Rotavirus NSP2: A Master Orchestrator of Early Viral Particle Assembly
Abstract
1. Introduction
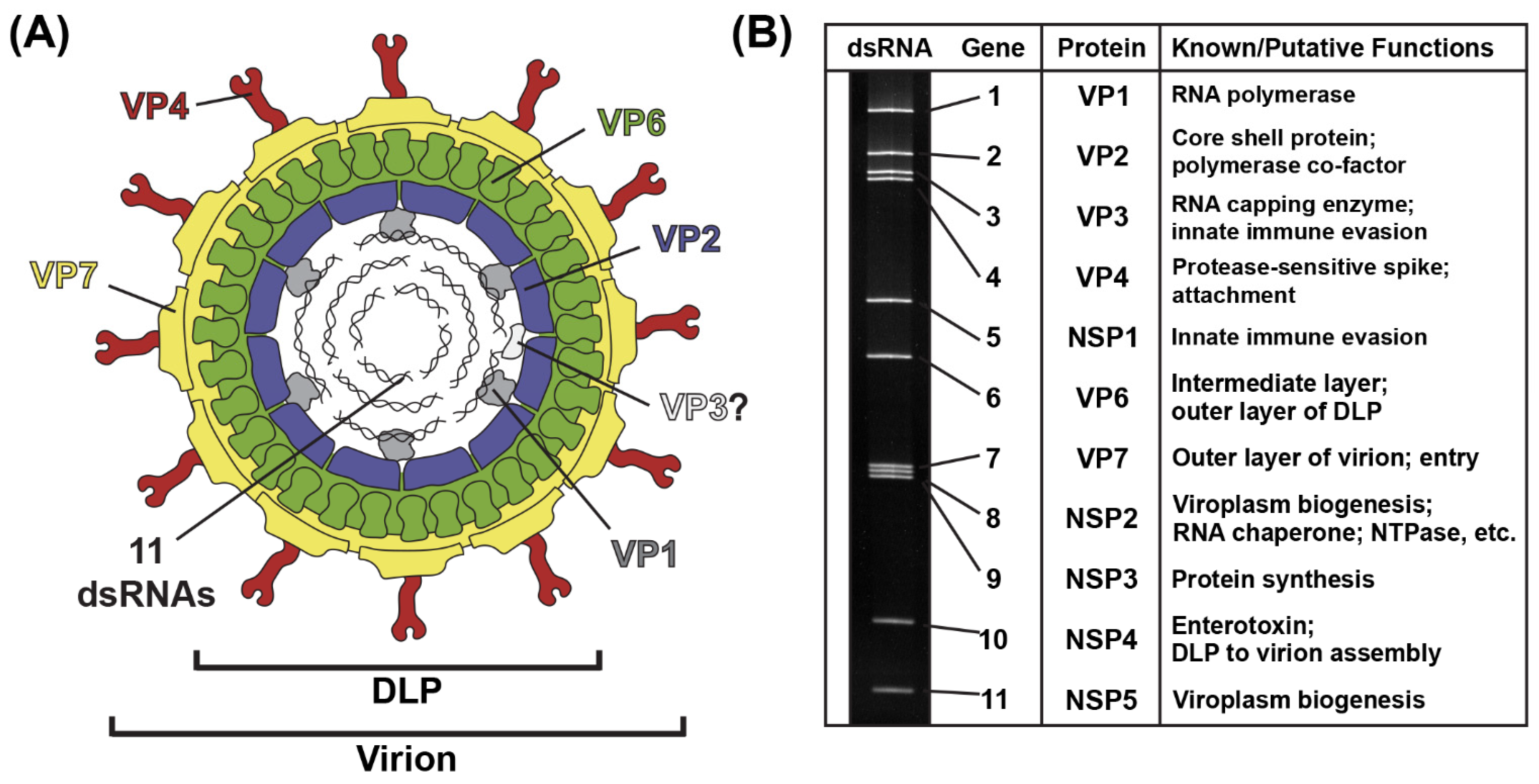
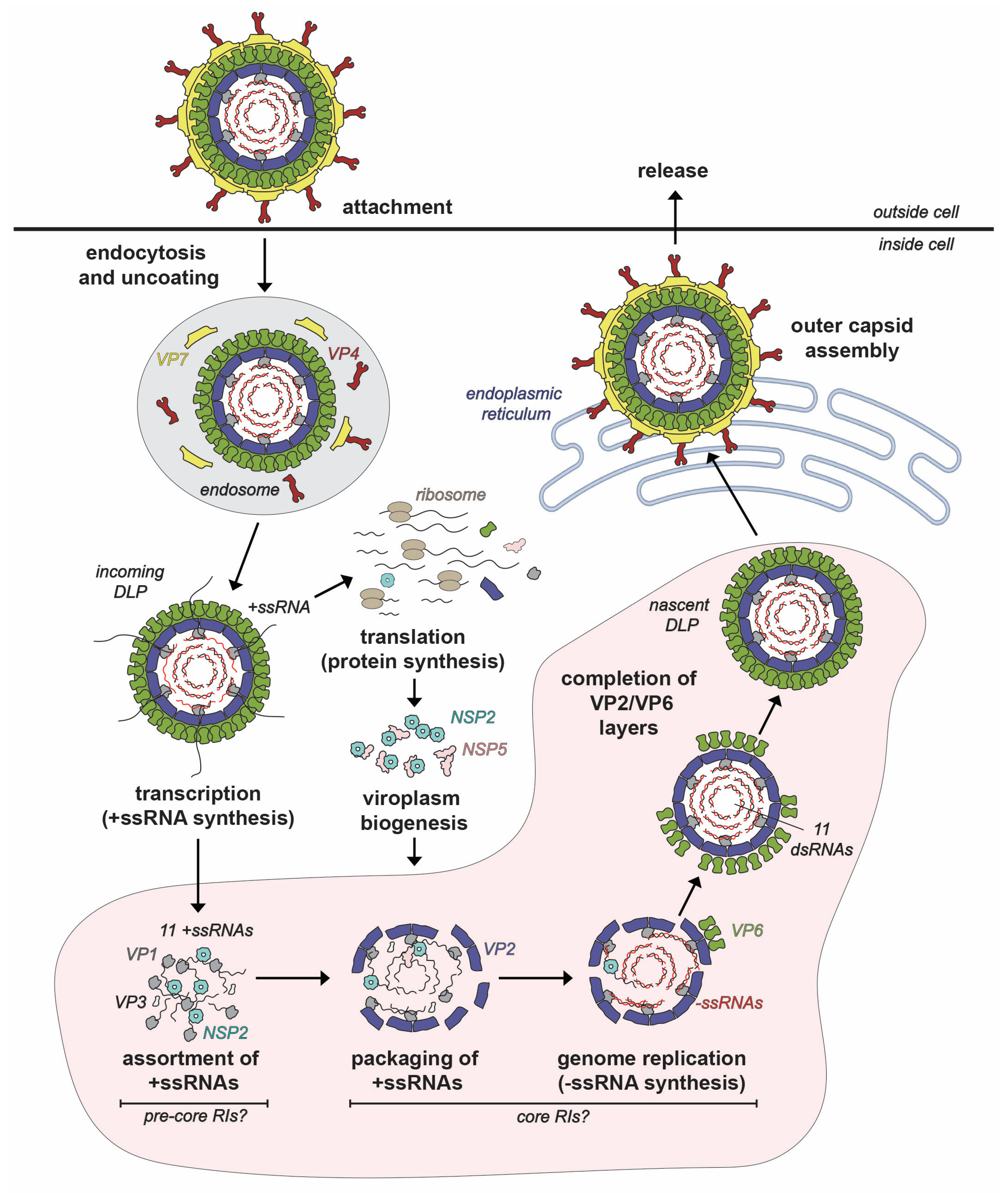
2. Virion, Genome and Replication Cycle
3. Discovery and Early Characterization of NSP2
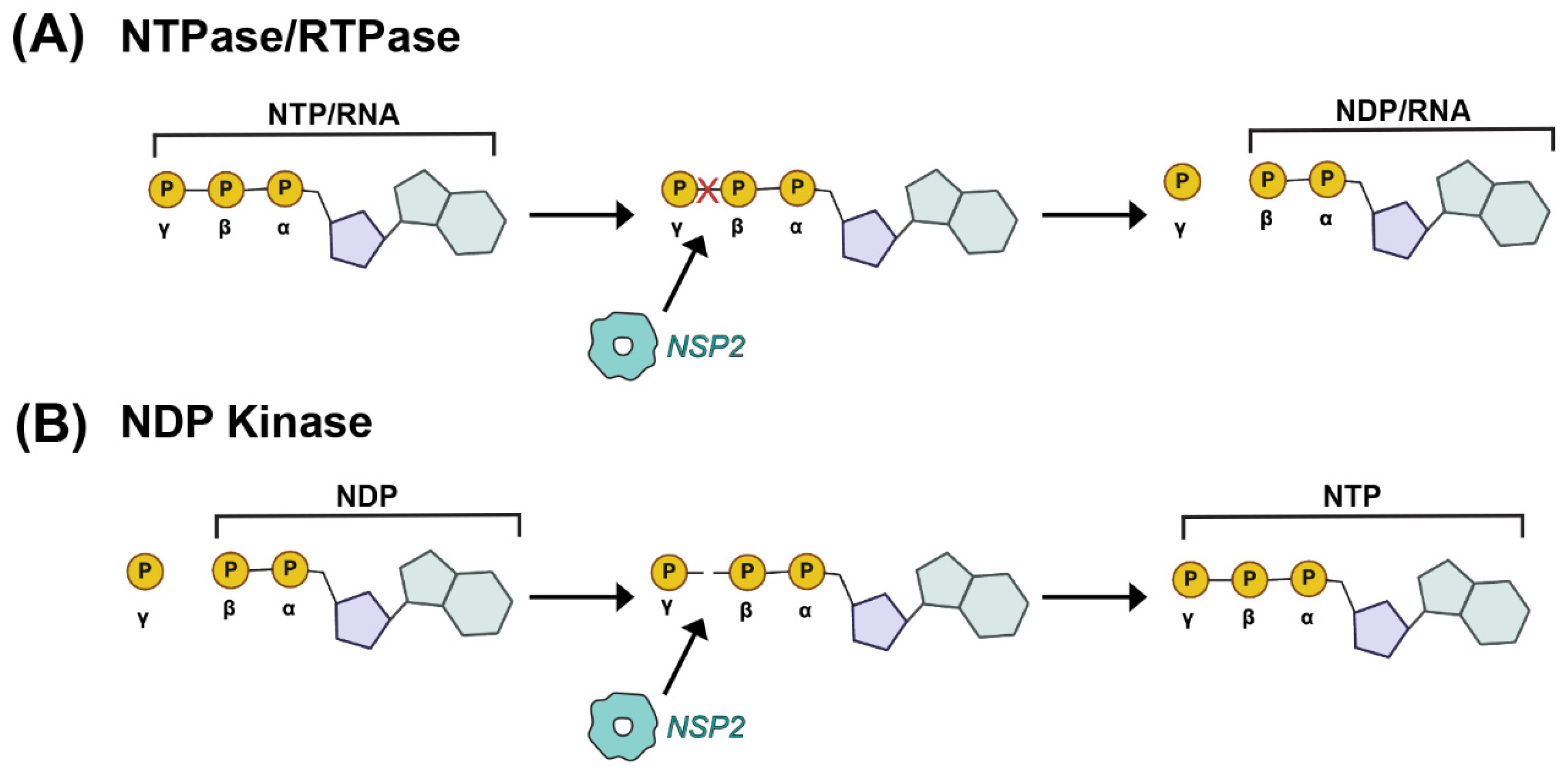
4. Structural and Enzymatic Studies of NSP2
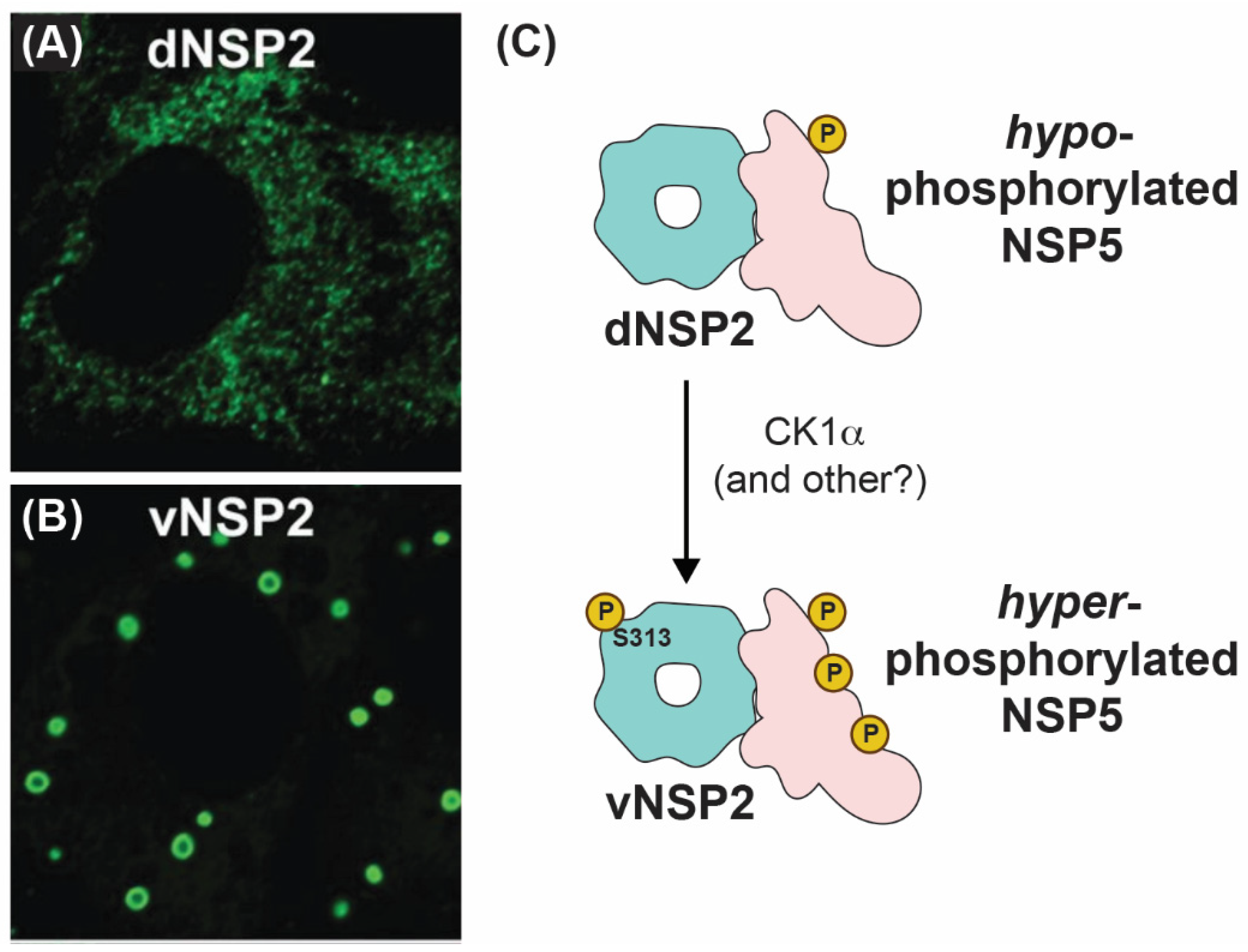
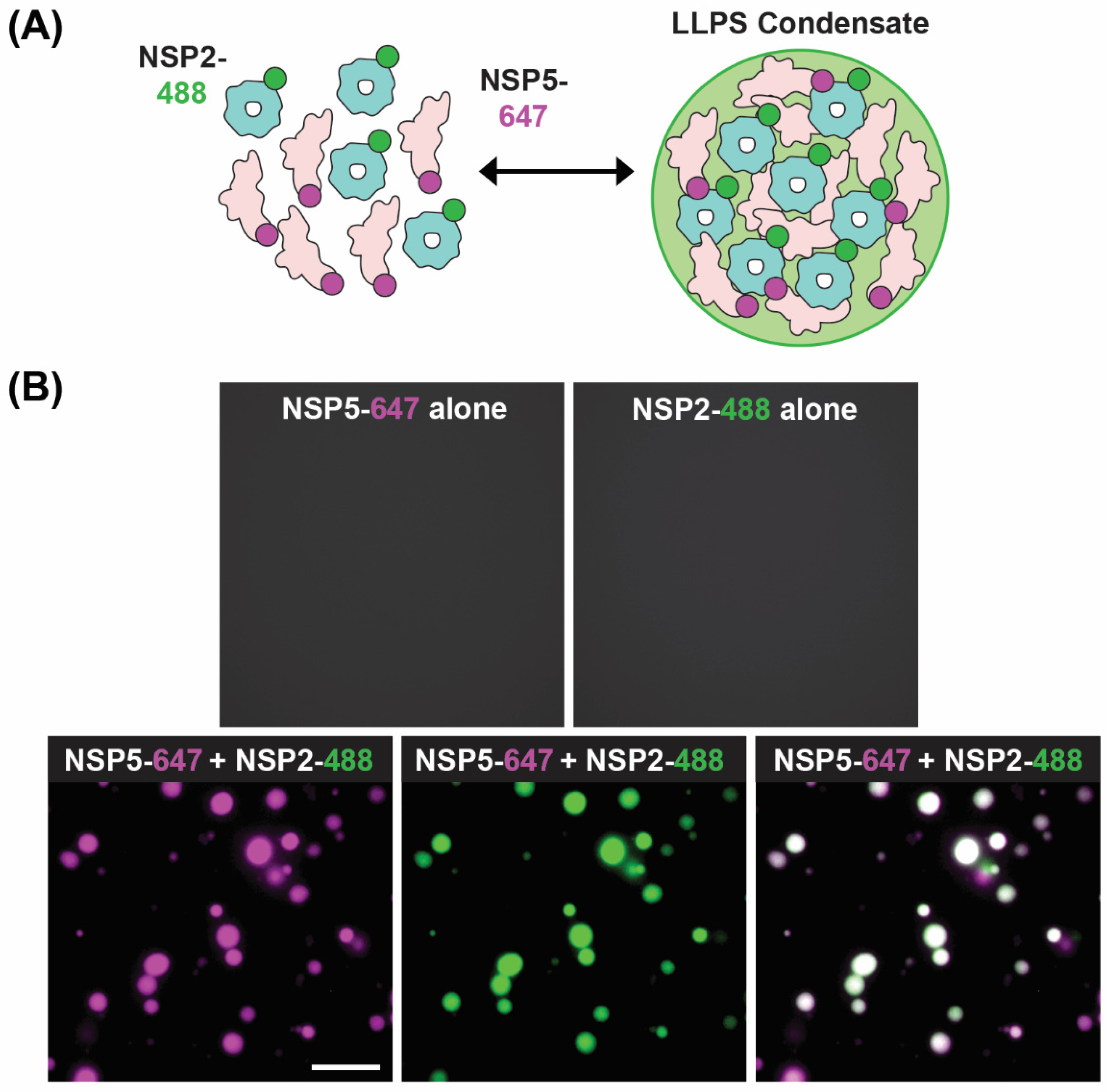
5. Role of NSP2 in Viroplasm Formation
6. Role of NSP2 during +ssRNA Assortment
7. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Matthijnssens, J.; Attoui, H.; Bányai, K.; Brussaard, C.P.; Danthi, P.; Del Vas, M.; Dermody, T.S.; Duncan, R.; Fang, Q.; Johne, R.; et al. ICTV Virus Taxonomy Profile: Sedoreoviridae 2022. J. Gen. Virol. 2022, 103. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Otto, P.H.; Ciarlet, M.; Desselberger, U.; Van Ranst, M.; Johne, R. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch. Virol. 2012, 157, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- Mihalov-Kovács, E.; Gellért, Á.; Marton, S.; Farkas, S.L.; Fehér, E.; Oldal, M.; Jakab, F.; Martella, V.; Bányai, K. Candidate new rotavirus species in sheltered dogs, hungary. Emerg. Infect. Dis. 2015, 21, 660–663. [Google Scholar] [CrossRef] [PubMed]
- Troeger, C.; Khalil, I.A.; Rao, P.C.; Cao, S.; Blacker, B.F.; Ahmed, T.; Armah, G.; Bines, J.E.; Brewer, T.G.; Colombara, D.V.; et al. Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea Among Children Younger Than 5 Years. JAMA Pediatr. 2018, 172, 958–965. [Google Scholar] [CrossRef]
- Díaz Alarcón, R.G.; Liotta, D.J.; Miño, S. Zoonotic RVA: State of the Art and Distribution in the Animal World. Viruses 2022, 14, 2554. [Google Scholar] [CrossRef]
- Arnold, M.; Patton, J.T.; McDonald, S.M. Culturing, storage, and quantification of rotaviruses. Curr. Protoc. Microbiol. 2009, 15. [Google Scholar] [CrossRef]
- Settembre, E.C.; Chen, J.Z.; Dormitzer, P.R.; Grigorieff, N.; Harrison, S.C. Atomic model of an infectious rotavirus particle. EMBO J. 2011, 30, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Jenni, S.; Salgado, E.N.; Herrmann, T.; Li, Z.; Grant, T.; Grigorieff, N.; Trapani, S.; Estrozi, L.F.; Harrison, S.C. In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase. J. Mol. Biol. 2019, 431, 3124–3138. [Google Scholar] [CrossRef]
- Dormitzer, P.R.; Greenberg, H.B.; Harrison, S.C. Proteolysis of monomeric recombinant rotavirus VP4 yields an oligomeric VP5* core. J. Virol. 2001, 75, 7339–7350. [Google Scholar] [CrossRef]
- Kumar, D.; Yu, X.; Crawford, S.E.; Moreno, R.; Jakana, J.; Sankaran, B.; Anish, R.; Kaundal, S.; Hu, L.; Estes, M.K.; et al. 2.7 A cryo-EM structure of rotavirus core protein VP3, a unique capping machine with a helicase activity. Sci. Adv. 2020, 6, eaay6410. [Google Scholar] [CrossRef]
- Ding, K.; Celma, C.C.; Zhang, X.; Chang, T.; Shen, W.; Atanasov, I.; Roy, P.; Zhou, Z.H. In situ structures of rotavirus polymerase in action and mechanism of mRNA transcription and release. Nat. Commun. 2019, 10, 2216. [Google Scholar] [CrossRef] [PubMed]
