Early-Life Demographic Factors Shape Gut Microbiome Patterns Associated with Rotavirus Gastroenteritis Severity
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants and Eligibility Criteria
2.2. Clinical Assessment
2.3. Sample Collection, PCR Amplification, and Sequencing of 16S rRNA
2.4. Bioinformatic Processing and Taxonomic Classification
2.5. Statistical Analysis
3. Results
3.1. Demographic and Clinical Characteristics of the Participants
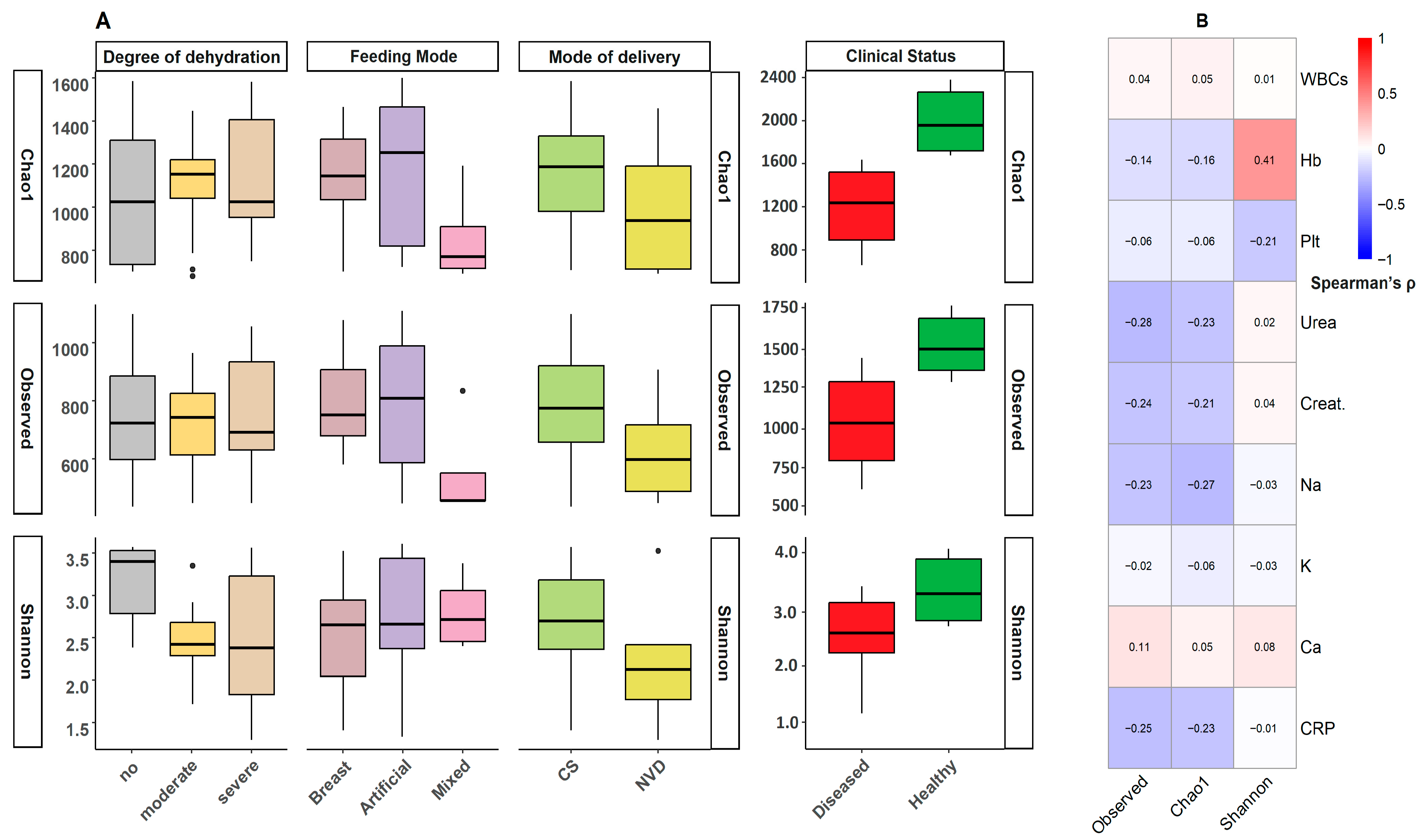
3.2. Bacterial Diversity Analysis
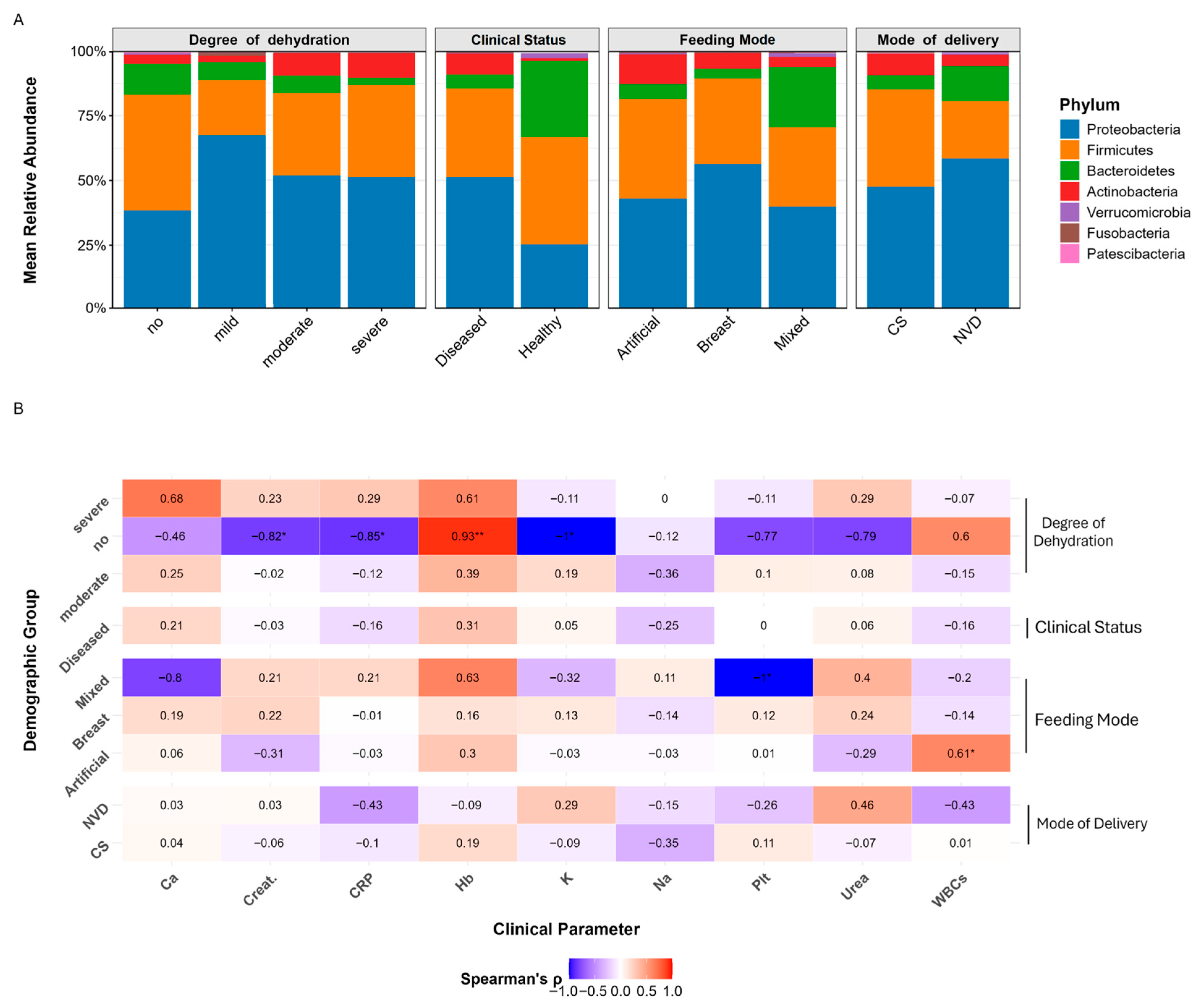
3.3. Taxonomic Profiling of RVGE Microbiomes: Phylum-Level Associations with Demographic and Clinical Variables
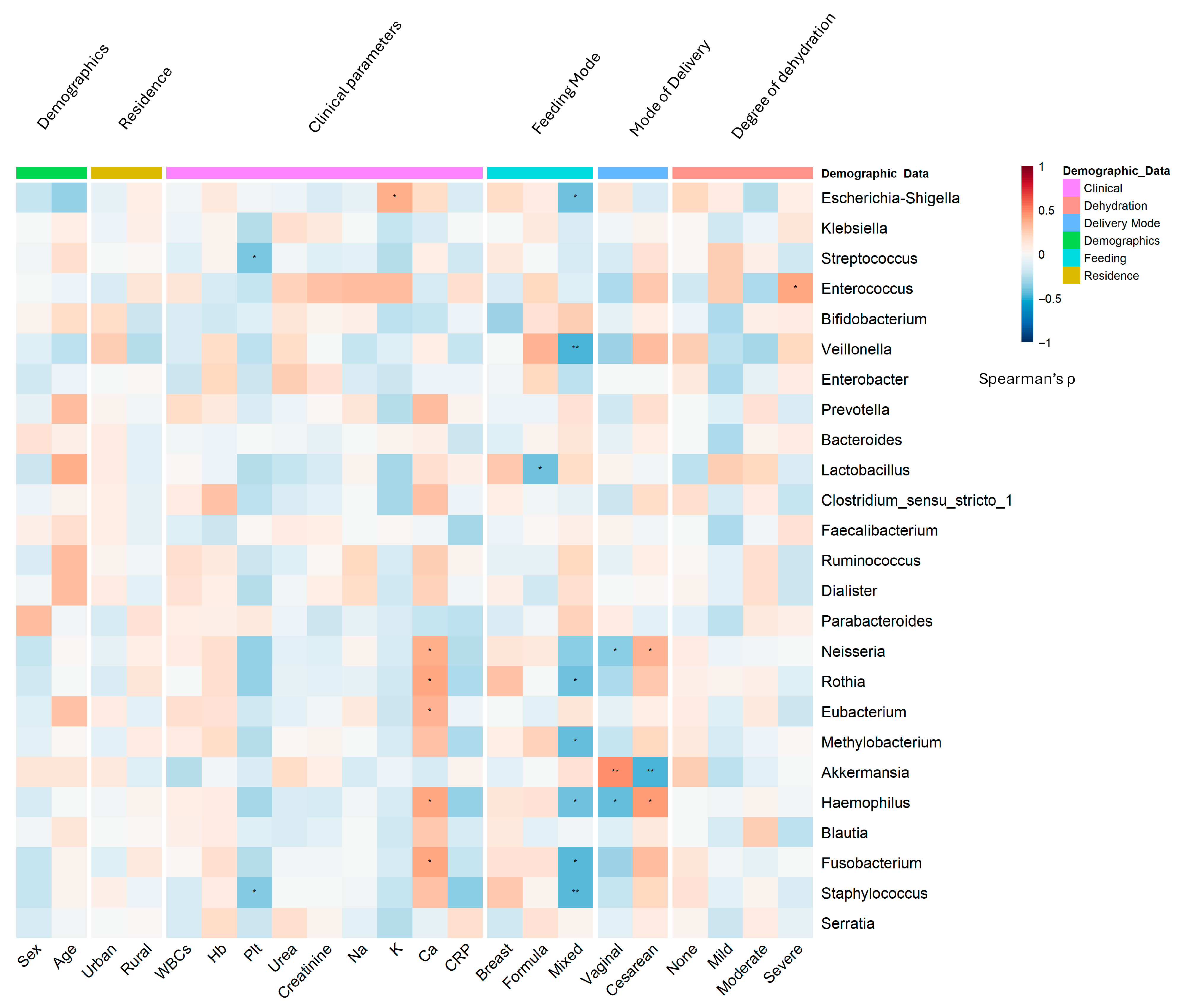
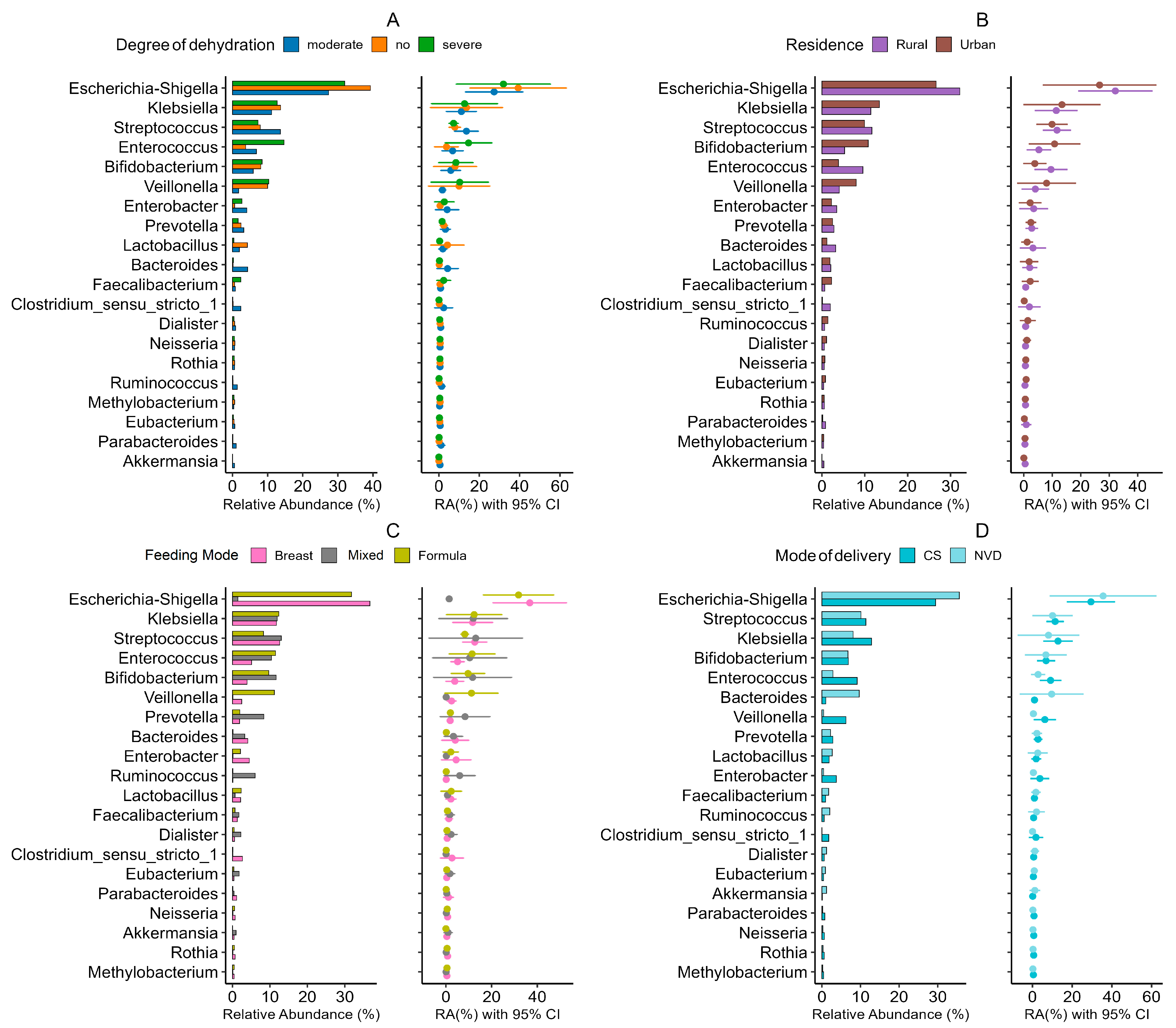
3.4. Microbial Signatures: Genus-Level Stratification Reveals Demographic Patterns in RVGE
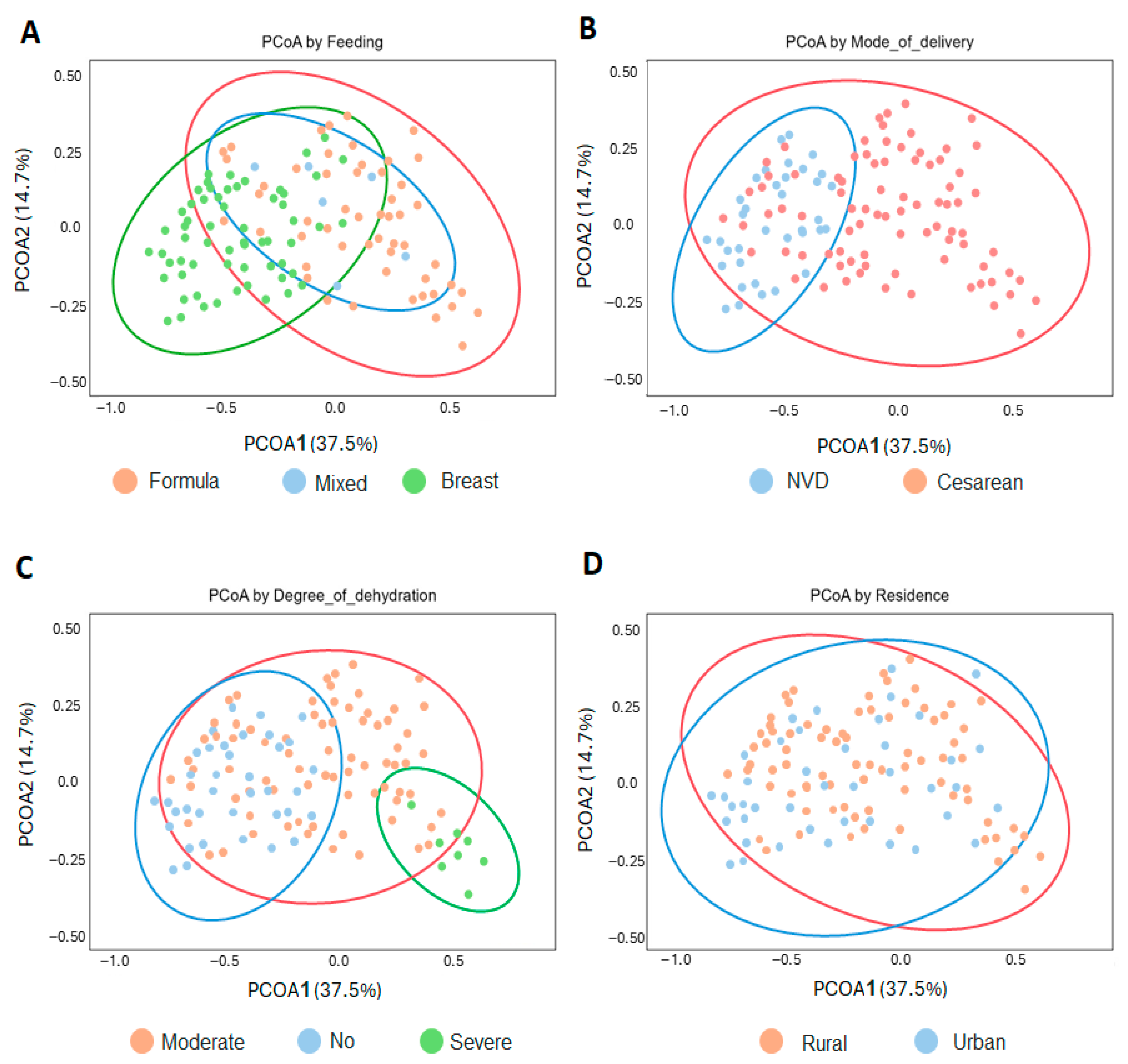
3.5. Influence of Demographic Factors on Microbiome Structure
