Isolation and Molecular Evidence of Tunisian Sheep-like Pestivirus (Pestivirus N) in Persistently Infected Sheep in Northern Italy, 2023
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
- -
- In November 2023, two blood samples were collected after the mountain pasture, respectively, with (i.e., ethylenediaminetetraacetic acid—EDTA anticoagulated) and without anticoagulated factors.
- -
- In January 2024, the ID77 sheep delivered at full term to a single, live, viable female lamb, which showed no clinical signs of any small ruminant disease. One week after delivery, a third sampling was performed, including two blood samples with and without anticoagulant, milk, nasal and rectal swabs of the sheep, and nasal and rectal swabs of the lamb (ID01).
2.2. Serological Analysis
2.3. Virological Analysis
2.4. Viral RNA Extraction and Real-Time RT-PCR for Pestiviruses Screening
2.5. PCR Amplification and Sequencing
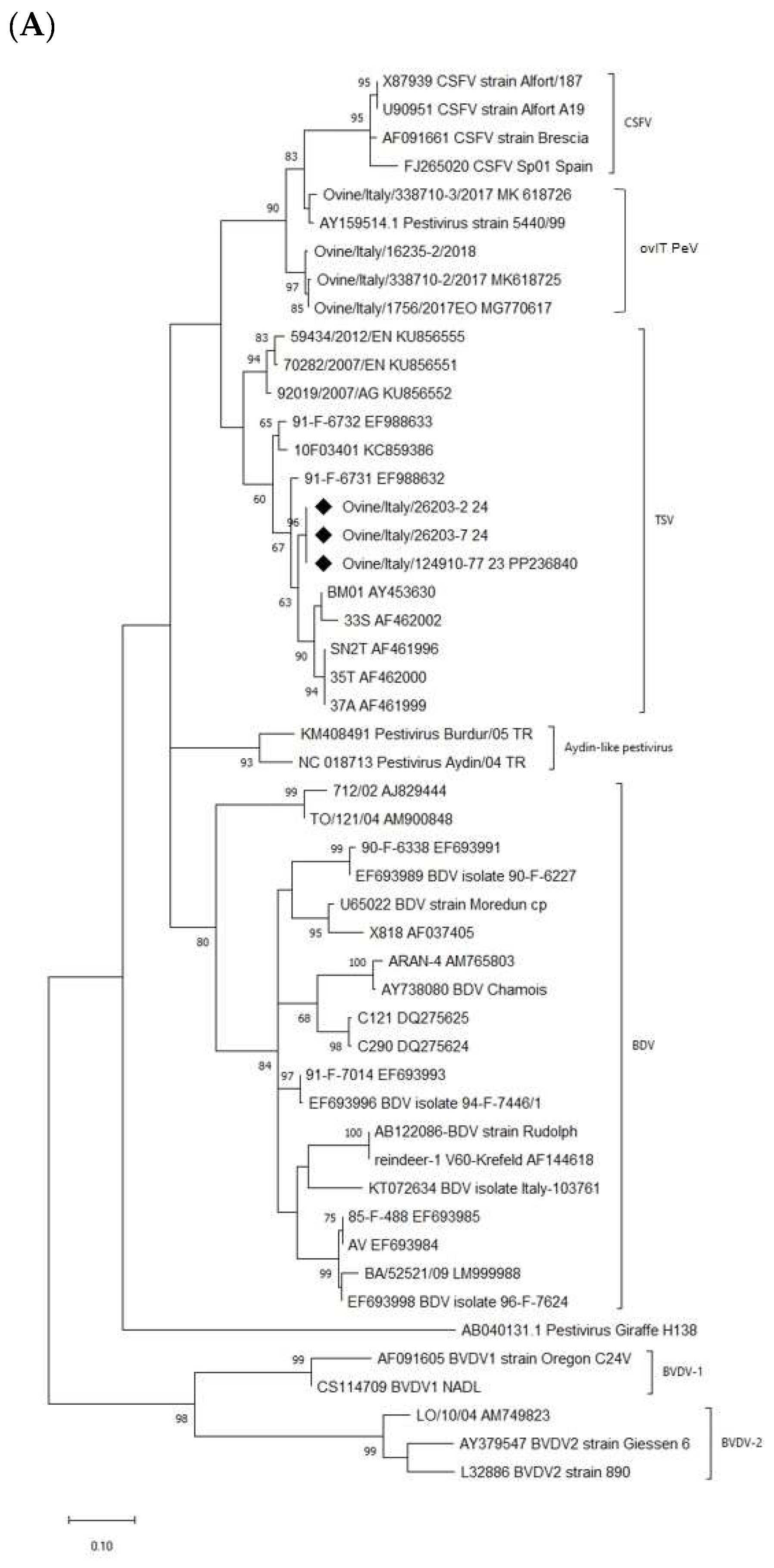
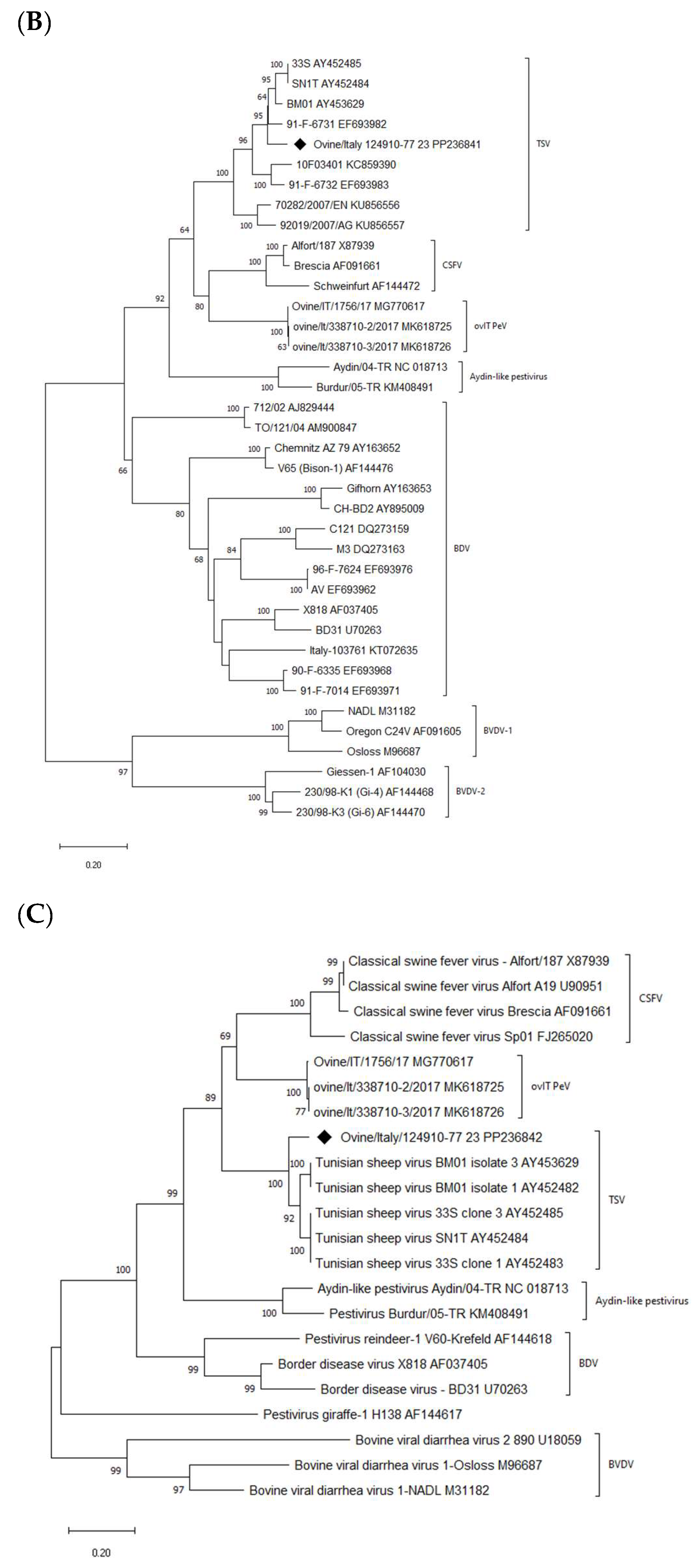
2.6. Phylogenetic Analysis
2.7. Nucleotide Sequence Accession Numbers
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Houe, H. Economic impact of BVDV infection in dairies. Biologicals 2003, 31, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Moennig, V.; Becher, P. Pestivirus control programs: How far have we come and where are we going? Anim. Health Res. Rev. 2015, 16, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Meyers, G.; Bukh, J.; Gould, E.A.; Monath, T.; Scott Muerhoff, A.; Pletnev, A.; Rico-Hesse, R.; Stapleton, J.T.; Simmonds, P.; et al. Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae. J. Gen. Virol. 2017, 98, 2106–2112. [Google Scholar] [CrossRef] [PubMed]
- Postel, A.; Smith, D.B.; Becher, P. Proposed Update to the Taxonomy of Pestiviruses: Eight Additional Species within the Genus Pestivirus, Family Flaviviridae. Viruses 2021, 13, 1542. [Google Scholar] [CrossRef] [PubMed]
- ICTV Report. Flaviviridae, Genus: Pestivirus. Available online: https://ictv.global/taxonomy/taxondetails?taxnode_id=202203153&taxon_name=Pestivirus%20bovis (accessed on 20 April 2024).