- Estes, M.; Kapikian, A.Z. Rotaviruses and Their Replication. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams and Wilkens: Philadelphia, PA, USA, 2007; pp. 1917–1974. [Google Scholar]
- Davidson, G.P.; Goller, I.; Bishop, R.F.; Townley, R.R.; Holmes, I.H.; Ruck, B.J. Immunofluorescence in duodenal mucosa of children with acute enteritis due to a new virus. J. Clin. Pathol. 1975, 28, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Starkey, W.G.; Collins, J.; Wallis, T.S.; Clarke, G.J.; Spencer, A.J.; Haddon, S.J.; Osborne, M.P.; Candy, D.C.A.; Stephen, J. Kinetics, tissue specificity and pathological changes in murine rotavirus infection of mice. J. Gen. Virol. 1986, 67 Pt 12, 2625–2634. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.F.; López, S. Rotavirus cell entry: Not so simple after all. Curr. Opin. Virol. 2021, 48, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Salinas, M.A.; Romero, P.; Espinosa, R.; Hoshino, Y.; López, S.; Arias, C.F. The Spike Protein VP4 Defines the Endocytic Pathway Used by Rotavirus To Enter MA104 Cells. J. Virol. 2013, 87, 1658–1663. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, M.; Isa, P.; Martin, C.S.-S.; Pérez-Vargas, J.; Espinosa, R.; Arias, C.F.; López, S. Different Rotavirus Strains Enter MA104 Cells through Different Endocytic Pathways: The Role of Clathrin-Mediated Endocytosis. J. Virol. 2010, 84, 9161–9169. [Google Scholar] [CrossRef] [PubMed]
- Trask, S.D.; McDonald, S.M.; Patton, J.T. Structural insights into the coupling of virion assembly and rotavirus replication. Nat. Rev. Microbiol. 2012, 10, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Salgado, E.N.; Rodriguez, B.G.; Narayanaswamy, N.; Krishnan, Y.; Harrison, S.C. Visualization of Calcium Ion Loss from Rotavirus during Cell Entry. J. Virol. 2018, 92, e01327-18. [Google Scholar] [CrossRef] [PubMed]
- Ludert, J.; Michelangeli, F.; Gil, F.; Liprandi, F.; Esparza, J. Penetration and uncoating of rotaviruses in cultured cells. Intervirology 1987, 27, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Lawton, J.A.; Estes, M.K.; Prasad, B.V. Mechanism of genome transcription in segmented dsRNA viruses. Adv. Virus Res. 2000, 55, 185–229. [Google Scholar]
- Patton, J.T.; Chen, D. RNA-binding and capping activities of proteins in rotavirus open cores. J. Virol. 1999, 73, 1382–1391. [Google Scholar] [CrossRef]
- Liu, M.; Mattion, N.M.; Estes, M.K. Rotavirus VP3 expressed in insect cells possesses guanylyltrans-ferase activity. Virology 1992, 188, 77–84. [Google Scholar] [CrossRef]
- Pizarro, J.L.; Sandino, A.M.; Pizarro, J.M.; Fernandez, J.; Spencer, E. Characterization of rotavirus guan-ylyltransferase activity associated with polypeptide VP3. J. Gen. Virol. 1991, 72 Pt 2, 325–332. [Google Scholar] [CrossRef]
- Vende, P.; Piron, M.; Castagne, N.; Poncet, D. Efficient translation of rotavirus mRNA requires simul-taneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J. Virol. 2000, 74, 7064–7071. [Google Scholar] [CrossRef]
- López, S.; Oceguera, A.; Sandoval-Jaime, C. Stress Response and Translation Control in Rotavirus Infection. Viruses 2016, 8, 162. [Google Scholar] [CrossRef] [PubMed]
