Restriction of Viral Glycoprotein Maturation by Cellular Protease Inhibitors
Abstract
1. Introduction: Proteolytic Maturation of Viral Glycoproteins
2. Endogenous Inhibitors of Viral Glycoprotein Cleavage
2.1. Serine Protease Inhibitors (Serpins)
2.1.1. Serpin E1/Plasminogen Activator Inhibitor 1 (PAI-1)
2.1.2. Serpin A1/Alpha1-Anti-Trypsin
2.1.3. Serpin C1/Anti-Thrombin III
2.2. α2-Macroglobulin
2.3. Protease-Activated Receptor 1 (PAR1)
2.4. Histatins 3 and 5
2.5. Alpha- and Beta-Soluble N-Ethylmaleimide-Sensitive Factor Attachment Protein (α-SNAP, β-SNAP)
2.6. Guanylate-Binding Proteins 2 and 5 (GBP2 and GBP5)
2.7. Membrane-Associated RING-CH 8 (MARCH8)
2.8. Serine Protease Inhibitor Kazal-Type 6 (SPINK6)
2.9. Secretory Leukocyte Protease Inhibitor (SLPI)
2.10. Pulmonary Surfactant
3. Viral Evasion and Counteraction Strategies
3.1. Evasion
3.1.1. Increased Ratio of Protease to Protease Inhibitor
3.1.2. Redundant Use of Multiple Proteases
3.1.3. Increased Cleavage Efficacy
3.1.4. Increased Viral Glycoprotein Production
3.2. Direct Counteraction
4. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Pasquato, A.; Ramos da Palma, J.; Galan, C.; Seidah, N.G.; Kunz, S. Viral envelope glycoprotein processing by proprotein convertases. Antivir. Res. 2013, 99, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 690–698. [Google Scholar] [CrossRef]
- Bertram, S.; Glowacka, I.; Steffen, I.; Kuhl, A.; Pohlmann, S. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev. Med. Virol. 2010, 20, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lee, K.H.; Steinhauer, D.A.; Stevens, D.J.; Skehel, J.J.; Wiley, D.C. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 1998, 95, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Kumar, R.; Ahmed, S.; Parray, H.A.; Samal, S. Efficiently cleaved HIV-1 envelopes: Can they be important for vaccine immunogen development? Ther. Adv. Vaccines Immunother. 2020, 8, 1–17. [Google Scholar] [CrossRef]
- Forsell, M.N.; Schief, W.R.; Wyatt, R.T. Immunogenicity of HIV-1 envelope glycoprotein oligomers. Curr. Opin. HIV AIDS 2009, 4, 380–387. [Google Scholar] [CrossRef]
- Weber, I.T.; Wang, Y.F.; Harrison, R.W. HIV Protease: Historical Perspective and Current Research. Viruses 2021, 13, 839. [Google Scholar] [CrossRef]
- Hu, Q.; Xiong, Y.; Zhu, G.H.; Zhang, Y.N.; Zhang, Y.W.; Huang, P.; Ge, G.B. The SARS-CoV-2 main protease (M(pro)): Structure, function, and emerging therapies for COVID-19. MedComm 2022, 3, e151. [Google Scholar] [CrossRef]
- Ullrich, S.; Nitsche, C. SARS-CoV-2 Papain-Like Protease: Structure, Function and Inhibition. Chembiochem 2022, 23, e202200327. [Google Scholar] [CrossRef]
- Decroly, E.; Wouters, S.; Di Bello, C.; Lazure, C.; Ruysschaert, J.M.; Seidah, N.G. Identification of the paired basic convertases implicated in HIV gp160 processing based on in vitro assays and expression in CD4(+) cell lines. J. Biol. Chem. 1996, 271, 30442–30450. [Google Scholar] [CrossRef]
- Vollenweider, F.; Benjannet, S.; Decroly, E.; Savaria, D.; Lazure, C.; Thomas, G.; Chretien, M.; Seidah, N.G. Comparative cellular processing of the human immunodeficiency virus (HIV-1) envelope glycoprotein gp160 by the mammalian subtilisin/kexin-like convertases. Biochem. J. 1996, 314 Pt 2, 521–532. [Google Scholar] [CrossRef]
- Bestle, D.; Heindl, M.R.; Limburg, H.; Van Lam van, T.; Pilgram, O.; Moulton, H.; Stein, D.A.; Hardes, K.; Eickmann, M.; Dolnik, O.; et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 2020, 3, e202000786. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Pohlmann, S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 2020, 78, 779–784.e775. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef]
- Panda, A.; Huang, Z.; Elankumaran, S.; Rockemann, D.D.; Samal, S.K. Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microb. Pathog. 2004, 36, 1–10. [Google Scholar] [CrossRef]
- Toyoda, T.; Sakaguchi, T.; Imai, K.; Inocencio, N.M.; Gotoh, B.; Hamaguchi, M.; Nagai, Y. Structural comparison of the cleavage-activation site of the fusion glycoprotein between virulent and avirulent strains of Newcastle disease virus. Virology 1987, 158, 242–247. [Google Scholar] [CrossRef]