- The random forest classifier demonstrated strong performance in discriminating infant feeding modes based on gut microbiome composition (Figure 5). Cross-validation revealed excellent discriminative ability for breastfed infants (AUC = 0.85, 95% CI: 0.79–0.91), very good discrimination for formula-fed infants (AUC = 0.82, 95% CI: 0.76–0.88), and good discrimination for mixed-fed infants (AUC = 0.78, 95% CI: 0.71–0.85). Overall multiclass classification achieved an AUC of 0.81 (95% CI: 0.75–0.87), indicating robust model performance (Supplementary Figure S2, Supplementary Table S4).
- Age and sex: The inflammatory genus Escherichia-Shigella was significantly enriched in male infants (r = 0.32, p = 0.003; DESeq2 log2FC = 2.3, p = 0.0026) and younger infants (<6 months; r = −0.41, p < 0.001), who also presented relatively high levels of Clostridium sensu stricto 1 (r = −0.35, p = 0.003) (Figure 5). In contrast, female infants and older infants presented greater abundances of the beneficial genera Bifidobacterium (r = −0.35, p < 0.001) and Bacteroides, respectively.
- Residence: Rural infants presented relatively high abundances of Clostridium sensu stricto 1 (LDA score = 3.5, p = 0.002) and Klebsiella (r = 0.38, p < 0.001) (Figure 5B).
- Delivery mode: Cesarean delivery was associated with an increased abundance of pathobionts such as Klebsiella (r = 0.45, p < 0.001) and Enterococcus, and a reduction in beneficial taxa such as Prevotella (MaAsLin2 coefficient = −0.89, p = 0.008) (Figure 5D). Vaginal delivery was associated with increased levels of Bifidobacterium (Supplementary Figure S3).
- Feeding Practices: Formula feeding was a major driver of dysbiosis, showing strong positive associations with Streptococcus (r = 0.47, p < 0.001) and Staphylococcus (r = 0.39, p = 0.002). Exclusive breastfeeding promoted a healthier microbiota enriched with Bifidobacterium (r = −0.42, p < 0.001) and Lactobacillus (r = −0.36, p = 0.004). Based on the AUC analysis, the Rikenellaceae RC9 gut group emerged as a notable discriminator for both the formula feeding (AUC = 0.758) and breastfeeding (AUC = 0.745) groups. Dickeya also exhibited strong performance for the formula-fed group (AUC = 0.682), whereas Fastidiosipila was significant for the breastfed group (AUC = 0.701) (Figure 5C; Supplementary Figure S4).