- Mitzel, D.N.; Weisend, C.M.; White, M.W.; Hardy, M.E. Translational regulation of rotavirus gene ex-pression. J. Gen. Virol. 2003, 84, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Papa, G.; Borodavka, A.; Desselberger, U. Viroplasms: Assembly and Functions of Rotavirus Replication Factories. Viruses 2021, 13, 1349. [Google Scholar] [CrossRef] [PubMed]
- Patton, J.T.; Silvestri, L.S.; Tortorici, M.A.; Vasquez-Del Carpio, R.; Taraporewala, Z.F. Rotavirus genome replication and morphogenesis: Role of the viroplasm. Curr. Top. Microbiol. Immunol. 2006, 309, 169–187. [Google Scholar] [PubMed]
- Taraporewala, Z.F.; Patton, J.T. Nonstructural proteins involved in genome packaging and replication of rotaviruses and other members of the Reoviridae. Virus Res. 2004, 101, 57–66. [Google Scholar] [CrossRef] [PubMed]
- McNulty, M.S.; Curran, W.L.; McFerran, J.B. The morphogenesis of a cytopathic bovine rotavirus in madin-darby bovine kidney cells. J. Gen. Virol. 1976, 33, 503–508. [Google Scholar] [CrossRef]
- Gardet, A.; Breton, M.; Fontanges, P.; Trugnan, G.; Chwetzoff, S. Rotavirus spike protein vp4 binds to and remodels actin bundles of the epithelial brush border into actin bodies. J. Virol. 2006, 80, 3947–3956. [Google Scholar] [CrossRef]
- Ericson, B.L.; Graham, D.Y.; Mason, B.B.; Estes, M.K. Identification, synthesis, and modifications of simian rotavirus SA11 polypeptides in infected cells. J. Virol. 1982, 42, 825–839. [Google Scholar] [CrossRef] [PubMed]
- Mason, B.B.; Graham, D.Y.; Estes, M.K. Biochemical mapping of the simian rotavirus SA11 genome. J. Virol. 1983, 46, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Both, G.W.; Bellamy, A.R.; Street, J.E.; Siegman, L.J. A general strategy for cloning double-stranded RNA: Nucleotide sequence of the Simian-11 rotavirus gene 8. Nucleic Acids Res. 1982, 10, 7075–7088. [Google Scholar] [CrossRef] [PubMed]
- Petrie, B.L.; Greenberg, H.B.; Graham, D.Y.; Estes, M.K. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res. 1984, 1, 133–152. [Google Scholar] [CrossRef] [PubMed]
- Patton, J.T.; Gallegos, C.O. Structure and protein composition of the rotavirus replicase particle. Virology 1988, 166, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Gallegos, C.O.; Patton, J.T. Characterization of rotavirus replication intermediates: A model for the assembly of single-shelled particles. Virology 1989, 172, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Aponte, C.; Poncet, D.; Cohen, J. Recovery and characterization of a replicase complex in rota-virus-infected cells by using a monoclonal antibody against NSP2. J. Virol. 1996, 70, 985–991. [Google Scholar] [CrossRef]
- Helmberger-Jones, M.; Patton, J.T. Characterization of subviral particles in cells infected with simian rotavirus SA11. Virology 1986, 155, 655–665. [Google Scholar] [CrossRef]
- McDonald, S.M.; Patton, J.T. Assortment and packaging of the segmented rotavirus genome. Trends Microbiol. 2011, 19, 136–144. [Google Scholar] [CrossRef]