- Kido, H.; Okumura, Y.; Takahashi, E.; Pan, H.Y.; Wang, S.; Yao, D.; Yao, M.; Chida, J.; Yano, M. Role of host cellular proteases in the pathogenesis of influenza and influenza-induced multiple organ failure. Biochim. Biophys. Acta 2012, 1824, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Regan, A.D.; Shraybman, R.; Cohen, R.D.; Whittaker, G.R. Differential role for low pH and cathepsin-mediated cleavage of the viral spike protein during entry of serotype II feline coronaviruses. Vet. Microbiol. 2008, 132, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Antoniak, S.; Owens, A.P., 3rd; Baunacke, M.; Williams, J.C.; Lee, R.D.; Weithauser, A.; Sheridan, P.A.; Malz, R.; Luyendyk, J.P.; Esserman, D.A.; et al. PAR-1 contributes to the innate immune response during viral infection. J. Clin. Invest. 2013, 123, 1310–1322. [Google Scholar] [CrossRef] [PubMed]
- Braun, E.; Hotter, D.; Koepke, L.; Zech, F.; Gross, R.; Sparrer, K.M.J.; Muller, J.A.; Pfaller, C.K.; Heusinger, E.; Wombacher, R.; et al. Guanylate-Binding Proteins 2 and 5 Exert Broad Antiviral Activity by Inhibiting Furin-Mediated Processing of Viral Envelope Proteins. Cell Rep. 2019, 27, 2092–2104.e10. [Google Scholar] [CrossRef]
- Medcalf, R.L. Fibrinolysis, inflammation, and regulation of the plasminogen activating system. J. Thromb. Haemost. 2007, 5 (Suppl. S1), 132–142. [Google Scholar] [CrossRef]
- Wang, J.; Luo, J.; Wen, Z.; Wang, X.; Shuai, L.; Zhong, G.; Wang, C.; Sun, Z.; Chen, W.; Ge, J.; et al. Alpha-Soluble NSF Attachment Protein Prevents the Cleavage of the SARS-CoV-2 Spike Protein by Functioning as an Interferon-Upregulated Furin Inhibitor. mBio 2022, 13, e0244321. [Google Scholar] [CrossRef]
- Kelly-Robinson, G.A.; Reihill, J.A.; Lundy, F.T.; McGarvey, L.P.; Lockhart, J.C.; Litherland, G.J.; Thornbury, K.D.; Martin, S.L. The Serpin Superfamily and Their Role in the Regulation and Dysfunction of Serine Protease Activity in COPD and Other Chronic Lung Diseases. Int. J. Mol. Sci. 2021, 22, 6351. [Google Scholar] [CrossRef]
- Spence, M.A.; Mortimer, M.D.; Buckle, A.M.; Minh, B.Q.; Jackson, C.J. A Comprehensive Phylogenetic Analysis of the Serpin Superfamily. Mol. Biol. Evol. 2021, 38, 2915–2929. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Snipas, S.; Orth, K.; Muzio, M.; Dixit, V.M.; Salvesen, G.S. Target protease specificity of the viral serpin CrmA. Analysis of five caspases. J. Biol. Chem. 1997, 272, 7797–7800. [Google Scholar] [CrossRef] [PubMed]
- Hunt, L.T.; Dayhoff, M.O. A surprising new protein superfamily containing ovalbumin, antithrombin-III, and alpha 1-proteinase inhibitor. Biochem. Biophys. Res. Commun. 1980, 95, 864–871. [Google Scholar] [CrossRef]
- Annand, R.R.; Dahlen, J.R.; Sprecher, C.A.; De Dreu, P.; Foster, D.C.; Mankovich, J.A.; Talanian, R.V.; Kisiel, W.; Giegel, D.A. Caspase-1 (interleukin-1beta-converting enzyme) is inhibited by the human serpin analogue proteinase inhibitor 9. Biochem. J. 1999, 342 Pt 3, 655–665. [Google Scholar] [CrossRef]
- Choi, Y.J.; Kim, S.; Choi, Y.; Nielsen, T.B.; Yan, J.; Lu, A.; Ruan, J.; Lee, H.R.; Wu, H.; Spellberg, B.; et al. SERPINB1-mediated checkpoint of inflammatory caspase activation. Nat. Immunol. 2019, 20, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Huntington, J.A.; Read, R.J.; Carrell, R.W. Structure of a serpin-protease complex shows inhibition by deformation. Nature 2000, 407, 923–926. [Google Scholar] [CrossRef] [PubMed]
- Gettins, P.G. Serpin structure, mechanism, and function. Chem. Rev. 2002, 102, 4751–4804. [Google Scholar] [CrossRef]
- Law, R.H.; Zhang, Q.; McGowan, S.; Buckle, A.M.; Silverman, G.A.; Wong, W.; Rosado, C.J.; Langendorf, C.G.; Pike, R.N.; Bird, P.I.; et al. An overview of the serpin superfamily. Genome Biol. 2006, 7, 216. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lucas, A.; Yaron, J.R.; Zhang, L.; Ambadapadi, S. Overview of Serpins and Their Roles in Biological Systems. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2018; Volume 1826, pp. 1–7. [Google Scholar] [CrossRef]
- Greene, C.M.; Marciniak, S.J.; Teckman, J.; Ferrarotti, I.; Brantly, M.L.; Lomas, D.A.; Stoller, J.K.; McElvaney, N.G. alpha1-Antitrypsin deficiency. Nat. Rev. Dis. Primers 2016, 2, 16051. [Google Scholar] [CrossRef] [PubMed]
