Soc. Bras. Med. Trop. 2019, 52, e20190266. [Google Scholar] [CrossRef]
- Pan American Health Organization (PAHO). With Rising Cases, Experts Discuss Chikungunya Spread in the Americas. 2023. Available online: https://www.publicnow.com/view/2BC67BD774BC52A6B9135FCF7AC0FCAED97C0409?1683233120 (accessed on 10 October 2025).
- De Souza, W.M.; Ribeiro, G.S.; de Lima, S.T.; de Jesus, R.; Moreira, F.R.; Whittaker, C.; Sallum, M.A.M.; Carrington, C.V.F.; Sabino, E.C.; Kitron, U.; et al. Chikungunya: A decade of burden in the Americas. Lancet Reg. Health–Am. 2024, 30, 100673. [Google Scholar] [CrossRef]
- Chaaithanya, I.K.; Muruganandam, N.; Surya, P.; Anwesh, M.; Alagarasu, K.; Vijayachari, P. Association of oligoadenylate synthetase gene cluster and DC-SIGN (CD209) gene polymorphisms with clinical symptoms in chikungunya virus infection. DNA Cell Biol. 2015, 35, 44–50. [Google Scholar] [CrossRef]
- Gotay, W.J.P.; Maciel, M.S.C.; Rodrigues, R.D.O.; Cardoso, C.C.; Oliveira, C.N.; Montenegro, A.F.L.; Yaochite, J.N.U. X-linked polymorphisms in TLR7 and TLR8 genes are associated with protection against Chikungunya fever. Mem. Inst. Oswaldo Cruz 2025, 120, e230224. [Google Scholar] [CrossRef]
- Gasque, P.; Courdec, T.; Lecuit, M.; Roques, P.; Ng, L.F.P. Chikungunya Virus Pathogenesis and Immunity. Vector-Borne Zoonotic Dis. 2015, 15, 241–249. [Google Scholar] [CrossRef]
- Ninla-Aesong, P.; Mitarnun, W.; Noipha, K. Proinflammatory cytokines and chemokines as biomarkers of persistent arthralgia and severe disease after chikungunya virus infection: A 5-year follow-up study in Southern Thailand. Viral Immunol. 2019, 32, 442–452. [Google Scholar] [CrossRef]
- Babu, N.; Mahilkar, S.; Jayaram, A.; Ibemgbo, S.A.; Mathur, G.; Shetty, U.; Sudandiradas, R.; Kumar, P.S.; Singh, S.; Paini, S.S.; et al. Cytokine profile, neutralisation potential and viral replication dynamics in sera of chikungunya patients in India: A cross-sectional study. Lancet Reg. Health-Southeast Asia 2023, 19, 100269. [Google Scholar] [CrossRef] [PubMed]
- Thanapati, S.; Ganu, M.; Giri, P.; Kulkarni, S.; Sharma, M.; Babar, P.; Ganu, A.; Tripathy, A.S. Impaired NK cell functionality and increased TNF-α production as biomarkers of chronic chikungunya arthritis and rheumatoid arthritis. Hum. Immunol. 2017, 78, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.Y.; Tritsch, S.; Reid, S.P.; Martins, K.; Encinales, L.; Pacheco, N.; Amdur, L.R.; Porras-Ramirez, A.; Rico-Mendoza, A.; Li, G.; et al. The cytokine profile in acute chikungunya infection is predictive of chronic arthritis 20 months post infection. Diseases 2018, 6, 95. [Google Scholar] [CrossRef]