- Mansell, E.A.; Patton, J.T. Rotavirus RNA replication: VP2, but not VP6, is necessary for viral replicase activity. J. Virol. 1990, 64, 4988–4996. [Google Scholar] [CrossRef] [PubMed]
- Patton, J.T. Rotavirus VP1 alone specifically binds to the 3′ end of viral mRNA, but the interaction is not sufficient to initiate minus-strand synthesis. J. Virol. 1996, 70, 7940–7947. [Google Scholar] [CrossRef] [PubMed]
- Patton, J.T.; Jones, M.T.; Kalbach, A.N.; He, Y.W.; Xiaobo, J. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J. Virol. 1997, 71, 9618–9626. [Google Scholar] [CrossRef] [PubMed]
- McDonald, S.M.; Patton, J.T. Rotavirus VP2 core shell regions critical for viral polymerase activation. J. Virol. 2011, 85, 3095–3105. [Google Scholar] [CrossRef] [PubMed]
- Long, C.P.; McDonald, S.M. Rotavirus genome replication: Some assembly required. PLoS Pathog. 2017, 13, e1006242. [Google Scholar] [CrossRef]
- Gombold, J.L.; Estes, M.K.; Ramig, R.F. Assignment of simian rotavirus SA11 temperature-sensitive mutant groups B and E to genome segments. Virology 1985, 143, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Ramig, R.F. Isolation and genetic characterization of temperature-sensitive mutants that define five additional recombination groups in simian rotavirus SA11. Virology 1983, 130, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Ramig, R.F.; Petrie, B.L. Characterization of temperature-sensitive mutants of simian rotavirus SA11: Protein synthesis and morphogenesis. J. Virol. 1984, 49, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Gombold, J.L.; Ramig, R.F. Intracellular RNA synthesis directed by temperature-sensitive mutants of simian rotavirus SA11. Virology 1990, 178, 143–151. [Google Scholar] [CrossRef]
- Kattoura, M.D.; Clapp, L.L.; Patton, J.T. The rotavirus nonstructural protein, NS35, possesses RNA-binding activity in vitro and in vivo. Virology 1992, 191, 698–708. [Google Scholar] [CrossRef]
- Kattoura, M.D.; Chen, X.; Patton, J.T. The rotavirus RNA-binding protein NS35 (NSP2) forms 10S multimers and interacts with the viral rna polymerase. Virology 1994, 202, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Taraporewala, Z.; Chen, D.; Patton, J.T. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J. Virol. 1999, 73, 9934–9943. [Google Scholar] [CrossRef]
- Taraporewala, Z.F.; Schuck, P.; Ramig, R.F.; Silvestri, L.; Patton, J.T. Analysis of a Temperature-sensitive mutant rotavirus indicates that NSP2 octamers are the functional form of the protein. J. Virol. 2002, 76, 7082–7093. [Google Scholar] [CrossRef] [PubMed]
- Schuck, P.; Taraporewala, Z.; McPhie, P.; Patton, J.T. Rotavirus nonstructural protein NSP2 self-assembles into octamers that undergo ligand-induced conformational changes. J. Biol. Chem. 2001, 276, 9679–9687. [Google Scholar] [CrossRef]
- Taraporewala, Z.F.; Patton, J.T. Identification and characterization of the Helix-destabilizing activity of rotavirus nonstructural protein NSP2. J. Virol. 2001, 75, 4519–4527. [Google Scholar] [CrossRef]
- Afrikanova, I.; Fabbretti, E.; Burrone, O.R.; Miozzo, M.C. Rotavirus NSP5 phosphorylation is up-regulated by interaction with NSP2. J. Gen. Virol. 1998, 79, 2679–2686. [Google Scholar] [CrossRef] [PubMed]