- Smith, N.; Warren, B.B.; Smith, J.; Jacobson, L.; Armstrong, J.; Kim, J.; Di Paola, J.; Manco-Johnson, M. Antithrombin deficiency: A pediatric disorder. Thromb. Res. 2021, 202, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Dittmann, M.; Hoffmann, H.H.; Scull, M.A.; Gilmore, R.H.; Bell, K.L.; Ciancanelli, M.; Wilson, S.J.; Crotta, S.; Yu, Y.; Flatley, B.; et al. A serpin shapes the extracellular environment to prevent influenza A virus maturation. Cell 2015, 160, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Esumi, M.; Ishibashi, M.; Yamaguchi, H.; Nakajima, S.; Tai, Y.; Kikuta, S.; Sugitani, M.; Takayama, T.; Tahara, M.; Takeda, M.; et al. Transmembrane serine protease TMPRSS2 activates hepatitis C virus infection. Hepatology 2015, 61, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Harbig, A.; Mernberger, M.; Bittel, L.; Pleschka, S.; Schughart, K.; Steinmetzer, T.; Stiewe, T.; Nist, A.; Bottcher-Friebertshauser, E. Transcriptome profiling and protease inhibition experiments identify proteases that activate H3N2 influenza A and influenza B viruses in murine airways. J. Biol. Chem. 2020, 295, 11388–11407. [Google Scholar] [CrossRef] [PubMed]
- Bernot, D.; Stalin, J.; Stocker, P.; Bonardo, B.; Scroyen, I.; Alessi, M.C.; Peiretti, F. Plasminogen activator inhibitor 1 is an intracellular inhibitor of furin proprotein convertase. J. Cell Sci. 2011, 124, 1224–1230. [Google Scholar] [CrossRef]
- Rosendal, E.; Mihai, I.S.; Becker, M.; Das, D.; Frangsmyr, L.; Persson, B.D.; Rankin, G.D.; Groning, R.; Trygg, J.; Forsell, M.; et al. Serine Protease Inhibitors Restrict Host Susceptibility to SARS-CoV-2 Infections. mBio 2022, 13, e0089222. [Google Scholar] [CrossRef]
- Azouz, N.P.; Klingler, A.M.; Callahan, V.; Akhrymuk, I.V.; Elez, K.; Raich, L.; Henry, B.M.; Benoit, J.L.; Benoit, S.W.; Noe, F.; et al. Alpha 1 Antitrypsin is an Inhibitor of the SARS-CoV-2-Priming Protease TMPRSS2. Pathog. Immun. 2021, 6, 55–74. [Google Scholar] [CrossRef]
- Wettstein, L.; Weil, T.; Conzelmann, C.; Muller, J.A.; Gross, R.; Hirschenberger, M.; Seidel, A.; Klute, S.; Zech, F.; Prelli Bozzo, C.; et al. Alpha-1 antitrypsin inhibits TMPRSS2 protease activity and SARS-CoV-2 infection. Nat. Commun. 2021, 12, 1726. [Google Scholar] [CrossRef]
- Wettstein, L.; Immenschuh, P.; Weil, T.; Conzelmann, C.; Almeida-Hernandez, Y.; Hoffmann, M.; Kempf, A.; Nehlmeier, I.; Lotke, R.; Petersen, M.; et al. Native and activated antithrombin inhibits TMPRSS2 activity and SARS-CoV-2 infection. J. Med. Virol. 2023, 95, e28124. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Zhang, X.Q.; Lo, C.W.; Liu, P.F.; Liu, Y.T.; Gallo, R.L.; Hsieh, M.F.; Schooley, R.T.; Huang, C.M. The essentiality of alpha-2-macroglobulin in human salivary innate immunity against new H1N1 swine origin influenza A virus. Proteomics 2010, 10, 2396–2401. [Google Scholar] [CrossRef] [PubMed]
- Aerts, L.; Hamelin, M.E.; Rheaume, C.; Lavigne, S.; Couture, C.; Kim, W.; Susan-Resiga, D.; Prat, A.; Seidah, N.G.; Vergnolle, N.; et al. Modulation of protease activated receptor 1 influences human metapneumovirus disease severity in a mouse model. PLoS ONE 2013, 8, e72529. [Google Scholar] [CrossRef]
- Kim, W.; Zekas, E.; Lodge, R.; Susan-Resiga, D.; Marcinkiewicz, E.; Essalmani, R.; Mihara, K.; Ramachandran, R.; Asahchop, E.; Gelman, B.; et al. Neuroinflammation-Induced Interactions between Protease-Activated Receptor 1 and Proprotein Convertases in HIV-Associated Neurocognitive Disorder. Mol. Cell. Biol. 2015, 35, 3684–3700. [Google Scholar] [CrossRef]
- Basak, A.; Ernst, B.; Brewer, D.; Seidah, N.G.; Munzer, J.S.; Lazure, C.; Lajoie, G.A. Histidine-rich human salivary peptides are inhibitors of proprotein convertases furin and PC7 but act as substrates for PC1. J. Pept. Res. 1997, 49, 596–603. [Google Scholar] [CrossRef]
- Groot, F.; Sanders, R.W.; ter Brake, O.; Nazmi, K.; Veerman, E.C.; Bolscher, J.G.; Berkhout, B. Histatin 5-derived peptide with improved fungicidal properties enhances human immunodeficiency virus type 1 replication by promoting viral entry. J. Virol. 2006, 80, 9236–9243. [Google Scholar] [CrossRef] [PubMed]
- Mesner, D.; Reuschl, A.K.; Whelan, M.V.X.; Bronzovich, T.; Haider, T.; Thorne, L.G.; Ragazzini, R.; Bonfanti, P.; Towers, G.J.; Jolly, C. SARS-CoV-2 evolution influences GBP and IFITM sensitivity. Proc. Natl. Acad. Sci. USA 2023, 120, e2212577120. [Google Scholar] [CrossRef] [PubMed]
- Srinivasachar Badarinarayan, S.; Shcherbakova, I.; Langer, S.; Koepke, L.; Preising, A.; Hotter, D.; Kirchhoff, F.; Sparrer, K.M.J.; Schotta, G.; Sauter, D. HIV-1 infection activates endogenous retroviral promoters regulating antiviral gene expression. Nucleic Acids Res. 2020, 48, 10890–10908. [Google Scholar] [CrossRef] [PubMed]