- Amr, K.; El-Awady, R.; Raslan, H. Assessment of the −74G/C (rs1800795) and −572G/C (rs1800796) interleukin 6 gene polymorphisms in Egyptian patients with rheumatoid arthritis. Open Access Maced. J. Med. Sci. 2016, 4, 574–577. [Google Scholar] [CrossRef] [PubMed]
- You, C.G.; Li, X.J.; Li, Y.M.; Wang, L.P.; Li, F.F.; Guo, X.L.; Gao, L.N. Association analysis of single nucleotide polymorphisms of proinflammatory cytokine and their receptors genes with rheumatoid arthritis in northwest Chinese Han population. Cytokine 2013, 61, 133–138. [Google Scholar] [CrossRef]
- Dettogni, R.S.; Tristão-Sá, R.; Dos Santos, M.; da Silva, F.F.; Louro, I.D. Single nucleotide polymorphisms in immune system genes and their association with clinical symptoms persistence in dengue-infected persons. Hum. Immunol. 2015, 76, 717–723. [Google Scholar] [CrossRef]
- Perez, A.B.; Sierra, B.; Garcia, G.; Aguirre, E.; Babel, N.; Alvarez, M.; Sanchez, L.; Valdes, L.; Volk, H.D.; Guzman, M.G. Tumor necrosis factor–alpha, transforming growth factor–β1, and interleukin-10 gene polymorphisms: Implication in protection or susceptibility to dengue hemorrhagic fever. Hum. Immunol. 2010, 71, 1135–1140. [Google Scholar] [CrossRef] [PubMed]
- Bucardo, F.; Reyes, Y.; Morales, M.; Briceño, R.; González, F.; Lundkvist, Å.; Svensson, L.; Nordgren, J. Association of Genetic Polymorphisms in DC-SIGN, Toll-Like Receptor 3, and Tumor Necrosis Factor α Genes and the Lewis-Negative Phenotype With Chikungunya Infection and Disease in Nicaragua. J. Infect. Dis. 2021, 223, 278–286. [Google Scholar] [CrossRef]
- Queiroz, J.L.; Maciel, M.S.C.; Teles, L.D.S.; Vieira, S.M.A.; Gomes, T.N.; Fernandes, H.F.; De Oliveira, J.S.; Ferreira, G.P.; Pereira, A.C.T.D.C. Prevalence and Influence of the −174 G/C Polymorphism in the Interleukin-6 Gene in Arboviruses Infections. Viral Immunol. 2021, 34, 559–566. [Google Scholar] [CrossRef]
- Chapman, S.J.; Hill, A.V. Human genetic susceptibility to infectious disease. Nat. Rev. Genet. 2012, 13, 175–188. [Google Scholar] [CrossRef]
- Briggs, M.; Closs, J.S. A descriptive study of the use of visual analogue scales and verbal rating scales for the assessment of postoperative pain in orthopedic patients. J. Pain Symptom Manag. 1999, 18, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Huits, R.; De Kort, J.; Van Den Berg, R.; Chong, L.; Tsoumanis, A.; Eggermont, K.; Bartholomeeusen, K.; Arieèn, K.K.; Jacobs, J.; Esbroeck, M.V.; et al. Chikungunya virus infection in Aruba: Diagnosis, clinical features and predictors of post-chikungunya chronic polyarthralgia. PLoS ONE 2018, 13, e0196630. [Google Scholar] [CrossRef]
- Hayd, R.L.N.; Moreno, M.R.; Naveca, F.; Amdur, R.; Suchowiecki, K.; Watson, H.; Firestein, G.S.; Simon, G.; Chang, A.Y. Persistent chikungunya arthritis in Roraima, Brazil. Clin. Rheumatol. 2020, 39, 2781–2787. [Google Scholar] [CrossRef]