- Fabbretti, E.; Afrikanova, I.; Vascotto, F.; Burrone, O.R. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 1999, 80, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Jayaram, H.; Taraporewala, Z.; Patton, J.T.; Prasad, B.V.V. Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold. Nature 2002, 417, 311–315. [Google Scholar] [CrossRef]
- Jiang, X.; Jayaram, H.; Kumar, M.; Ludtke, S.J.; Estes, M.K.; Prasad, B.V.V. Cryoelectron microscopy structures of rotavirus NSP2-NSP5 and NSP2-RNA complexes: Implications for genome replication. J. Virol. 2006, 80, 10829–10835. [Google Scholar] [CrossRef]
- Kumar, M.; Jayaram, H.; Vasquez-Del Carpio, R.; Jiang, X.; Taraporewala, Z.F.; Jacobson, R.H.; Patton, J.T.; Prasad, B.V. Crystallographic and biochemical analysis of rotavirus NSP2 with nucleotides reveals a nucleoside diphosphate kinase-like activity. J. Virol. 2007, 81, 12272–12284. [Google Scholar] [CrossRef]
- Hu, L.; Chow, D.-C.; Patton, J.T.; Palzkill, T.; Estes, M.K.; Prasad, B.V.V. Crystallographic Analysis of Rotavirus NSP2-RNA Complex Reveals Specific Recognition of 5′ GG Sequence for RTPase Activity. J. Virol. 2012, 86, 10547–10557. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Criglar, J.M.; Anish, R.; Hu, L.; Crawford, S.E.; Sankaran, B.; Prasad, B.V.V.; Estes, M.K. Phosphorylation cascade regulates the formation and maturation of rotaviral replication factories. Proc. Natl. Acad. Sci. USA 2018, 115, E12015–E12023. [Google Scholar] [CrossRef] [PubMed]
- Bravo, J.P.K.; Bartnik, K.; Venditti, L.; Acker, J.; Gail, E.H.; Colyer, A.; Davidovich, C.; Lamb, D.C.; Tuma, R.; Calabrese, A.N.; et al. Structural basis of rotavirus RNA chaperone displacement and RNA annealing. Proc. Natl. Acad. Sci. USA 2021, 118, e2100198118. [Google Scholar] [CrossRef] [PubMed]
- Carpio, R.V.-D.; Gonzalez-Nilo, F.D.; Riadi, G.; Taraporewala, Z.F.; Patton, J.T. Histidine triad-like motif of the rotavirus NSP2 Octamer mediates both RTPase and NTPase activities. J. Mol. Biol. 2006, 362, 539–554. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.; Duarte, M.; Lepault, J.; Poncet, D. Sequestration of free tubulin molecules by the viral protein NSP2 induces microtubule depolymerization during rotavirus infection. J. Virol. 2010, 84, 2522–2532. [Google Scholar] [CrossRef] [PubMed]
- Eichwald, C.; Rodriguez, J.F.; Burrone, O.R. Characterization of rotavirus NSP2/NSP5 interactions and the dynamics of viroplasm formation. J. Gen. Virol. 2004, 85, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Carreño-Torres, J.J.; Gutiérrez, M.; Arias, C.F.; López, S.; Isa, P. Characterization of viroplasm formation during the early stages of rotavirus infection. Virol. J. 2010, 7, 350. [Google Scholar] [CrossRef]
- Eichwald, C.; Arnoldi, F.; Laimbacher, A.S.; Schraner, E.M.; Fraefel, C.; Wild, P.; Burrone, O.R.; Ackermann, M. Rotavirus viroplasm fusion and perinuclear localization are dynamic processes requiring stabilized microtubules. PLoS ONE 2012, 7, e47947. [Google Scholar] [CrossRef] [PubMed]