- Thabti, F.; Fronzaroli, L.; Dlissi, E.; Guibert, J.-M.; Hammami, S.; Pepin, M.; Russo, P. Experimental model of border disease virus infection in lambs: Comparative pathogenicity of pestiviruses isolated in France and Tunisia. Vet. Res. 2002, 33, 35–45. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thabti, F.; Letellier, C.; Hammami, S.; Pepin, M.; Ribiere, M.; Mesplede, A.; Kerkhofs, P.; Russo, P. Detection of a novel border disease virus subgroup in Tunisian sheep. Arch. Virol. 2004, 150, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.; Postel, A.; Wiedemann, A.; Nur Cagatay, G.; Ciulli, S.; Guercio, A.; Becher, P. Comparative Analysis of Tunisian Sheep-like Virus, Bungowannah Virus and Border Disease Virus Infection in the Porcine Host. Viruses 2021, 13, 1539. [Google Scholar] [CrossRef]
- Dubois, E.; Russo, P.; Prigent, M.; Thiéry, R. Genetic characterization of ovine pestiviruses isolated in France, between 1985 and 2006. Vet. Microbiol. 2008, 130, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Ciulli, S.; Purpari, G.; Agnello, S.; Di Marco, P.; Di Bella, S.; Volpe, E.; Mira, F.; de Aguiar Saldanha Pinheiro, A.C.; Vullo, S.; Guercio, A. Evidence for Tunisian-like pestiviruses presence in small ruminants in Italy since 2007. Transbound. Emerg. Dis. 2016, 64, 1243–1253. [Google Scholar] [CrossRef]
- Postel, A.; Schmeiser, S.; Oguzoglu, T.C.; Indenbirken, D.; Alawi, M.; Fischer, N.; Grundhoff, A.; Becher, P. Close relationship of ruminant pestiviruses and classical Swine Fever virus. Emerg. Infect. Dis. 2015, 21, 668–672. [Google Scholar] [CrossRef]
- Bohórquez, J.A.; Sozzi, E.; Wang, M.; Alberch, M.; Abad, X.; Gaffuri, A.; Lelli, D.; Rosell, R.; Pérez, L.J.; Moreno, A.; et al. The new emerging ovine pestivirus can infect pigs and confers strong protection against classical swine fever virus. Transbound. Emerg. Dis. 2022, 69, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Dlissi, E.; Nettleton, P.F. Border disease and abortion in small ruminants. In Proceedings of the Mondial Vet Lyon’99, 26th World Veterinary Association, Lyon, France, 23–26 September 1999. [Google Scholar]