- Bertolotti, A.; Thioune, M.; Abel, S.; Belrose, G.; Calmont, I.; Césaire, R.; Cervantes, M.; Fagour, L.; Javelle, E.; Lebris, C.; et al. Prevalence of chronic chikungunya and associated risks factors in the French West Indies (La Martinique): A prospective cohort study. PLoS Neglected Trop. Dis. 2020, 14, e0007327. [Google Scholar] [CrossRef]
- Elsinga, J.; Grobusch, M.P.; Tami, A.; Gerstenbluth, I.; Bailey, A. Health-related impact on quality of life and coping strategies for chikungunya: A qualitative study in Curaçao. PLoS Neglected Trop. Dis. 2017, 11, e0005987. [Google Scholar] [CrossRef]
- Lozano-Parra, A.; Herrera, V.; Calderón, C.; Badillo, R.; Gélvez Ramírez, R.M.; Estupiñán Cárdenas, M.I.; Jiménez, J.F.L.; Villar, L.Á.; Rojas Garrido, E.M. Chronic rheumatologic disease in chikungunya virus fever: Results from a cohort study conducted in Piedecuesta, Colombia. Trop. Med. Infect. Dis. 2024, 9, 247. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.M.; Kikuti, M.; Anjos, R.O.; Portilho, M.M.; Santos, V.C.; Gonçalves, T.S.; Tauro, L.B.; Moreira, P.S.S.; Jacob-Nascimento, L.C.; Santana, P.M. Risk of chronic arthralgia and impact of pain on daily activities in a cohort of patients with chikungunya virus infection from Brazil. Int. J. Infect. Dis. 2021, 105, 608–616. [Google Scholar] [CrossRef]
- Tiozzo, G.; de Roo, A.M.; Gurgel do Amaral, G.S.; Hofstra, H.; Vondeling, G.T.; Postma, M.J. Assessing chikungunya’s economic burden and impact on health-related quality of life: Two systematic literature reviews. PLOS Neglected Trop. Dis. 2025, 19, e0012990. [Google Scholar] [CrossRef]
- Essackjee, K.; Goorah, S.; Ramchurn, S.K.; Cheeneebash, J.; Walker-Bone, K. Prevalence of and risk factors for chronic arthralgia and rheumatoid-like polyarthritis more than 2 years after infection with chikungunya virus. Postgrad. Med. J. 2013, 89, 440–447. [Google Scholar] [CrossRef]
- Noor, F.M.; Hossain, M.B.; Islam, Q.T. Prevalence of and risk factors for long-term disabilities following chikungunya virus disease: A meta-analysis. Travel Med. Infect. Dis. 2020, 35, 101618. [Google Scholar] [CrossRef] [PubMed]
- Vicente, C.R.; Louro, L.S.; de Jesus, N.R.; Lopes, D.T.D.S.; Cabidelle, A.S.A.; Cerutti Junior, C.; Miranda, A.E.B.; Louro, I.D.; Meira, D.D.; Chan, K.R. Factors Associated with Chronic Chikungunya in Vitória, Espírito Santo State, Brazil, Between 2016 and 2020. Viruses 2024, 16, 1679. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, A.C.M.; de Moura, E.L.; Ferreira, J.M.; de Moura, A.W.A.; Ferreira, A.D.S.; Bezerra, R.P.; Figueiredo, D.d.S.; Farias, K.F.; Andrade, T.G.d.A.; Figueiredo, E.V.M.d.S. Association of TNFA (−308G/A), IFNG (+874 A/T) and IL-10 (−819 C/T) polymorphisms with protection and susceptibility to dengue in Brazilian population. Asian Pac. J. Trop. Med. 2017, 10, 1065–1071. [Google Scholar] [CrossRef]