- Nichols, S.L.; Nilsson, E.M.; Brown-Harding, H.; LaConte, L.E.W.; Acker, J.; Borodavka, A.; Esstman, S.M. Flexibility of the Rotavirus NSP2 C-Terminal Region Supports Factory Formation via Liquid-Liquid Phase Separation. J. Virol. 2023, 97, e0003923. [Google Scholar] [CrossRef]
- Geiger, F.; Acker, J.; Papa, G.; Wang, X.; Arter, W.E.; Saar, K.L.; Erkamp, N.A.; Qi, R.; Bravo, J.P.; Strauss, S.; et al. Liquid-liquid phase separation underpins the formation of replication factories in rotaviruses. EMBO J. 2021, 40, e107711. [Google Scholar] [CrossRef]
- Strauss, S.; Acker, J.; Papa, G.; Desiro, D.; Schueder, F.; Borodavka, A.; Jungmann, R. Principles of RNA recruitment to viral ribonucleoprotein condensates in a segmented dsRNA virus. eLife 2023, 12, e68670. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, P.; Tandra, V.N.; Chorghade, S.G.; Namsa, N.D.; Sahoo, L.; Rao, C.D. Cytoplasmic Relocalization and Colocalization with Viroplasms of Host Cell Proteins, and Their Role in Rotavirus Infection. J. Virol. 2018, 92, e00612-18. [Google Scholar] [CrossRef] [PubMed]
- López, T.; Rojas, M.; Ayala-Bretón, C.; López, S.; Arias, C.F. Reduced expression of the rotavirus NSP5 gene has a pleiotropic effect on virus replication. J. Gen. Virol. 2005, 86, 1609–1617. [Google Scholar] [CrossRef] [PubMed]
- Silvestri, L.S.; Taraporewala, Z.F.; Patton, J.T. Rotavirus replication: Plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J. Virol. 2004, 78, 7763–7774. [Google Scholar] [CrossRef]
- Carpio, R.V.-D.; González-Nilo, F.D.; Jayaram, H.; Spencer, E.; Prasad, B.V.V.; Patton, J.T.; Taraporewala, Z.F. Role of the histidine triad-like motif in nucleotide hydrolysis by the rotavirus RNA-packaging protein NSP2. J. Biol. Chem. 2004, 279, 10624–10633. [Google Scholar] [CrossRef] [PubMed]
- Taraporewala, Z.F.; Jiang, X.; Vasquez-Del Carpio, R.; Jayaram, H.; Prasad, B.V.; Patton, J.T. Structure-function analysis of rotavirus NSP2 octamer by using a novel complementation system. J. Virol. 2006, 80, 7984–7994. [Google Scholar] [CrossRef]
- Criglar, J.M.; Hu, L.; Crawford, S.E.; Hyser, J.M.; Broughman, J.R.; Prasad, B.V.V.; Estes, M.K. A novel form of rotavirus NSP2 and phosphorylation-dependent NSP2-NSP5 interactions are associated with viroplasm assembly. J. Virol. 2014, 88, 786–798. [Google Scholar] [CrossRef] [PubMed]
- Criglar, J.M.; Crawford, S.E.; Zhao, B.; Smith, H.G.; Stossi, F.; Estes, M.K. A Genetically Engineered Rotavirus NSP2 Phosphorylation Mutant Impaired in Viroplasm Formation and Replication Shows an Early Interaction between vNSP2 and Cellular Lipid Droplets. J. Virol. 2020, 94, e00972-20. [Google Scholar] [CrossRef]
- Eichwald, C.; Jacob, G.; Muszynski, B.; Allende, J.E.; Burrone, O.R. Uncoupling substrate and activation functions of rotavirus NSP5: Phosphorylation of Ser-67 by casein kinase 1 is essential for hyperphos-phorylation. Proc. Natl. Acad. Sci. USA 2004, 101, 16304–16309. [Google Scholar] [CrossRef]
- Criglar, J.M.; Estes, M.K.; Crawford, S.E. Rotavirus-Induced Lipid Droplet Biogenesis Is Critical for Virus Replication. Front. Physiol. 2022, 13, 836870. [Google Scholar] [CrossRef]