- Hoffmann, B.; Depner, K.; Schirrmeier, H.; Beer, M. A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J. Virol. Methods 2006, 136, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Sozzi, E.; Righi, C.; Boldini, M.; Bazzucchi, M.; Pezzoni, G.; Gradassi, M.; Petrini, S.; Lelli, D.; Ventura, G.; Pierini, I.; et al. Cross-Reactivity Antibody Response after Vaccination with Modified Live and Killed Bovine Viral Diarrhoea Virus (BVD) Vaccines. Vaccines 2020, 8, 374. [Google Scholar] [CrossRef] [PubMed]
- Rios, L.; Coronado, L.; Naranjo-Feliciano, D.; Martínez-Pérez, O.; Perera, C.L.; Hernandez-Alvarez, L.; Díaz de Arce, H.; Núñez, J.I.; Ganges, L.; Pérez, L.J. Deciphering the emergence, genetic diversity and evolution of classical swine fever virus. Sci. Rep. 2017, 7, 17887. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sozzi, E.; Bohórquez, J.A.; Alberch, M.; Pujols, J.; Cantero, G.; Gaffuri, A.; Lelli, D.; Rosell, R.; Bensaid, A.; et al. Decrypting the Origin and Pathogenesis in Pregnant Ewes of a New Ovine Pestivirus Closely Related to Classical Swine Fever Virus. Viruses 2020, 12, 775. [Google Scholar] [CrossRef] [PubMed]
- Letellier, C.; Kerkhofs, P.; Wellemans, G.; Vanopdenbosch, E. Detection and genotyping of bovine diarrhea virus by reverse transcription-polymerase chain amplification of the 5′ untranslated region. Vet. Microbiol. 1999, 64, 155–167. [Google Scholar] [CrossRef]
- Vilček, Š.; Belák, S. Genetic identification of pestivirus strain Frijters as a border disease virus from pigs. J. Virol. Methods 1996, 60, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Lang, Y.; Gao, S.; Du, J.; Shao, J.; Cong, G.; Lin, T.; Zhao, F.; Liu, L.; Chang, H. Polymorphic genetic characterization of E2 gene of bovine viral diarrhea virus in China. Vet. Microbiol. 2014, 174, 554–559. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kirkland, P.D.; Read, A.J.; Frost, M.J.; Finlaison, D.S. Bungowannah virus a probable new species of pestivirus what have we found in the last 10 years? Anim. Health Res. Rev. 2015, 16, 60–63. [Google Scholar] [CrossRef]