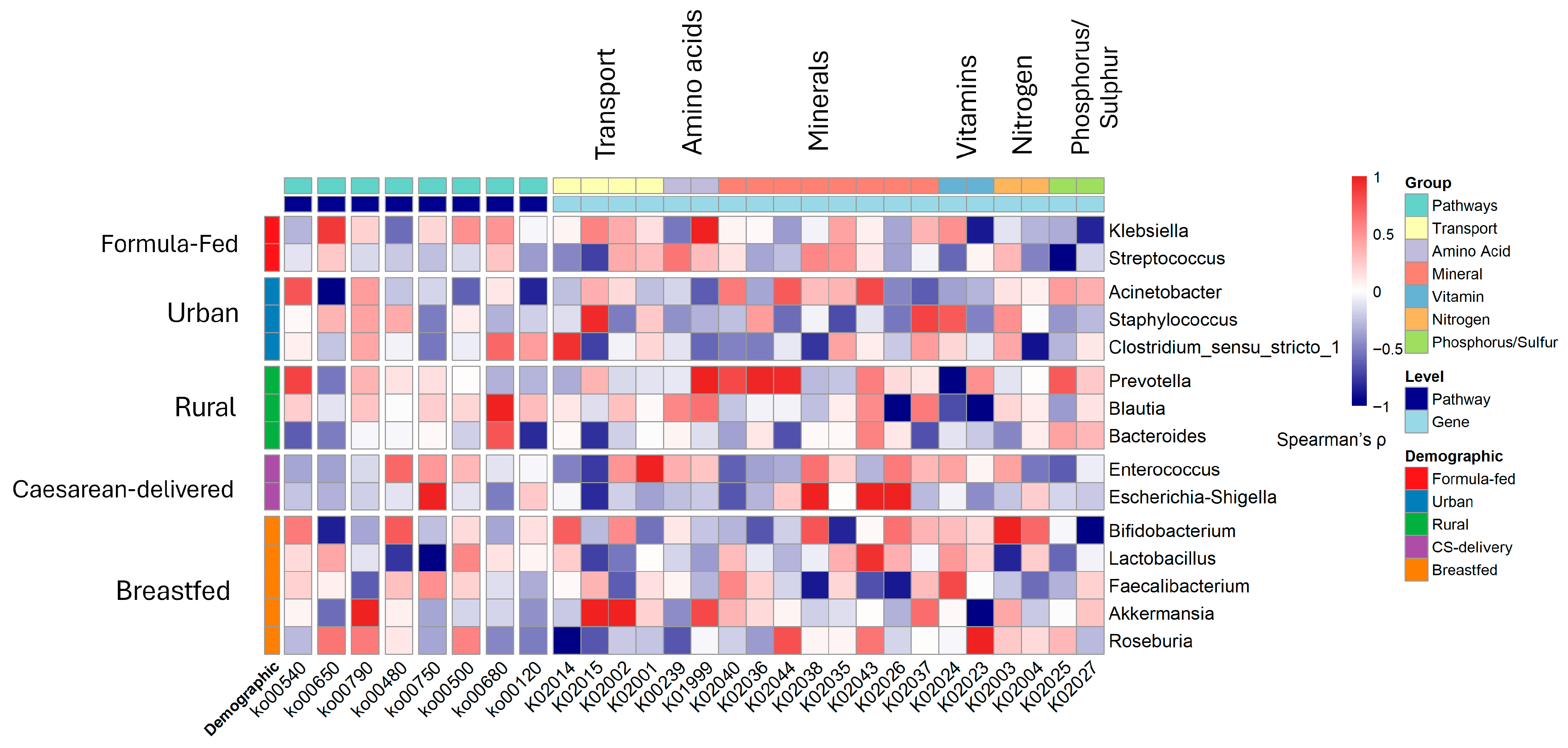
3.6. Synergistic Associations and Clinical Correlations with Gut Microbiomes
3.7. Metabolic Pathway Alterations and Functional Profiles
3.8. Clinical and Inflammatory Correlations
4. Discussion
4.1. Influence of Age and Sex on Microbiome and Severity
4.2. Impact of Geographic Residence and Local Epidemiology
4.3. The Critical Role of Feeding Practices
4.4. The Potential Impact of Delivery Mode and Synergistic Risk Factors
4.5. Core Taxonomic and Functional Dysbiosis in RVGE
4.6. Underlying Metabolic Shifts and Clinical Implications
4.7. Clinical Relevance and Therapeutic Implications
4.8. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Troeger, C.; Forouzanfar, M.; Rao, P.C.; Khalil, I.; Brown, A.; Reiner, R.C.; Fullman, N.; Thompson, R.L.; Abajobir, A.; Ahmed, M.; et al. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect. Dis. 2017, 17, 909–948. [Google Scholar] [CrossRef]
- Tosisa, W.; Regassa, B.T.; Eshetu, D.; Irenso, A.A.; Mulu, A.; Hundie, G.B. Rotavirus infections and their genotype distribution pre- and post-vaccine introduction in Ethiopia: A systemic review and meta-analysis. BMC Infect. Dis. 2024, 24, 836. [Google Scholar] [CrossRef]
- Harris, V.; Ali, A.; Fuentes, S.; Korpela, K.; Kazi, M.; Tate, J.; Parashar, U.; Wiersinga, W.J.; Giaquinto, C.; de Weerth, C.; et al. Rotavirus vaccine response correlates with the infant gut microbiota composition in Pakistan. Gut Microbes 2018, 9, 93–101. [Google Scholar] [CrossRef]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Chibuye, M.; Mende, D.R.; Spijker, R.; Simuyandi, M.; Luchen, C.C.; Bosomprah, S.; Chilengi, R.; Schultsz, C.; Harris, V.C. Systematic review of associations between gut microbiome composition and stunting in under-five children. NPJ Biofilms Microbiomes 2024, 10, 46. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S. Factors influencing development of the infant microbiota: From prenatal period to early infancy. Clin. Exp. Pediatr. 2022, 65, 439–447. [Google Scholar] [CrossRef]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K.; et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017, 171, 647–654. [Google Scholar] [CrossRef]
- Aziz, T.; Hussain, N.; Hameed, Z.; Lin, L. Elucidating the role of diet in maintaining gut health to reduce the risk of obesity, cardiovascular and other age-related inflammatory diseases: Recent challenges and future recommendations. Gut Microbes 2024, 16, 2297864. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Godoy-Vitorino, F.; Knight, R.; Blaser, M.J. Role of the microbiome in human development. Gut 2019, 68, 1108–1114. [Google Scholar] [CrossRef] [PubMed]
- Stinson, L.F.; Norrish, I.; Mhembere, F.; Cheema, A.S.; Mullally, C.A.; Payne, M.S.; Geddes, D.T. Seeding and feeding: Nutrition and birth-associated exposures shape gut microbiome assembly in breastfed infants. Gut Microbes 2025, 17, 2557981. [Google Scholar] [CrossRef]
- Heston, S.M.; Hurst, J.H.; Kelly, M.S. Understanding the influence of the microbiome on childhood infections. Expert Rev. Anti-Infect. Ther. 2024, 22, 529–545. [Google Scholar] [CrossRef]
- Campbell, D.E.; Li, Y.; Ingle, H.; Baldridge, M.T. Impact of the Microbiota on Viral Infections. Annu. Rev. Virol. 2023, 10, 371–395. [Google Scholar] [CrossRef] [PubMed]
- Engevik, M.A.; Banks, L.D.; Engevik, K.A.; Chang-Graham, A.L.; Perry, J.L.; Hutchinson, D.S.; Ajami, N.J.; Petrosino, J.F.; Hyser, J.M. Rotavirus infection induces glycan availability to promote ileum-specific changes in the microbiome aiding rotavirus virulence. Gut Microbes 2020, 11, 1324–1347. [Google Scholar] [CrossRef]
- Ramani, S.; Stewart, C.J.; Laucirica, D.R.; Ajami, N.J.; Robertson, B.; Autran, C.A.; Shinge, D.; Rani, S.; Anandan, S.; Hu, L.; et al. Human milk oligosaccharides, milk microbiome and infant gut microbiome modulate neonatal rotavirus infection. Nat. Commun. 2018, 9, 5010. [Google Scholar] [CrossRef]
- Omatola, C.A.; Olaniran, A.O. Rotaviruses: From Pathogenesis to Disease Control-A Critical Review. Viruses 2022, 14, 875. [Google Scholar] [CrossRef]
- Vangay, P.; Johnson, A.J.; Ward, T.L.; Al-Ghalith, G.A.; Shields-Cutler, R.R.; Hillmann, B.M.; Lucas, S.K.; Beura, L.K.; Thompson, E.A.; Till, L.M.; et al. US immigration westernizes the human gut microbiome. Cell 2018, 175, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Hayes, R.B. Environmental Influences on the Human Microbiome and Implications for Noncommunicable Disease. Annu. Rev. Public Health 2021, 42, 277–292. [Google Scholar] [CrossRef] [PubMed]
- Flandroy, L.; Poutahidis, T.; Berg, G.; Clarke, G.; Dao, M.C.; Decaestecker, E.; Furman, E.; Haahtela, T.; Massart, S.; Plovier, H.; et al. The impact of human activities and lifestyles on the interlinked microbiota and health of humans and of ecosystems. Sci. Total Environ. 2018, 627, 1018–1038. [Google Scholar] [CrossRef]
- Castaño-Rodríguez, N.; Underwood, A.P.; Merif, J.; Riordan, S.M.; Rawlinson, W.D.; Mitchell, H.M.; Kaakoush, N.O. Gut Microbiome Analysis Identifies Potential Etiological Factors in Acute Gastroenteritis. Infect. Immun. 2018, 86, e00060-18. [Google Scholar] [CrossRef]
- Huo, W.; Qiao, Y.; Li, E.; Li, M.; Che, L. Interplay between nutrition, microbiota, and immunity in rotavirus infection: Insights from human and animal models. Front. Vet. Sci. 2025, 12, 1680448. [Google Scholar] [CrossRef]
- Kim, A.H.; Hogarty, M.P.; Harris, V.C.; Baldridge, M.T. The Complex Interactions Between Rotavirus and the Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 586751. [Google Scholar] [CrossRef]
- Guarino, A.; Guandalini, S.; Vecchio, A.L. Probiotics for prevention and treatment of diarrhea. J. Clin. Gastroenterol. 2011, 45, S149–S153. [Google Scholar] [CrossRef]
- Han, Z.; Min, Y.; Pang, K.; Wu, D. Therapeutic Approach Targeting Gut Microbiome in Gastrointestinal Infectious Diseases. Int. J. Mol. Sci. 2023, 24, 15654. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Fan, X.; Mao, X.; Yu, B.; He, J.; Yan, H.; Wang, J. The protective role of prebiotics and probiotics on diarrhea and gut damage in the rotavirus-infected piglets. J. Anim. Sci. Biotechnol. 2024, 15, 61. [Google Scholar] [CrossRef]