- Lun, C.M.; Waheed, A.A.; Majadly, A.; Powell, N.; Freed, E.O. Mechanism of Viral Glycoprotein Targeting by Membrane-Associated RING-CH Proteins. mBio 2021, 12, e00219-21. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Li, S.; Zhang, X.; Khan, I.; Ahmad, I.; Zhou, Y.; Li, S.; Shi, J.; Wang, Y.; Zheng, Y.H. MARCH8 Inhibits Ebola Virus Glycoprotein, Human Immunodeficiency Virus Type 1 Envelope Glycoprotein, and Avian Influenza Virus H5N1 Hemagglutinin Maturation. mBio 2020, 11, e01882-20. [Google Scholar] [CrossRef] [PubMed]
- Umthong, S.; Lynch, B.; Timilsina, U.; Waxman, B.; Ivey, E.B.; Stavrou, S. Elucidating the Antiviral Mechanism of Different MARCH Factors. mBio 2021, 12, e03264-20. [Google Scholar] [CrossRef]
- Meyer-Hoffert, U.; Wu, Z.; Kantyka, T.; Fischer, J.; Latendorf, T.; Hansmann, B.; Bartels, J.; He, Y.; Glaser, R.; Schroder, J.M. Isolation of SPINK6 in human skin: Selective inhibitor of kallikrein-related peptidases. J. Biol. Chem. 2010, 285, 32174–32181. [Google Scholar] [CrossRef]
- Wang, D.; Li, C.; Chiu, M.C.; Yu, Y.; Liu, X.; Zhao, X.; Huang, J.; Cheng, Z.; Yuan, S.; Poon, V.; et al. SPINK6 inhibits human airway serine proteases and restricts influenza virus activation. EMBO Mol. Med. 2022, 14, e14485. [Google Scholar] [CrossRef] [PubMed]
- Kido, H.; Sakai, K.; Kishino, Y.; Tashiro, M. Pulmonary surfactant is a potential endogenous inhibitor of proteolytic activation of Sendai virus and influenza A virus. FEBS Lett. 1993, 322, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Heit, C.; Jackson, B.C.; McAndrews, M.; Wright, M.W.; Thompson, D.C.; Silverman, G.A.; Nebert, D.W.; Vasiliou, V. Update of the human and mouse SERPIN gene superfamily. Hum. Genom. 2013, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Ellis, V.; Scully, M.; MacGregor, I.; Kakkar, V. Inhibition of human factor Xa by various plasma protease inhibitors. Biochim. Biophys. Acta 1982, 701, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Hippensteel, J.; Leavitt, A.; Maloney, J.P.; Beckham, D.; Garcia, C.; Li, Q.; Freed, B.M.; Ordway, D.; Sandhaus, R.A.; et al. Hypothesis: Alpha-1-antitrypsin is a promising treatment option for COVID-19. Med. Hypotheses 2021, 146, 110394. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, C.; Das, C.; Ghosh, A.; Singh, A.K.; Mukherjee, S.; Majumder, P.P.; Basu, A.; Biswas, N.K. SARS-CoV-2 mutation 614G creates an elastase cleavage site enhancing its spread in high AAT-deficient regions. Infect. Genet. Evol. 2021, 90, 104760. [Google Scholar] [CrossRef] [PubMed]
- Faria, N.; Ines Costa, M.; Gomes, J.; Sucena, M. Alpha-1 antitrypsin deficiency severity and the risk of COVID-19: A Portuguese cohort. Respir. Med. 2021, 181, 106387. [Google Scholar] [CrossRef]
- Shapira, G.; Shomron, N.; Gurwitz, D. Ethnic differences in alpha-1 antitrypsin deficiency allele frequencies may partially explain national differences in COVID-19 fatality rates. FASEB J. 2020, 34, 14160–14165. [Google Scholar] [CrossRef]
- Yoshikura, H. Epidemiological correlation between COVID-19 epidemic and prevalence of alpha-1 antitrypsin deficiency in the world. Glob. Health Med. 2021, 3, 73–81. [Google Scholar] [CrossRef]
- Nygren, D.; Molstad, U.; Thulesius, H.; Hillman, M.; Broman, L.M.; Tanash, H.; Landin-Olsson, M.; Rasmussen, M.; Thunander, M. Low Prevalence of Mild Alpha-1-Antitrypsin Deficiency in Hospitalized COVID-19-Patients. Int. J. Gen. Med. 2022, 15, 5843–5848. [Google Scholar] [CrossRef]
- Schneider, C.V.; Strnad, P. SARS-CoV-2 infection in alpha1-antitrypsin deficiency. Respir. Med. 2021, 184, 106466. [Google Scholar] [CrossRef]
- Guttman, O.; Baranovski, B.M.; Schuster, R.; Kaner, Z.; Freixo-Lima, G.S.; Bahar, N.; Kalay, N.; Mizrahi, M.I.; Brami, I.; Ochayon, D.E.; et al. Acute-phase protein alpha1-anti-trypsin: Diverting injurious innate and adaptive immune responses from non-authentic threats. Clin. Exp. Immunol. 2015, 179, 161–172. [Google Scholar] [CrossRef]
- McElvaney, O.J.; McEvoy, N.L.; McElvaney, O.F.; Carroll, T.P.; Murphy, M.P.; Dunlea, D.M.; Ni Choileain, O.; Clarke, J.; O’Connor, E.; Hogan, G.; et al. Characterization of the Inflammatory Response to Severe COVID-19 Illness. Am. J. Respir. Crit. Care Med. 2020, 202, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Bertram, S.; Glowacka, I.; Blazejewska, P.; Soilleux, E.; Allen, P.; Danisch, S.; Steffen, I.; Choi, S.Y.; Park, Y.; Schneider, H.; et al. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 2010, 84, 10016–10025. [Google Scholar] [CrossRef]