- Ribbens, S.; Dewulf, J.; Koenen, F.; Laevens, H.; De Kruif, A. Transmission of classical swine fever. A review. Vet. Q. 2004, 26, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Houe, H. Epidemiology of Bovine Viral Diarrhea Virus. Vet. Clin. N. Am. Food Anim. Pract. 1995, 11, 521–547. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Duquesne, V.; Adam, G.; Belleau, E.; Gauthier, D.; Champion, J.L.; Saegerman, C.; Thiéry, R.; Dubois, E. Pestiviruses infections at the wild and domestic ruminants interface in the French Southern Alps. Vet. Microbiol. 2015, 175, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Russell, G.C. Pestivirus infection of sheep—Testing times? Vet. Rec. 2018, 183, 217–219. [Google Scholar] [CrossRef]
- Hurtado, A.; García-Pérez, A.L.; Aduriz, G.; Juste, R.A. Genetic diversity of ruminant pestiviruses from Spain. Virus Res. 2003, 92, 67–73. [Google Scholar] [CrossRef]
| PREVIOUS TAXONOMY | NEW TAXONOMY | |||
|---|---|---|---|---|
| Family | Genus | Species | Abbreviation | Species |
| Flaviviridae | Pestivirus | Pestivirus A | BVDV-1 | Pestivirus bovis |
| Flaviviridae | Pestivirus | Pestivirus B | BVDV-2 | Pestivirus tauri |
| Flaviviridae | Pestivirus | Pestivirus C | CSFV | Pestivirus suis |
| Flaviviridae | Pestivirus | Pestivirus D | BDV | Pestivirus ovis |
| Flaviviridae | Pestivirus | Pestivirus E | PHV | Pestivirus antilocaprae |
| Flaviviridae | Pestivirus | Pestivirus F | BuPV | Pestivirus australiaense |
| Flaviviridae | Pestivirus | Pestivirus G | - | Pestivirus giraffae |
| Flaviviridae | Pestivirus | Pestivirus H | HoBi | Pestivirus brazilense |
| Flaviviridae | Pestivirus | Pestivirus I | - | Pestivirus aydinense |
| Flaviviridae | Pestivirus | Pestivirus J | NrPV | Pestivirus ratti |
| Flaviviridae | Pestivirus | Pestivirus K | APPV | Pestivirus scrofae |
| Isolation on Cell Culture | ||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | Host | Specimen | Date of Sampling | Real Time RT-PCR (Ct Value) | MDBK | Primary Bovine Foetal Kidney | Lki | ST |
| 124910/77 | sheep ID77 | serum | April 2023 | 22.29 | no | no | n.d | n.d |
| 373570-1 | sheep ID77 | serum | November 2023 | 35.33 | yes | no | n.d | no |