- Mederle, A.L.; Dima, M.; Stoicescu, E.R.; Căpăstraru, B.F.; Levai, C.M.; Hațegan, O.A.; Maghiari, A.L. Impact of Gut Microbiome Interventions on Glucose and Lipid Metabolism in Metabolic Diseases: A Systematic Review and Meta-Analysis. Life 2024, 14, 1485. [Google Scholar] [CrossRef] [PubMed]
- Steyer, A.; Mičetić-Turk, D.; Fijan, S. The Efficacy of Probiotics as Antiviral Agents for the Treatment of Rotavirus Gastrointestinal Infections in Children: An Updated Overview of Literature. Microorganisms 2022, 10, 2392. [Google Scholar] [CrossRef]
- Allen, S.J.; Martinez, E.G.; Gregorio, G.V.; Dans, L.F. Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst. Rev. 2010, 2010, Cd003048. [Google Scholar] [CrossRef]
- Săsăran, M.O.; Mărginean, C.O.; Adumitrăchioaiei, H.; Meliț, L.E. Pathogen-Specific Benefits of Probiotic and Synbiotic Use in Childhood Acute Gastroenteritis: An Updated Review of the Literature. Nutrients 2023, 15, 643. [Google Scholar] [CrossRef]
- Esona, M.D.; McDonald, S.; Kamili, S.; Kerin, T.; Gautam, R.; Bowen, M.D. Comparative evaluation of commercially available manual and automated nucleic acid extraction methods for rotavirus RNA detection in stools. J. Virol. Methods 2013, 194, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Guarino, A.; Ashkenazi, S.; Gendrel, D.; Lo Vecchio, A.; Shamir, R.; Szajewska, H. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition/European Society for Pediatric Infectious Diseases evidence-based guidelines for the management of acute gastroenteritis in children in Europe: Update 2014. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 132–152. [Google Scholar] [CrossRef]
- Schnadower, D.; Tarr, P.I.; Gorelick, M.H.; O’Connell, K.; Roskind, C.G.; Powell, E.C.; Rao, J.; Bhatt, S.; Freedman, S.B. Validation of the Modified Vesikari Score in Children With Gastroenteritis in 5 US Emergency Departments. J. Pediatr. Gastroenterol. Nutr. 2013, 57, 514–519. [Google Scholar] [CrossRef]
- Lacey, J.; Corbett, J.; Forni, L.; Hooper, L.; Hughes, F.; Minto, G.; Moss, C.; Price, S.; Whyte, G.; Woodcock, T.; et al. A multidisciplinary consensus on dehydration: Definitions, diagnostic methods and clinical implications. Ann. Med. 2019, 51, 232–251. [Google Scholar] [CrossRef]
- King, C.K.; Glass, R.; Bresee, J.S.; Duggan, C.; Centers for Disease Control and Prevention. Managing acute gastroenteritis among children. MMWR Recomm. Rep. 2003, 52, 16. [Google Scholar]
- Kelly, J.R.; Kennedy, P.J.; Cryan, J.F.; Dinan, T.G.; Clarke, G.; Hyland, N.P. Breaking down the barriers: The gut microbiome, intestinal permeability and stress-related psychiatric disorders. Front. Cell. Neurosci. 2015, 9, 392. [Google Scholar] [CrossRef]
- Vandeputte, D.; Kathagen, G.; D’hoe, K.; Vieira-Silva, S.; Valles-Colomer, M.; Sabino, J.; Wang, J.; Tito, R.Y.; De Commer, L.; Darzi, Y.; et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 2017, 551, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Mattiello, F.; Verbist, B.; Faust, K.; Raes, J.; Shannon, W.D.; Bijnens, L.; Thas, O. A web application for sample size and power calculation in case-control microbiome studies. Bioinformatics 2016, 32, 2038–2040. [Google Scholar] [CrossRef]
- Ramadan, M.; Hetta, H.F.; Saleh, M.M.; Ali, M.E.; Ahmed, A.A.; Salah, M. Alterations in skin microbiome mediated by radiotherapy and their potential roles in the prognosis of radiotherapy-induced dermatitis: A pilot study. Sci. Rep. 2021, 11, 5179. [Google Scholar] [CrossRef] [PubMed]
- Hurst, S.A. Declaration of Helsinki and protection for vulnerable research participants. JAMA 2014, 311, 1252. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, M.; Solyman, S.; Yones, M.; Abdallah, Y.; Halaby, H.; Hanora, A. Skin Microbiome Differences in Atopic Dermatitis and Healthy Controls in Egyptian Children and Adults, and Association with Serum Immunoglobulin E. Omics J. Integr. Biol. 2019, 23, 247–260. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Pasolli, E.; Truong, D.T.; Malik, F.; Waldron, L.; Segata, N. Machine learning meta-analysis of large metagenomic datasets: Tools and biological insights. PLoS Comput. Biol. 2016, 12, e1004977. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Knights, D.; Costello, E.K.; Knight, R. Supervised classification of human microbiota. FEMS Microbiol. Rev. 2011, 35, 343–359. [Google Scholar] [CrossRef]
- Aßhauer, K.P.; Wemheuer, B.; Daniel, R.; Meinicke, P. Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 2015, 31, 2882–2884. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef] [PubMed]
- Wise, J.L.; Cummings, B.P. The 7-α-dehydroxylation pathway: An integral component of gut bacterial bile acid metabolism and potential therapeutic target. Front. Microbiol. 2023, 13, 1093420. [Google Scholar] [CrossRef]
- Ingram, J.; Jorgensen, S.E.; Fath, B.D. Berger–Parker Index. Encycl. Ecol. 2008, 2008, 332–334. [Google Scholar]
- Anderson, M.J.; Walsh, D.C. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: What null hypothesis are you testing? Ecol. Monogr. 2013, 83, 557–574. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- R Core Team, R. R: A Language and Environment for Statistical; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Hurst, J.H.; Heston, S.M.; Kelly, M.S. Host microbiome-pathogen interactions in pediatric infections. Curr. Opin. Infect. Dis. 2023, 36, 399–404. [Google Scholar] [CrossRef]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Parashar, U.D.; World Health Organization–Coordinated Global Rotavirus Surveillance Network. Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000–2013. Clin. Infect. Dis. 2016, 62, S96–S105. [Google Scholar] [CrossRef]
- Kumbhare, S.V.; Patangia, D.V.; Patil, R.H.; Shouche, Y.S.; Patil, N.P. Factors influencing the gut microbiome in children: From infancy to childhood. J. Biosci. 2019, 44, 49. [Google Scholar] [CrossRef]