- Bryan, C.L.; Beard, K.S.; Pott, G.B.; Rahkola, J.; Gardner, E.M.; Janoff, E.N.; Shapiro, L. HIV infection is associated with reduced serum alpha-1-antitrypsin concentrations. Clin. Invest. Med. 2010, 33, E384–E389. [Google Scholar] [CrossRef]
- Shapiro, L.; Pott, G.B.; Ralston, A.H. Alpha-1-antitrypsin inhibits human immunodeficiency virus type 1. FASEB J. 2001, 15, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Shapiro, L.; Fellingham, G.; Willardson, B.M.; Burton, G.F. HIV replication in CD4+ T lymphocytes in the presence and absence of follicular dendritic cells: Inhibition of replication mediated by alpha-1-antitrypsin through altered IkappaBalpha ubiquitination. J. Immunol. 2011, 186, 3148–3155. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, Z.; Zhang, J.; Adelsberger, J.W.; Yang, J.; Burton, G.F. Alpha-1-antitrypsin interacts with gp41 to block HIV-1 entry into CD4+ T lymphocytes. BMC Microbiol. 2016, 16, 172. [Google Scholar] [CrossRef] [PubMed]
- Grover, S.P.; Mackman, N. Anticoagulant SERPINs: Endogenous Regulators of Hemostasis and Thrombosis. Front. Cardiovasc. Med. 2022, 9, 878199. [Google Scholar] [CrossRef] [PubMed]
- Sottrup-Jensen, L.; Stepanik, T.M.; Kristensen, T.; Wierzbicki, D.M.; Jones, C.M.; Lonblad, P.B.; Magnusson, S.; Petersen, T.E. Primary structure of human alpha 2-macroglobulin. V. The complete structure. J. Biol. Chem. 1984, 259, 8318–8327. [Google Scholar] [CrossRef] [PubMed]
- Out, T.A.; Jansen, H.M.; van Steenwijk, R.P.; de Nooijer, M.J.; van de Graaf, E.A.; Zuijderhoudt, F.M. ELISA of ceruloplasmin and alpha-2-macroglobulin in paired bronchoalveolar lavage fluid and serum samples. Clin. Chim. Acta 1987, 165, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Pederson, E.D.; Stanke, S.R.; Whitener, S.J.; Sebastiani, P.T.; Lamberts, B.L.; Turner, D.W. Salivary levels of alpha 2-macroglobulin, alpha 1-antitrypsin, C-reactive protein, cathepsin G and elastase in humans with or without destructive periodontal disease. Arch. Oral. Biol. 1995, 40, 1151–1155. [Google Scholar] [CrossRef] [PubMed]
- Borth, W. Alpha 2-macroglobulin, a multifunctional binding protein with targeting characteristics. FASEB J. 1992, 6, 3345–3353. [Google Scholar] [CrossRef] [PubMed]
- Sottrup-Jensen, L. Alpha-macroglobulins: Structure, shape, and mechanism of proteinase complex formation. J. Biol. Chem. 1989, 264, 11539–11542. [Google Scholar] [CrossRef] [PubMed]
- Enghild, J.J.; Thogersen, I.B.; Roche, P.A.; Pizzo, S.V. A conserved region in alpha-macroglobulins participates in binding to the mammalian alpha-macroglobulin receptor. Biochemistry 1989, 28, 1406–1412. [Google Scholar] [CrossRef]
- Pritchett, T.J.; Paulson, J.C. Basis for the potent inhibition of influenza virus infection by equine and guinea pig alpha 2-macroglobulin. J. Biol. Chem. 1989, 264, 9850–9858. [Google Scholar] [CrossRef]
- Rogers, G.N.; Pritchett, T.J.; Lane, J.L.; Paulson, J.C. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor of infection: Selection of receptor specific variants. Virology 1983, 131, 394–408. [Google Scholar] [CrossRef]
- Ryan-Poirier, K.A.; Kawaoka, Y. Alpha 2-macroglobulin is the major neutralizing inhibitor of influenza A virus in pig serum. Virology 1993, 193, 974–976. [Google Scholar] [CrossRef]
- Athauda, S.B.; Ido, E.; Arakawa, H.; Nishigai, M.; Kyushiki, H.; Yoshinaka, Y.; Takahashi, T.; Ikai, A.; Tang, J.; Takahashi, K. Entrapment and inhibition of human immunodeficiency virus proteinase by alpha 2-macroglobulin and structural changes in the inhibitor. J. Biochem. 1993, 113, 742–746. [Google Scholar] [CrossRef]
- Kisselev, A.F.; von der Helm, K. Human immunodeficiency virus type 1 proteinase is rapidly and efficiently inactivated in human plasma by alpha 2-macroglobulin. Biol. Chem. Hoppe Seyler 1994, 375, 711–714. [Google Scholar] [PubMed]
- Meier, U.C.; Billich, A.; Mann, K.; Schramm, H.J.; Schramm, W. alpha 2-Macroglobulin is cleaved by HIV-1 protease in the bait region but not in the C-terminal inter-domain region. Biol. Chem. Hoppe Seyler 1991, 372, 1051–1056. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, S.R.; Seatter, M.J.; Kanke, T.; Hunter, G.D.; Plevin, R. Proteinase-activated receptors. Pharmacol. Rev. 2001, 53, 245–282. [Google Scholar] [PubMed]