- Crawford, S.E.; Desselberger, U. Lipid droplets form complexes with viroplasms and are crucial for rotavirus replication. Curr. Opin. Virol. 2016, 19, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Cheung, W.; Gill, M.; Esposito, A.; Kaminski, C.F.; Courousse, N.; Chwetzoff, S.; Trugnan, G.; Keshavan, N.; Lever, A.; Desselberger, U. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J. Virol. 2010, 84, 6782–6798. [Google Scholar] [CrossRef] [PubMed]
- Campagna, M.; Marcos-Villar, L.; Arnoldi, F.; de la Cruz-Herrera, C.F.; Gallego, P.; González-Santamaría, J.; González, D.; Lopitz-Otsoa, F.; Rodriguez, M.S.; Burrone, O.R.; et al. Rotavirus viroplasm proteins interact with the cellular sumoylation system: Implications for viroplasm-like structure formation. J. Virol. 2013, 87, 807–817. [Google Scholar] [CrossRef] [PubMed]
- Contin, R.; Arnoldi, F.; Campagna, M.; Burrone, O.R. Rotavirus NSP5 orchestrates recruitment of viroplasmic proteins. J. Gen. Virol. 2010, 91, 1782–1793. [Google Scholar] [CrossRef] [PubMed]
- Mohan, K.V.K.; Muller, J.; Atreya, C.D.; Campbell, T.B.; Schneider, K.; Wrin, T.; Petropoulos, C.J.; Connick, E. The N- and C-terminal regions of rotavirus NSP5 are the critical determinants for the formation of viroplasm-like structures independent of NSP2. J. Virol. 2003, 77, 12105–12112. [Google Scholar] [CrossRef] [PubMed]
- Buttafuoco, A.; Michaelsen, K.; Tobler, K.; Ackermann, M.; Fraefel, C.; Eichwald, C. Conserved Rotavirus NSP5 and VP2 Domains Interact and Affect Viroplasm. J. Virol. 2020, 94, e01965-19. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.; Ouldali, M.; Ménétrey, J.; Poncet, D. Structural organisation of the rotavirus nonstructural protein NSP5. J. Mol. Biol. 2011, 413, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Afrikanova, I.; Miozzo, M.C.; Giambiagi, S.; Burrone, O. Phosphorylation generates different forms of rotavirus NSP5. J. Gen. Virol. 1996, 77 Pt 9, 2059–2065. [Google Scholar] [CrossRef] [PubMed]
- Eichwald, C.; Vascotto, F.; Fabbretti, E.; Burrone, O.R. Rotavirus NSP5: Mapping phosphorylation sites and kinase activation and viroplasm localization domains. J. Virol. 2002, 76, 3461–3470. [Google Scholar] [CrossRef]
- Poncet, D.; Lindenbaum, P.; L’Haridon, R.; Cohen, J. In vivo and in vitro phosphorylation of rotavirus NSP5 correlates with its localization in viroplasms. J. Virol. 1997, 71, 34–41. [Google Scholar] [CrossRef]
- Papa, G.; Venditti, L.; Arnoldi, F.; Schraner, E.M.; Potgieter, C.; Borodavka, A.; Eichwald, C.; Burrone, O.R. Recombinant Rotaviruses Rescued by Reverse Genetics Reveal the Role of NSP5 Hyperphosphorylation in the Assembly of Viral Factories. J. Virol. 2019, 94, e01110-19. [Google Scholar] [CrossRef] [PubMed]
- Campagna, M.; Budini, M.; Arnoldi, F.; Desselberger, U.; Allende, J.E.; Burrone, O.R. Impaired hyper-phosphorylation of rotavirus NSP5 in cells depleted of casein kinase 1alpha is associated with the for-mation of viroplasms with altered morphology and a moderate decrease in virus replication. J. Gen. Virol. 2007, 88, 2800–2810. [Google Scholar] [CrossRef] [PubMed]