- Angel, J.; Franco, M.A.; Greenberg, H.B. Rotavirus immune responses and correlates of protection. Curr. Opin. Virol. 2012, 2, 419–425. [Google Scholar] [CrossRef]
- Mantadakis, E.; Chatzimichael, E.; Zikidou, P. Iron Deficiency Anemia in Children Residing in High and Low-Income Countries: Risk Factors, Prevention, Diagnosis and Therapy. Mediterr. J. Hematol. Infect. Dis. 2020, 12, e2020041. [Google Scholar] [CrossRef] [PubMed]
- Chung The, H.; Le, S.H. Dynamic of the human gut microbiome under infectious diarrhea. Curr. Opin. Microbiol. 2022, 66, 79–85. [Google Scholar] [CrossRef]
- Karampatsas, K.; Osborne, L.; Seah, M.L.; Tong, C.Y.W.; Prendergast, A.J. Clinical characteristics and complications of rotavirus gastroenteritis in children in east London: A retrospective case-control study. PLoS ONE 2018, 13, e0194009. [Google Scholar] [CrossRef]
- Aliabadi, N.; Tate, J.E.; Haynes, A.K.; Parashar, U.D.; Centers for Disease Control and Prevention (CDC). Sustained decrease in laboratory detection of rotavirus after implementation of routine vaccination—United States, 2000–2014. MMWR Morb. Mortal. Wkly. Rep. 2015, 64, 337–342. [Google Scholar]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Kelly, J.C.; Nolan, L.S.; Good, M. Vaginal seeding after cesarean birth: Can we build a better infant microbiome? Med 2021, 2, 889–891. [Google Scholar] [CrossRef] [PubMed]
- Vandenplas, Y.; Berger, B.; Carnielli, V.P.; Ksiazyk, J.; Lagström, H.; Sanchez Luna, M.; Migacheva, N.; Mosselmans, J.-M.; Picaud, J.-C.; Possner, M.; et al. Human milk oligosaccharides: 2′-fucosyllactose (2′-FL) and lacto-N-neotetraose (LNnT) in infant formula. Nutrients 2018, 10, 1161. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Burrows, M.; Khan, A.A.; Graham, L.; Volchkov, P.; Becker, L.; Antonopoulos, D.; Umesaki, Y.; Chervonsky, A.V. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013, 39, 400–412. [Google Scholar] [CrossRef]
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529, 212–215. [Google Scholar] [CrossRef]
- Rook, G.A.; Raison, C.L.; Lowry, C.A. Microbial ‘old friends’, immunoregulation and socioeconomic status. Clin. Exp. Immunol. 2014, 177, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bode, L.; Jantscher-Krenn, E. Structure-function relationships of human milk oligosaccharides. Adv. Nutr. 2012, 3, 383S–391S. [Google Scholar] [CrossRef]
- Henrick, B.M.; Rodriguez, L. Bifidobacteria-mediated immune system imprinting. Cell Rep. 2021, 35, 109054. [Google Scholar] [CrossRef]
- Li, Y.; Xia, S.; Jiang, X.; Feng, C.; Gong, S.; Ma, J.; Fang, Z.; Yin, J.; Yin, Y. Gut Microbiota and Diarrhea: An Updated Review. Front. Cell. Infect. Microbiol. 2021, 11, 625210. [Google Scholar] [CrossRef]
- Greenfield, K.G.; Harlow, O.S.; Witt, L.T.; Dziekan, E.M.; Tamar, C.R.; Meier, J.; Brumbaugh, J.E.; Levy, E.R.; Knoop, K.A. Neonatal intestinal colonization of Streptococcus agalactiae and the multiple modes of protection limiting translocation. Gut Microbes 2024, 16, 2379862. [Google Scholar] [CrossRef]
- Guo, Q.; Goldenberg, J.Z.; Humphrey, C.; El Dib, R.; Johnston, B.C. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst. Rev. 2019, 4, Cd004827. [Google Scholar] [CrossRef]
- Victora, C.G.; Bahl, R.; Barros, A.J.; França, G.V.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C.; et al. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef]
- Ho, T.T.B.; Groer, M.W.; Kane, B.; Yee, A.L.; Torres, B.A.; Gilbert, J.A.; Maheshwari, A. Dichotomous development of the gut microbiome in preterm infants. Microbiome 2018, 6, 157. [Google Scholar] [CrossRef] [PubMed]
- Inchingolo, F.; Inchingolo, A.M.; Latini, G.; Ferrante, L.; de Ruvo, E.; Campanelli, M.; Longo, M.; Palermo, A.; Inchingolo, A.D.; Dipalma, G. Difference in the Intestinal Microbiota between Breastfeed Infants and Infants Fed with Artificial Milk: A Systematic Review. Pathogens 2024, 13, 533. [Google Scholar] [CrossRef] [PubMed]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. BioMed Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef]
- Mizutani, T.; Aboagye, S.Y.; Ishizaka, A.; Afum, T.; Mensah, G.I.; Asante-Poku, A.; Asandem, D.A.; Parbie, P.K.; Abana, C.Z.-Y.; Kushitor, D.; et al. Gut microbiota signature of pathogen-dependent dysbiosis in viral gastroenteritis. Sci. Rep. 2021, 11, 13945. [Google Scholar] [CrossRef] [PubMed]
- Hugouvieux-Cotte-Pattat, N.; Pédron, J.; Van Gijsegem, F. Insight into biodiversity of the recently rearranged genus Dickeya. Front. Plant Sci. 2023, 14, 1168480. [Google Scholar] [CrossRef]
- Van Gijsegem, F.; van der Wolf, J.M.; Toth, I.K. Plant Diseases Caused by Dickeya and Pectobacterium Species; Springer: Cham, Switzerland, 2021. [Google Scholar]
- Dong, L.-N.; Wang, M.; Guo, J.; Wang, J.-P. Role of intestinal microbiota and metabolites in inflammatory bowel disease. Chin. Med. J. 2019, 132, 1610–1614. [Google Scholar] [CrossRef]
- Ferrer-Picón, E.; Dotti, I.; Corraliza, A.M.; Mayorgas, A.; Esteller, M.; Perales, J.C.; Ricart, E.; Masamunt, M.C.; Carrasco, A.; Tristán, E.; et al. Intestinal Inflammation Modulates the Epithelial Response to Butyrate in Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2020, 26, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Chen, C.; Kang, S.; Kwon, J.-I.; Jin, J.; Che, H. Effect of different feeding methods and gut microbiota on premature infants and clinical outcomes. Front. Nutr. 2022, 9, 888304. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Depommier, C.; Derrien, M.; Everard, A.; de Vos, W.M. Akkermansia muciniphila: Paradigm for next-generation beneficial microorganisms. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 625–637. [Google Scholar] [CrossRef]
- Oraby, D. Studying Factors Associated with Increased Prevalence of Caesarean Section in Cairo and Gharbia Governorates. 2023. Available online: https://egypt.unfpa.org/sites/default/files/pub-pdf/final_cs_report.pdf (accessed on 12 November 2025).