- Vu, T.K.; Hung, D.T.; Wheaton, V.I.; Coughlin, S.R. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell 1991, 64, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Trejo, J.; Hammes, S.R.; Coughlin, S.R. Termination of signaling by protease-activated receptor-1 is linked to lysosomal sorting. Proc. Natl. Acad. Sci. USA 1998, 95, 13698–13702. [Google Scholar] [CrossRef] [PubMed]
- Khoufache, K.; Berri, F.; Nacken, W.; Vogel, A.B.; Delenne, M.; Camerer, E.; Coughlin, S.R.; Carmeliet, P.; Lina, B.; Rimmelzwaan, G.F.; et al. PAR1 contributes to influenza A virus pathogenicity in mice. J. Clin. Invest. 2013, 123, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Le, V.B.; Riteau, B.; Alessi, M.C.; Couture, C.; Jandrot-Perrus, M.; Rheaume, C.; Hamelin, M.E.; Boivin, G. Protease-activated receptor 1 inhibition protects mice against thrombin-dependent respiratory syncytial virus and human metapneumovirus infections. Br. J. Pharmacol. 2018, 175, 388–403. [Google Scholar] [CrossRef]
- Rovai, E.S.; Alves, T.; Holzhausen, M. Protease-activated receptor 1 as a potential therapeutic target for COVID-19. Exp. Biol. Med. 2021, 246, 688–694. [Google Scholar] [CrossRef]
- Oppenheim, F.G.; Xu, T.; McMillian, F.M.; Levitz, S.M.; Diamond, R.D.; Offner, G.D.; Troxler, R.F. Histatins, a novel family of histidine-rich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Candida albicans. J. Biol. Chem. 1988, 263, 7472–7477. [Google Scholar] [CrossRef]
- Gusman, H.; Grogan, J.; Kagan, H.M.; Troxler, R.F.; Oppenheim, F.G. Salivary histatin 5 is a potent competitive inhibitor of the cysteine proteinase clostripain. FEBS Lett. 2001, 489, 97–100. [Google Scholar] [CrossRef]
- Gusman, H.; Travis, J.; Helmerhorst, E.J.; Potempa, J.; Troxler, R.F.; Oppenheim, F.G. Salivary histatin 5 is an inhibitor of both host and bacterial enzymes implicated in periodontal disease. Infect. Immun. 2001, 69, 1402–1408. [Google Scholar] [CrossRef]
- Nishikata, M.; Kanehira, T.; Oh, H.; Tani, H.; Tazaki, M.; Kuboki, Y. Salivary histatin as an inhibitor of a protease produced by the oral bacterium Bacteroides gingivalis. Biochem. Biophys. Res. Commun. 1991, 174, 625–630. [Google Scholar] [CrossRef]
- Ruissen, A.L.; Groenink, J.; Helmerhorst, E.J.; Walgreen-Weterings, E.; Van’t Hof, W.; Veerman, E.C.; Nieuw Amerongen, A.V. Effects of histatin 5 and derived peptides on Candida albicans. Biochem. J. 2001, 356, 361–368. [Google Scholar] [CrossRef]
- Stenbeck, G. Soluble NSF-attachment proteins. Int. J. Biochem. Cell Biol. 1998, 30, 573–577. [Google Scholar] [CrossRef]
- Zhou, A.; Martin, S.; Lipkind, G.; LaMendola, J.; Steiner, D.F. Regulatory roles of the P domain of the subtilisin-like prohormone convertases. J. Biol. Chem. 1998, 273, 11107–11114. [Google Scholar] [CrossRef]
- Kutsch, M.; Coers, J. Human guanylate binding proteins: Nanomachines orchestrating host defense. FEBS J. 2021, 288, 5826–5849. [Google Scholar] [CrossRef] [PubMed]
- Krapp, C.; Hotter, D.; Gawanbacht, A.; McLaren, P.J.; Kluge, S.F.; Sturzel, C.M.; Mack, K.; Reith, E.; Engelhart, S.; Ciuffi, A.; et al. Guanylate Binding Protein (GBP) 5 Is an Interferon-Inducible Inhibitor of HIV-1 Infectivity. Cell Host Microbe 2016, 19, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Hotter, D.; Sauter, D.; Kirchhoff, F. Guanylate binding protein 5: Impairing virion infectivity by targeting retroviral envelope glycoproteins. Small GTPases 2017, 8, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Qu, X.; Liu, X.; Huan, C.; Wang, H.; Zhao, Z.; Yang, X.; Hua, S.; Zhang, W. GBP5 Is an Interferon-Induced Inhibitor of Respiratory Syncytial Virus. J. Virol. 2020, 94, e01407-20. [Google Scholar] [CrossRef] [PubMed]
- Lenz, O.; ter Meulen, J.; Klenk, H.D.; Seidah, N.G.; Garten, W. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. USA 2001, 98, 12701–12705. [Google Scholar] [CrossRef]
- Tada, T.; Zhang, Y.; Koyama, T.; Tobiume, M.; Tsunetsugu-Yokota, Y.; Yamaoka, S.; Fujita, H.; Tokunaga, K. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins. Nat. Med. 2015, 21, 1502–1507. [Google Scholar] [CrossRef]
- Zhang, Y.; Tada, T.; Ozono, S.; Yao, W.; Tanaka, M.; Yamaoka, S.; Kishigami, S.; Fujita, H.; Tokunaga, K. Membrane-associated RING-CH (MARCH) 1 and 2 are MARCH family members that inhibit HIV-1 infection. J. Biol. Chem. 2019, 294, 3397–3405. [Google Scholar] [CrossRef]