- Cabral-Romero, C.; Padilla-Noriega, L. Association of rotavirus viroplasms with microtubules through NSP2 and NSP5. Memórias Inst. Oswaldo Cruz 2006, 101, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Jing, Z.; Shi, H.; Chen, J.; Shi, D.; Liu, J.; Guo, L.; Tian, J.; Wu, Y.; Dong, H.; Zhang, J.; et al. Rotavirus Viroplasm Biogenesis Involves Microtubule-Based Dynein Transport Mediated by an Interaction between NSP2 and Dynein Intermediate Chain. J. Virol. 2021, 95, e0124621. [Google Scholar] [CrossRef] [PubMed]
- Borodavka, A.; Dykeman, E.C.; Schrimpf, W.; Lamb, D.C. Protein-mediated RNA folding governs sequence-specific interactions between rotavirus genome segments. eLife 2017, 6, e27453. [Google Scholar] [CrossRef]
- Coria, A.; Wienecke, A.; Knight, M.L.; Desiro, D.; Laederach, A.; Borodavka, A. Rotavirus RNA chap-erone mediates global transcriptome-wide increase in RNA backbone flexibility. Nucleic Acids Res. 2022, 50, 10078–10092. [Google Scholar] [CrossRef]
- Boudreaux, C.E.; Kelly, D.F.; McDonald, S.M. Electron microscopic analysis of rotavirus assembly-replication intermediates. Virology 2015, 477, 32–41. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Imai, M.; Akatani, K.; Ikegami, N.; Furuichi, Y. Capped and conserved terminal structures in human rotavirus genome double-stranded RNA segments. J. Virol. 1983, 47, 125–136. [Google Scholar] [CrossRef]
- McCrae, M.; McCorquodale, J. Molecular biology of rotaviruses V. terminal structure of viral RNA species. Virology 1983, 126, 204–212. [Google Scholar] [CrossRef]
- Viskovska, M.; Anish, R.; Hu, L.; Chow, D.-C.; Hurwitz, A.M.; Brown, N.G.; Palzkill, T.; Estes, M.K.; Prasad, B.V.V. Probing the sites of interactions of rotaviral proteins involved in replication. J. Virol. 2014, 88, 12866–12881. [Google Scholar] [CrossRef]
- Vende, P.; Tortorici, M.; Taraporewala, Z.F.; Patton, J.T. Rotavirus NSP2 interferes with the core lattice protein VP2 in initiation of minus-strand synthesis. Virology 2003, 313, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Komoto, S.; Kawagishi, T.; Nouda, R.; Nagasawa, N.; Onishi, M.; Matsuura, Y.; Taniguchi, K.; Kobayashi, T. Entirely plasmid-based reverse genetics system for rotaviruses. Proc. Natl. Acad. Sci. USA 2017, 114, 2349–2354. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nichols, S.L.; Haller, C.; Borodavka, A.; Esstman, S.M. Rotavirus NSP2: A Master Orchestrator of Early Viral Particle Assembly. Viruses 2024, 16, 814. https://doi.org/10.3390/v16060814
Nichols SL, Haller C, Borodavka A, Esstman SM. Rotavirus NSP2: A Master Orchestrator of Early Viral Particle Assembly. Viruses. 2024; 16(6):814. https://doi.org/10.3390/v16060814
Chicago/Turabian StyleNichols, Sarah L., Cyril Haller, Alexander Borodavka, and Sarah M. Esstman. 2024. "Rotavirus NSP2: A Master Orchestrator of Early Viral Particle Assembly" Viruses 16, no. 6: 814. https://doi.org/10.3390/v16060814
APA StyleNichols, S. L., Haller, C., Borodavka, A., & Esstman, S. M. (2024). Rotavirus NSP2: A Master Orchestrator of Early Viral Particle Assembly. Viruses, 16(6), 814. https://doi.org/10.3390/v16060814