- Elnakib, S.; Abdel-Tawab, N.; Orbay, D.; Hassanein, N. Medical and non-medical reasons for cesarean section delivery in Egypt: A hospital-based retrospective study. BMC Pregnancy Childbirth 2019, 19, 411. [Google Scholar] [CrossRef]
- Mitchell, C.M.; Mazzoni, C.; Hogstrom, L.; Bryant, A.; Bergerat, A.; Cher, A.; Pochan, S.; Herman, P.; Carrigan, M.; Sharp, K.; et al. Delivery Mode Affects Stability of Early Infant Gut Microbiota. Cell Rep. Med. 2020, 1, 100156. [Google Scholar] [CrossRef] [PubMed]
- Gleizes, O.; Desselberger, U.; Tatochenko, V.; Rodrigo, C.; Salman, N.; Mezner, Z.; Giaquinto, C.; Grimprel, E. Nosocomial rotavirus infection in European countries: A review of the epidemiology, severity and economic burden of hospital-acquired rotavirus disease. Pediatr. Infect. Dis. J. 2006, 25, S12–S21. [Google Scholar] [CrossRef]
- Valles-Colomer, M.; Falony, G.; Darzi, Y.; Tigchelaar, E.F.; Wang, J.; Tito, R.Y.; Schiweck, C.; Kurilshikov, A.; Joossens, M.; Wijmenga, C.; et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019, 4, 623–632. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef]
- Dinh, D.M.; Ramadass, B.; Kattula, D.; Sarkar, R.; Braunstein, P.; Tai, A.; Wanke, C.A.; Hassoun, S.; Kane, A.V.; Naumova, E.N.; et al. Longitudinal analysis of the intestinal microbiota in persistently stunted young children in South India. PLoS ONE 2016, 11, e0155405. [Google Scholar] [CrossRef]
- Yan, Q.; Chen, Y.; Gao, E.-B.; Lu, Y.; Wu, J.; Qiu, H. The characteristics of intestinal microflora in infants with rotavirus enteritis, changes in microflora before and after treatment and their clinical values. Sci. Rep. 2025, 15, 4312. [Google Scholar] [CrossRef] [PubMed]
- Bull, M.J.; Plummer, N.T. Part 1: The Human Gut Microbiome in Health and Disease. Integr. Med. 2014, 13, 17–22. [Google Scholar]
- Bernaola Aponte, G.; Bada Mancilla, C.A.; Carreazo, N.Y.; Rojas Galarza, R.A. Probiotics for treating persistent diarrhoea in children. Cochrane Database Syst. Rev. 2013, 2013, Cd007401. [Google Scholar] [CrossRef] [PubMed]
- Longmore, D.K.; Batch, J.A.; McMahon, S.K.; Conwell, L.S. Klebsiella pneumoniae bacteraemia complicating rotavirus gastroenteritis in two infants with glucocorticoid deficiency. J. Pediatr. Endocrinol. Metab. 2010, 23, 293–295. [Google Scholar] [CrossRef]
- Jeon, S.G.; Kayama, H.; Ueda, Y.; Takahashi, T.; Asahara, T.; Tsuji, H.; Tsuji, N.M.; Kiyono, H.; Ma, J.S.; Kusu, T.; et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 2012, 8, e1002714. [Google Scholar] [CrossRef]
- Kassai, S.; de Vos, P. Gastrointestinal barrier function, immunity, and neurocognition: The role of human milk oligosaccharide (hMO) supplementation in infant formula. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13271. [Google Scholar] [CrossRef]
- Zhu, W.; Winter, M.G.; Byndloss, M.X.; Spiga, L.; Duerkop, B.A.; Hughes, E.R.; Büttner, L.; De Lima Romão, E.; Behrendt, C.L.; Lopez, C.A.; et al. Precision editing of the gut microbiota ameliorates colitis. Nature 2018, 553, 208–211. [Google Scholar] [CrossRef]
- Miquel, S.; Martín, R.; Rossi, O.; Bermúdez-Humarán, L.G.; Chatel, J.M.; Sokol, H.; Thomas, M.; Wells, J.M.; Langella, P. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 2013, 16, 255–261. [Google Scholar] [CrossRef]
- Kho, Z.Y.; Lal, S.K. The human gut microbiome—A potential controller of wellness and disease. Front. Microbiol. 2018, 9, 356589. [Google Scholar] [CrossRef]
- Levy, M.; Thaiss, C.A.; Zeevi, D.; Dohnalová, L.; Zilberman-Schapira, G.; Mahdi, J.A.; David, E.; Savidor, A.; Korem, T.; Herzig, Y.; et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 2017, 168, 389–402. [Google Scholar] [CrossRef] [PubMed]
- D’Aimmo, M.R.; Satti, M.; Scarafile, D.; Modesto, M.; Pascarelli, S.; Biagini, S.A.; Luiselli, D.; Mattarelli, P.; Andlid, T. Folate-producing bifidobacteria: Metabolism, genetics, and relevance. Microbiome Res. Rep. 2024, 3, 11. [Google Scholar] [CrossRef]
- Ríos-Covian, D.; Langella, P.; Martín, R. From Short- to Long-Term Effects of C-Section Delivery on Microbiome Establishment and Host Health. Microorganisms 2021, 9, 2122. [Google Scholar] [CrossRef]
- Kerr, C.A.; Grice, D.M.; Tran, C.D.; Bauer, D.C.; Li, D.; Hendry, P.; Hannan, G.N. Early life events influence whole-of-life metabolic health via gut microflora and gut permeability. Crit. Rev. Microbiol. 2015, 41, 326–340. [Google Scholar] [CrossRef]
- Bolte, E.E.; Moorshead, D.; Aagaard, K.M. Maternal and early life exposures and their potential to influence development of the microbiome. Genome Med. 2022, 14, 4. [Google Scholar] [CrossRef] [PubMed]
- Biasucci, G.; Benenati, B.; Morelli, L.; Bessi, E.; Boehm, G. Cesarean Delivery May Affect the Early Biodiversity of Intestinal Bacteria1,2. J. Nutr. 2008, 138, 1796S–1800S. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Diaz, J.; Ruiz-Ojeda, F.J.; Gil-Campos, M.; Gil, A. Mechanisms of action of probiotics. Adv. Nutr. 2019, 10, S49–S66. [Google Scholar] [CrossRef]
- Jia, W.; Xie, G.; Jia, W. Bile acid–microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef]