- Zhang, Y.; Tada, T.; Ozono, S.; Kishigami, S.; Fujita, H.; Tokunaga, K. MARCH8 inhibits viral infection by two different mechanisms. eLife 2020, 9, e57763. [Google Scholar] [CrossRef]
- Zhang, Y.; Ozono, S.; Tada, T.; Tobiume, M.; Kameoka, M.; Kishigami, S.; Fujita, H.; Tokunaga, K. MARCH8 Targets Cytoplasmic Lysine Residues of Various Viral Envelope Glycoproteins. Microbiol. Spectr. 2022, 10, e0061821. [Google Scholar] [CrossRef]
- Hamilton, B.S.; Whittaker, G.R. Cleavage activation of human-adapted influenza virus subtypes by kallikrein-related peptidases 5 and 12. J. Biol. Chem. 2013, 288, 17399–17407. [Google Scholar] [CrossRef]
- Magnen, M.; Gueugnon, F.; Guillon, A.; Baranek, T.; Thibault, V.C.; Petit-Courty, A.; de Veer, S.J.; Harris, J.; Humbles, A.A.; Si-Tahar, M.; et al. Kallikrein-Related Peptidase 5 Contributes to H3N2 Influenza Virus Infection in Human Lungs. J. Virol. 2017, 91, e00421-17. [Google Scholar] [CrossRef]
- Bottcher, E.; Matrosovich, T.; Beyerle, M.; Klenk, H.D.; Garten, W.; Matrosovich, M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 2006, 80, 9896–9898. [Google Scholar] [CrossRef] [PubMed]
- Majchrzak-Gorecka, M.; Majewski, P.; Grygier, B.; Murzyn, K.; Cichy, J. Secretory leukocyte protease inhibitor (SLPI), a multifunctional protein in the host defense response. Cytokine Growth Factor Rev. 2016, 28, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.C.; Ohlsson, K. Isolation, properties, and complete amino acid sequence of human secretory leukocyte protease inhibitor, a potent inhibitor of leukocyte elastase. Proc. Natl. Acad. Sci. USA 1986, 83, 6692–6696. [Google Scholar] [CrossRef] [PubMed]
- Beppu, Y.; Imamura, Y.; Tashiro, M.; Towatari, T.; Ariga, H.; Kido, H. Human mucus protease inhibitor in airway fluids is a potential defensive compound against infection with influenza A and Sendai viruses. J. Biochem. 1997, 121, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Kido, H.; Okumura, Y.; Yamada, H.; Mizuno, D.; Higashi, Y.; Yano, M. Secretory leukoprotease inhibitor and pulmonary surfactant serve as principal defenses against influenza A virus infection in the airway and chemical agents up-regulating their levels may have therapeutic potential. Biol. Chem. 2004, 385, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, T.S.; Roghanian, A.; Simpson, A.J.; Sallenave, J.M. WAP domain proteins as modulators of mucosal immunity. Biochem. Soc. Trans. 2011, 39, 1409–1415. [Google Scholar] [CrossRef] [PubMed]
- Ying, Q.L.; Kemme, M.; Simon, S.R. Functions of the N-terminal domain of secretory leukoprotease inhibitor. Biochemistry 1994, 33, 5445–5450. [Google Scholar] [CrossRef] [PubMed]
- McNeely, T.B.; Dealy, M.; Dripps, D.J.; Orenstein, J.M.; Eisenberg, S.P.; Wahl, S.M. Secretory leukocyte protease inhibitor: A human saliva protein exhibiting anti-human immunodeficiency virus 1 activity in vitro. J. Clin. Invest. 1995, 96, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Shugars, D.C.; Sauls, D.L.; Weinberg, J.B. Secretory leukocyte protease inhibitor blocks infectivity of primary monocytes and mononuclear cells with both monocytotropic and lymphocytotropic strains of human immunodeficiency virus type I. Oral. Dis. 1997, 3 (Suppl. S1), S70–S72. [Google Scholar] [CrossRef] [PubMed]
- Wahl, S.M.; McNeely, T.B.; Janoff, E.N.; Shugars, D.; Worley, P.; Tucker, C.; Orenstein, J.M. Secretory leukocyte protease inhibitor (SLPI) in mucosal fluids inhibits HIV-I. Oral. Dis. 1997, 3 (Suppl. S1), S64–S69. [Google Scholar] [CrossRef]
- McNeely, T.B.; Shugars, D.C.; Rosendahl, M.; Tucker, C.; Eisenberg, S.P.; Wahl, S.M. Inhibition of human immunodeficiency virus type 1 infectivity by secretory leukocyte protease inhibitor occurs prior to viral reverse transcription. Blood 1997, 90, 1141–1149. [Google Scholar] [CrossRef]
- Crouch, E.; Hartshorn, K.; Ofek, I. Collectins and pulmonary innate immunity. Immunol. Rev. 2000, 173, 52–65. [Google Scholar] [CrossRef]
- Yang, B.; Yao, D.F.; Ohuchi, M.; Ide, M.; Yano, M.; Okumura, Y.; Kido, H. Ambroxol suppresses influenza-virus proliferation in the mouse airway by increasing antiviral factor levels. Eur. Respir. J. 2002, 19, 952–958. [Google Scholar] [CrossRef]
- Tashiro, M.; Beppu, Y.; Sakai, K.; Kido, H. Inhibitory effect of pulmonary surfactant on Sendai virus infection in rat lungs. Arch. Virol. 1996, 141, 1571–1577. [Google Scholar] [CrossRef]