- Fishman, D.; Faulds, G.; Jeffery, R.; Mohamed-Ali, V.; Yudkin, J.S.; Humphries, S.; Woo, P. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J. Clin. Investig. 1998, 102, 1369–1376. [Google Scholar] [CrossRef]
- Braga, M.P.; Maciel, S.M.; de Moraes Marchiori, L.L.; Poli-Frederico, R.C. Association between interleukin-6 polymorphism in the −174 G/C region and hearing loss in the elderly with a history of occupational noise exposure. Braz. J. Otorhinolaryngol. 2014, 80, 373–378. [Google Scholar] [CrossRef]
- Pawlik, A.; Wrzesniewska, J.; Florczak, M.; Gawronska-Szklarz, B.; Herczynska, M. IL-6 promoter polymorphism in patients with rheumatoid arthritis. Scand. J. Rheumatol. 2005, 34, 109–113. [Google Scholar] [CrossRef]
- Moreira, S.T.; Cardoso, D.M.; Visentainer, J.E.; Fonzar, U.J.V.; Moliterno, R.A. The Possible Protective Role of the IL6 -174 GC Genotype in Dengue Fever. Open Trop. Med. J. 2008, 1, 87–91. [Google Scholar] [CrossRef]
- Toonen, E.J.M.; Barrera, P.; Fransen, J.; De Brouwer, A.P.M.; Eijsbouts, A.M.; Miossec, P.; Marotte, H.; Scheffer, H.; Van Riel, P.L.C.M.; Franke, B.; et al. Meta-analysis identified the TNFA -308G > A promoter polymorphism as a risk factor for disease severity in patients with rheumatoid arthritis. Arthritis Res. Ther. 2012, 14, R264. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, Y.; Yu, J.; Shen, J. Association between tumor necrosis factor alpha rs1800629 polymorphism and risk of osteoarthritis in a Chinese population. Braz. J. Med. Biol. Res. 2018, 51, e7311. [Google Scholar] [CrossRef]
- Rizvi, S.T.F.; Arif, A.; Azhar, A. TNF gene promoter region polymorphisms and association with young-onset rheumatoid arthritis. Pak. J. Pharm. Sci 2019, 32, 2295–2297. [Google Scholar]
- Fernandez-Mestre, M.T.; Gendzekhadze, K.; Rivas-Vetencourt, P.; Layrisse, Z. TNF-α-308A allele, a possible severity risk factor of hemorrhagic manifestation in dengue fever patients. Tissue Antigens 2004, 64, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Souza, A.M.D.; Resende, S.S.; Sousa, T.N.D.; Brito, C.F.A.D. A systematic scoping review of the genetic ancestry of the Brazilian population. Genet. Mol. Biol. 2019, 42, 495–508. [Google Scholar] [CrossRef]
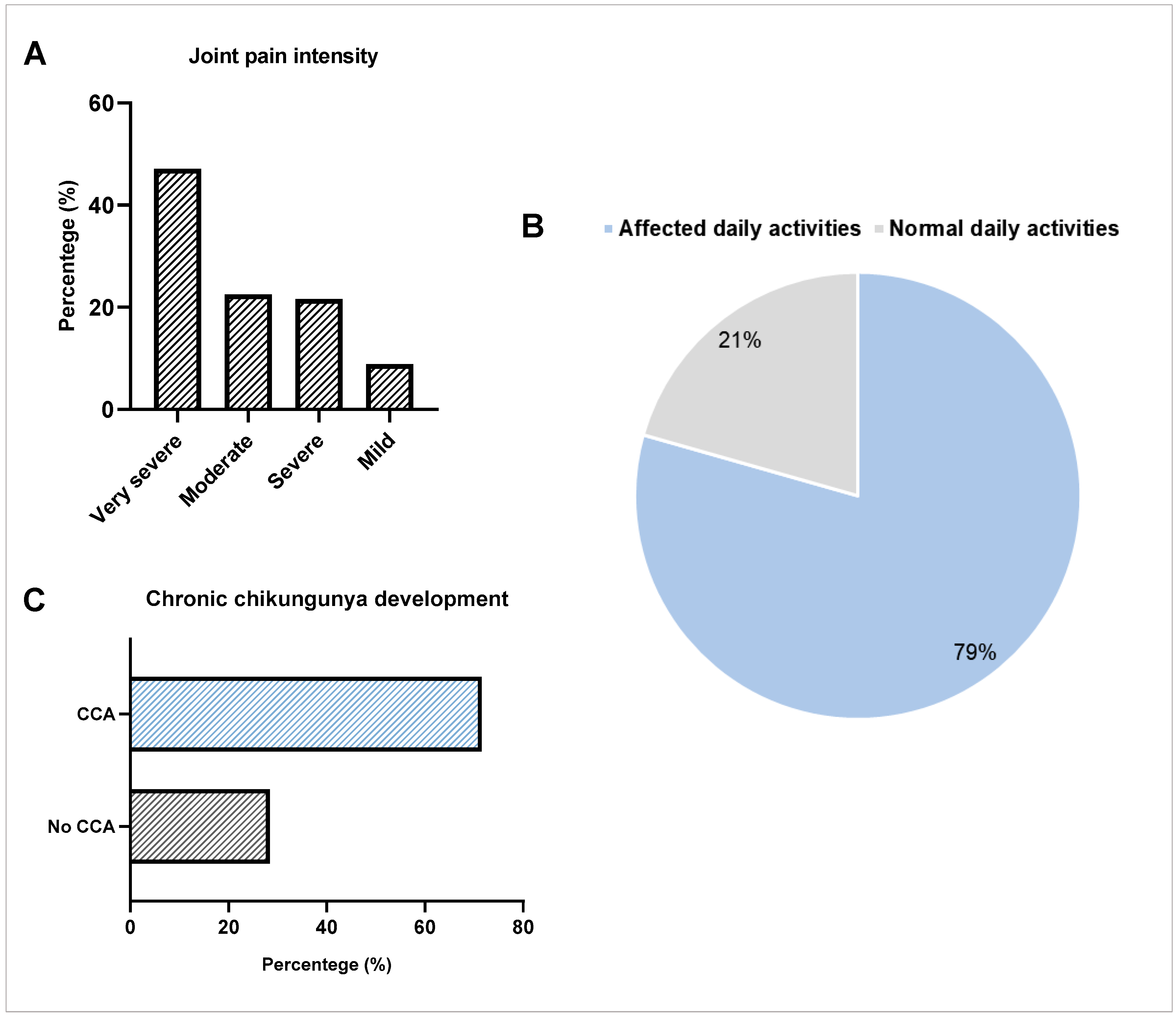
| Clinical Manifestations | Number of Observations n = 102 (%) |
|---|---|
| Arthralgia | 102 (100) |
| Fever | 87 (85.3) |
| Myalgia | 87 (85.3) |
| Headache | 73 (71.6) |
| Edema | 65 (63.7) |
| Prostration | 53 (52) |
| Retro-orbital pain | 42 (41.2) |
| Spots on the skin | 39 (38.2) |