- Claus, S.P.; Guillou, H.; Ellero-Simatos, S. The gut microbiota: A major player in the toxicity of environmental pollutants? npj Biofilms Microbiomes 2016, 2, 16003. [Google Scholar] [CrossRef]
- Plaza-Díaz, J.; Ruiz-Ojeda, F.J.; Vilchez-Padial, L.M.; Gil, A. Evidence of the anti-inflammatory effects of probiotics and synbiotics in intestinal chronic diseases. Nutrients 2017, 9, 555. [Google Scholar] [CrossRef]
- Wahlström, A.; Sayin, S.I.; Marschall, H.-U.; Bäckhed, F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Sohail, M.U.; Al Khatib, H.A.; Al Thani, A.A.; Al Ansari, K.; Yassine, H.M.; Al-Asmakh, M. Microbiome profiling of rotavirus infected children suffering from acute gastroenteritis. Gut Pathog. 2021, 13, 21. [Google Scholar] [CrossRef]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.O.; Sarkar, A.; McSkimming, D.I. Gut Microbiota and Immune System Interactions. Microorganisms 2020, 8, 1587. [Google Scholar] [CrossRef]
- Hiippala, K.; Jouhten, H.; Ronkainen, A.; Hartikainen, A.; Kainulainen, V.; Jalanka, J.; Satokari, R. The Potential of Gut Commensals in Reinforcing Intestinal Barrier Function and Alleviating Inflammation. Nutrients 2018, 10, 988. [Google Scholar] [CrossRef]
- Kukov, A.; Peruhova, M.; Syarov, A.; Altankova, I.; Yurukova, N.; Goncharov, A.; Vazharova, R.; Mihova, A.; Velikova, T. Alterations of gut bacteria Akkermansia muciniphila and Faecalibacterium prausnitzii in late post-transplant period after liver transplantation. Iberoam. J. Med. 2022, 4, 45–51. [Google Scholar] [CrossRef]
- Bozzi Cionci, N.; Baffoni, L.; Gaggìa, F.; Di Gioia, D. Therapeutic Microbiology: The Role of Bifidobacterium breve as Food Supplement for the Prevention/Treatment of Paediatric Diseases. Nutrients 2018, 10, 1723. [Google Scholar] [CrossRef] [PubMed]
- Park, M.S.; Kwon, B.; Ku, S.; Ji, G.E. The efficacy of Bifidobacterium longum BORI and Lactobacillus acidophilus AD031 probiotic treatment in infants with rotavirus infection. Nutrients 2017, 9, 887. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Ma, Y.; Liu, X.; Tian, C.; Zhong, X.; Zhao, L. A systematic review and meta-analysis: Lactobacillus acidophilus for treating acute gastroenteritis in children. Nutrients 2022, 14, 682. [Google Scholar] [CrossRef] [PubMed]
| Variable | Category | RVGE (n = 120) | Healthy (n = 45) | Effect Size (95% CI) | Test Statistic | p-Value |
|---|---|---|---|---|---|---|
| Demographic Factors | ||||||
| Sex, n (%) | Male | 45 (37.5%) | 21 (46.7%) | OR = 0.68 (0.34–1.35) | χ2 = 1.12 | 0.29 |
| Female | 75 (62.5%) | 24 (53.3%) | ||||
| Age (months) | Mean ± SD | 5.8 ± 3.2 | 6.9 ± 3.8 | MD = −1.1 (−2.3–0.1) | t = −1.82 | 0.071 |
| Residence, n (%) | Rural | 48 (40.0%) | 22 (48.9%) | OR = 0.70 (0.35–1.39) | χ2 = 1.06 | 0.30 |
| Urban | 72 (60.0%) | 23 (51.1%) | ||||
| Feeding Mode, n (%) | Formula | 56 (46.7%) | 9 (20.0%) | -- | χ2 = 9.87 | 0.002 |
| Breast | 58 (48.3%) | 31 (68.9%) | ||||
| Mixed | 6 (5.0%) | 5 (11.1%) | ||||
| Delivery Mode, n (%) | Cesarean | 85 (70.8%) | 34 (75.6%) | OR = 0.79 (0.36–1.75) | χ2 = 0.31 | 0.58 |
| Vaginal | 35 (29.2%) | 11 (24.4%) | ||||
| Clinical and Laboratory Parameters | ||||||
| WBC (×103/µL) | Mean ± SD | 11.2 ± 3.1 | 6.1 ± 2.0 | MD = 5.1 (4.2–6.0) | t = 11.2 | <0.001 |
| Hemoglobin (g/dL) | Mean ± SD | 11.0 ± 1.3 | 12.3 ± 0.8 | MD = −1.3 (−1.7–−0.9) | t = −6.87 | <0.001 |
| Platelets (×103/µL) | Mean ± SD | 510 ± 98 | 425 ± 50 | MD = 85 (56–114) | t = 5.78 | <0.001 |
| CRP (mg/L) | Median [IQR] | 1.6 [0.9–2.2] | 0.5 [0.3–0.7] | -- | U = 985.5 | <0.001 |
| Dehydration Status, n (%) | None | 36 (30.0%) | 45 (100%) | -- | χ2 = 72.0 | <0.001 |
| Moderate | 76 (63.3%) | 0 (0%) | ||||
| Severe | 8 (6.7%) | 0 (0%) | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdelbary, E.R.; Ramadan, M.; Amin, I.A.; Abd-Elsamea, F.S.; Elsaghier, A.M.; Abd-Alrahman, E.A.; Ozbak, H.A.; Hemeg, H.A.; Almutawif, Y.A.; Zaki, S.A.; et al. Early-Life Demographic Factors Shape Gut Microbiome Patterns Associated with Rotavirus Gastroenteritis Severity. Viruses 2025, 17, 1542. https://doi.org/10.3390/v17121542
Abdelbary ER, Ramadan M, Amin IA, Abd-Elsamea FS, Elsaghier AM, Abd-Alrahman EA, Ozbak HA, Hemeg HA, Almutawif YA, Zaki SA, et al. Early-Life Demographic Factors Shape Gut Microbiome Patterns Associated with Rotavirus Gastroenteritis Severity. Viruses. 2025; 17(12):1542. https://doi.org/10.3390/v17121542
Chicago/Turabian StyleAbdelbary, Eman R., Mohammed Ramadan, Ibrahim A. Amin, Fatma S. Abd-Elsamea, Ashraf Mohamed Elsaghier, Eman Ahmed Abd-Alrahman, Hani A. Ozbak, Hassan A. Hemeg, Yahya A. Almutawif, Shadi A. Zaki, and et al. 2025. "Early-Life Demographic Factors Shape Gut Microbiome Patterns Associated with Rotavirus Gastroenteritis Severity" Viruses 17, no. 12: 1542. https://doi.org/10.3390/v17121542
APA StyleAbdelbary, E. R., Ramadan, M., Amin, I. A., Abd-Elsamea, F. S., Elsaghier, A. M., Abd-Alrahman, E. A., Ozbak, H. A., Hemeg, H. A., Almutawif, Y. A., Zaki, S. A., Abdelrahman, A. A., & Salah, M. (2025). Early-Life Demographic Factors Shape Gut Microbiome Patterns Associated with Rotavirus Gastroenteritis Severity. Viruses, 17(12), 1542. https://doi.org/10.3390/v17121542