| 373570-2 | sheep ID77 | EDTA-blood | November 2023 | 35.35 | n.d | n.d | n.d | n.d |
| 26203-1 | sheep ID77 | EDTA-blood | January 2024 | 21.95 | no | no | yes | yes |
| 26203-2 | sheep ID77 | serum | January 2024 | 20.26 | no | no | yes | yes |
| 26203-3 | sheep ID77 | milk | January 2024 | 24.88 | n.d | n.d | n.d | n.d |
| 26203-4 | sheep ID77 | nasal swab | January 2024 | 27.34 | no | no | yes | yes |
| 26203-5 | sheep ID77 | rectal swab | January 2024 | 32.99 | no | no | no | no |
| 26203-6 | lamb ID01 | nasal swab | January 2024 | 25.58 | no | no | no | no |
| 26203-7 | lamb ID01 | rectal swab | January 2024 | 24.19 | no | no | yes | yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sozzi, E.; Leo, G.; Lamcja, F.; Lazzaro, M.; Salogni, C.; Lelli, D.; Bertasio, C.; Magagna, G.; Moreno, A.; Alborali, G.L.; et al. Isolation and Molecular Evidence of Tunisian Sheep-like Pestivirus (Pestivirus N) in Persistently Infected Sheep in Northern Italy, 2023. Viruses 2024, 16, 815. https://doi.org/10.3390/v16060815
Sozzi E, Leo G, Lamcja F, Lazzaro M, Salogni C, Lelli D, Bertasio C, Magagna G, Moreno A, Alborali GL, et al. Isolation and Molecular Evidence of Tunisian Sheep-like Pestivirus (Pestivirus N) in Persistently Infected Sheep in Northern Italy, 2023. Viruses. 2024; 16(6):815. https://doi.org/10.3390/v16060815
Chicago/Turabian StyleSozzi, Enrica, Gabriele Leo, Fatbardha Lamcja, Massimiliano Lazzaro, Cristian Salogni, Davide Lelli, Cristina Bertasio, Giulia Magagna, Ana Moreno, Giovanni Loris Alborali, and et al. 2024. "Isolation and Molecular Evidence of Tunisian Sheep-like Pestivirus (Pestivirus N) in Persistently Infected Sheep in Northern Italy, 2023" Viruses 16, no. 6: 815. https://doi.org/10.3390/v16060815
APA StyleSozzi, E., Leo, G., Lamcja, F., Lazzaro, M., Salogni, C., Lelli, D., Bertasio, C., Magagna, G., Moreno, A., Alborali, G. L., Bazzucchi, M., & Lavazza, A. (2024). Isolation and Molecular Evidence of Tunisian Sheep-like Pestivirus (Pestivirus N) in Persistently Infected Sheep in Northern Italy, 2023. Viruses, 16(6), 815. https://doi.org/10.3390/v16060815