| Nausea | 33 (32,3) |
| Fatigue | 24 (23.5) |
| Abdominal pain | 23 (22.5) |
| Dizziness | 23 (22.5) |
| Back pain | 22 (21.6) |
| Vomiting | 22 (21.6) |
| Diarrhea | 18 (17.6) |
| Joints affected | |
| Knees | 77 (75.5) |
| Ankles | 71 (69.6) |
| Elbow | 67 (65.7) |
| Shoulders | 54 (52.9) |
| Wrists | 50 (49) |
| Hips | 41 (40.2) |
| Characteristics | Non-CCA | CCA | p | OR (95% CI) |
|---|---|---|---|---|
| n = 29 (%) | n = 73 (%) | |||
| Sex (%) | ||||
| Female | 15 (51.7) | 56 (76.7) | 0.018 | 3.08 (1.29–7.41) |
| Male | 14 (48.3) | 17 (23.3) | ||
| Mild Pain | ||||
| Yes | 6 (20.7) | 3 (4.1) | 0.015 | 0.16 (0.04–0.63) |
| No | 23 (79.3) | 70 (95.9) | ||
| Moderate Pain | ||||
| Yes | 11 (37.9) | 12 (16.4) | 0.034 | 0.32 (0.12–0.89) |
| No | 18 (62.1) | 61 (83.6) | ||
| Severe Pain | ||||
| Yes | 7 (24.1) | 15 (20.5) | 0.791 | 0.81 (0.29–2.11) |
| No | 22 (75.9) | 58 (79.5) | ||
| Very Severe Pain | ||||
| Yes | 5 (17.2) | 43 (58.9) | <0.001 | 6.88 (2.29–17.7) |
| No | 24 (82.8) | 30 (41.1) |
| IL-6 -174 G/C | Case n = 102 (%) | Control n = 182 (%) | p | OR (95% CI) |
|---|---|---|---|---|
| Genotypes | ||||
| GG | 56 (54.9) | 87 (47.8) | - | (Reference) |
| GC | 39 (38.2) | 80 (43.9) | 0.284 | 0.76 (0.45–1.25) |
| CC | 7 (6.9) | 15 (8.2) | 0.509 | 0.72 (0.28–1.94) |
| GC + GG | 95 (93.1) | 167 (91.7) | 0.563 | 0.88 (0.59–1.34) |
| GC + CC | 46 (45.1) | 95 (52.2) | 0.250 | 0.75 (0.46–1.22) |
| Alleles | ||||
| G | 151(74.0) | 254 (69.8) | - | (Reference) |
| C | 53 (26.0) | 110 (30.2) | 0.283 | 0.81 (0.55–1.18) |
| IL-6 -174 G/C | ASY n = 29 (%) | Case n = 102 (%) | p | OR (95% CI) | ASY n = 29 (%) | Control n = 182 (%) | p | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|
| Genotypes | ||||||||
| GG | 17 (58.6) | 56 (54.9) | - | (Reference) | 17 (58.6) | 87 (47.8) | - | (Reference) |
| GC | 11 (37.9) | 39 (38.2) | 0.867 | 0.93 (0.40–2.16) | 11 (37.9) | 80 (43.9) | 0.397 | 0.70 (0.33–1.64) |
| CC | 1 (3.4) | 7 (6.9) | 0.676 | 0.47 (0.04–3.07) | 1 (3.4) | 15 (8.2) | 0.461 | 0.34 (0.03–2.10) |
| GC + GG | 28 (96.5) | 95 (93.1) | 0.932 | 0.97 (0.50–1.87) | 28 (96.5) | 167 (91.7) | 0.647 | 0.86 (0.45–1.68) |
| GC + CC | 12 (41.4) | 46 (45.1) | 0.722 | 0.86 (0.38–1.92) | 12 (41.4) | 95 (52.7) | 0.279 | 0.65 (0.29–1.45) |
| Alleles | ||||||||
| G | 45 (77.6) | 151 (74.0) | - | (Reference) | 45 (77.6) | 254 (69.8) | - | (Reference) |
| C | 13 (22.4) | 53 (26.0) | 0.580 | 0.82 (0.40–1.62) | 13 (22.4) | 110 (30.2) | 0.224 | 0.67 (0.34–1.27) |
| TNFα -308 G/A | Case n = 102 (%) | Control n = 182 (%) | p | OR (95% CI) |
|---|---|---|---|---|
| Genotypes | ||||
| GG | 73 (71.6) | 142 (78.0) | - | (Reference) |
| GA | 29 (28.4) | 39 (21.4) | 0.193 | 1.45 (0.84–2.53) |
| AA | 0 (0.0) | 1 (0.5) | 1.000 | 0.00 |
| GA + GG | 102 (100) | 181 (99.4) | 0.628 | 1.10 (0.75–1.58) |
| GA + AA | 29 (28.4) | 40 (22) | 0.223 | 1.41 (0.82–2.46) |
| Alleles | ||||
| G | 175 (85.8) | 323 (88.7) | - | (Reference) |
| A | 29 (14.2) | 41 (11.3) | 0.304 | 1.31 (0.79–2.15) |
| TNFα -308 G/A | ASY n = 29 (%) | Case n = 102 (%) | p | OR (95% CI) | ASY n = 29 (%) | Control n = 182 (%) | p | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|
| Genotypes | ||||||||
| GG | 23 (79.3) | 73 (71.6) | - | (Reference) | 23 (79.3) | 142 (78.0) | - | (Reference) |
| GA | 6 (20.7) | 29 (28.4) | 0.405 | 0.66 (0.24–1.78) | 6 (20.7) | 39 (21.4) | 0.916 | 0.95 (0.37–2.39) |
| AA | 0 (0.0) | 0 (0.0) | 1.000 | 0.00 | 0 (0.0) | 1 (0.5) | 1.000 | 0.00 |
| GA + GG | 29 (100) | 102 (100) | 0.747 | 0.90 (0.49–1.70) | 29 (100) | 181 (99.4) | 0.971 | 0.99 (0.55–1.74) |
| GA + AA | 6 (20.7) | 29 (28.4) | 0.405 | 0.66 (0.24–1.78) | 6 (20.7) | 40 (22.0) | 0.876 | 0.93 (0.36–2.32) |
| Alleles | ||||||||
| G | 52 (89.6) | 175 (85.8) | - | (Reference) | 52 (89.6) | 323 (88.7) | - | (Reference) |
| A | 6 (10.3) | 29 (14.2) | 0.444 | 0.70 (0.29–1.74) | 6 (10.3) | 41 (11.3) | 0.836 | 0.91 (0.39–2.26) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maciel, M.S.C.; Martins, C.D.d.B.; dos Santos, A.G.M.; Oliveira, C.N.; Fernandes, H.F.; Rodrigues, R.d.O.; Yaochite, J.N.U. Chronic Chikungunya Arthritis in Northeastern Brazil: An Association with Very Severe Joint Pain and Lack of Correlation with IL-6 and TNFα Gene Polymorphisms. Viruses 2025, 17, 1543. https://doi.org/10.3390/v17121543
Maciel MSC, Martins CDdB, dos Santos AGM, Oliveira CN, Fernandes HF, Rodrigues RdO, Yaochite JNU. Chronic Chikungunya Arthritis in Northeastern Brazil: An Association with Very Severe Joint Pain and Lack of Correlation with IL-6 and TNFα Gene Polymorphisms. Viruses. 2025; 17(12):1543. https://doi.org/10.3390/v17121543
Chicago/Turabian StyleMaciel, Mariella Sousa Coêlho, Catharina Diniz de Brito Martins, Alan Gleison Moreira dos Santos, Caroline Nobre Oliveira, Hygor Ferreira Fernandes, Raphael de Oliveira Rodrigues, and Juliana Navarro Ueda Yaochite. 2025. "Chronic Chikungunya Arthritis in Northeastern Brazil: An Association with Very Severe Joint Pain and Lack of Correlation with IL-6 and TNFα Gene Polymorphisms" Viruses 17, no. 12: 1543. https://doi.org/10.3390/v17121543
APA StyleMaciel, M. S. C., Martins, C. D. d. B., dos Santos, A. G. M., Oliveira, C. N., Fernandes, H. F., Rodrigues, R. d. O., & Yaochite, J. N. U. (2025). Chronic Chikungunya Arthritis in Northeastern Brazil: An Association with Very Severe Joint Pain and Lack of Correlation with IL-6 and TNFα Gene Polymorphisms. Viruses, 17(12), 1543. https://doi.org/10.3390/v17121543