- Alfaifi, A.; Sultan, A.S.; Montelongo-Jauregui, D.; Meiller, T.F.; Jabra-Rizk, M.A. Long-Term Post-COVID-19 Associated Oral Inflammatory Sequelae. Front. Cell. Infect. Microbiol. 2022, 12, 831744. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, Z.; Zhanapiya, A.; Kalbacher, H.; Burster, T. Neutrophil Elastase and Proteinase 3 Cleavage Sites Are Adjacent to the Polybasic Sequence within the Proteolytic Sensitive Activation Loop of the SARS-CoV-2 Spike Protein. ACS Omega 2021, 6, 7181–7185. [Google Scholar] [CrossRef] [PubMed]
- Braun, E.; Sauter, D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunol. 2019, 8, e1073. [Google Scholar] [CrossRef] [PubMed]
- Klein-Szanto, A.J.; Bassi, D.E. Proprotein convertase inhibition: Paralyzing the cell’s master switches. Biochem. Pharmacol. 2017, 140, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Johnson, B.A.; Xia, H.; Ku, Z.; Schindewolf, C.; Widen, S.G.; An, Z.; Weaver, S.C.; Menachery, V.D.; et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 2022, 39, 110829. [Google Scholar] [CrossRef] [PubMed]
- Galvan, J.R.; de Vries, M.; Fisher, A.; Prescott, R.A.; Crosse, K.M.; Duerr, R.; Dittmann, M. In silico docking screen identifies airway host protease targets for human SERPINs. bioRxiv 2022. [Google Scholar] [CrossRef]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef] [PubMed]
- Mantzourani, C.; Vasilakaki, S.; Gerogianni, V.E.; Kokotos, G. The discovery and development of transmembrane serine protease 2 (TMPRSS2) inhibitors as candidate drugs for the treatment of COVID-19. Expert. Opin. Drug Discov. 2022, 17, 231–246. [Google Scholar] [CrossRef]
- Kaur, U.; Chakrabarti, S.S.; Ojha, B.; Pathak, B.K.; Singh, A.; Saso, L.; Chakrabarti, S. Targeting Host Cell Proteases to Prevent SARS-CoV-2 Invasion. Curr. Drug Targets 2021, 22, 192–201. [Google Scholar] [CrossRef]
- Liu, T.; Luo, S.; Libby, P.; Shi, G.P. Cathepsin L-selective inhibitors: A potentially promising treatment for COVID-19 patients. Pharmacol. Ther. 2020, 213, 107587. [Google Scholar] [CrossRef]
- Dahlen, J.R.; Jean, F.; Thomas, G.; Foster, D.C.; Kisiel, W. Inhibition of soluble recombinant furin by human proteinase inhibitor 8. J. Biol. Chem. 1998, 273, 1851–1854. [Google Scholar] [CrossRef]
- Petersen, M.; Lotke, R.; Hopfensperger, K.; Victoria, S.; Haussmann, I.; Burster, T.; Baldauf, H.M.; Sauter, D. Inhibition of Infectious HIV-1 Production by Rerouting the Cellular Furin Inhibitor Serpin B8. J. Virol. 2023, 97, e0029423. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.D.; Thomas, L.; Hayflick, J.S.; Thomas, G. Inhibition of HIV-1 gp160-dependent membrane fusion by a furin-directed alpha 1-antitrypsin variant. J. Biol. Chem. 1993, 268, 24887–24891. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Hirano, A.; Stenglein, S.; Nelson, J.; Thomas, G.; Wong, T.C. Engineered serine protease inhibitor prevents furin-catalyzed activation of the fusion glycoprotein and production of infectious measles virus. J. Virol. 1995, 69, 3206–3210. [Google Scholar] [CrossRef] [PubMed]
- Bahbouhi, B.; Bendjennat, M.; Guetard, D.; Seidah, N.G.; Bahraoui, E. Effect of alpha-1 antitrypsin Portland variant (alpha 1-PDX) on HIV-1 replication. Biochem. J. 2000, 352 Pt 1, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Hada, K.; Isshiki, K.; Matsuda, S.; Yuasa, K.; Tsuji, A. Engineering of alpha1-antitrypsin variants with improved specificity for the proprotein convertase furin using site-directed random mutagenesis. Protein Eng. Des. Sel. 2013, 26, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Izaguirre, G.; Arciniega, M.; Quezada, A.G. Specific and Selective Inhibitors of Proprotein Convertases Engineered by Transferring Serpin B8 Reactive-Site and Exosite Determinants of Reactivity to the Serpin alpha1PDX. Biochemistry 2019, 58, 1679–1688. [Google Scholar] [CrossRef]
- Maisa, A.; Stroher, U.; Klenk, H.D.; Garten, W.; Strecker, T. Inhibition of Lassa virus glycoprotein cleavage and multicycle replication by site 1 protease-adapted alpha(1)-antitrypsin variants. PLoS Negl. Trop. Dis. 2009, 3, e446. [Google Scholar] [CrossRef] [PubMed]
- Van Rompaey, L.; Proost, P.; Van den Berghe, H.; Marynen, P. Design of a new protease inhibitor by the manipulation of the bait region of alpha 2-macroglobulin: Inhibition of the tobacco etch virus protease by mutant alpha 2-macroglobulin. Biochem. J. 1995, 312 Pt 1, 191–195. [Google Scholar] [CrossRef]
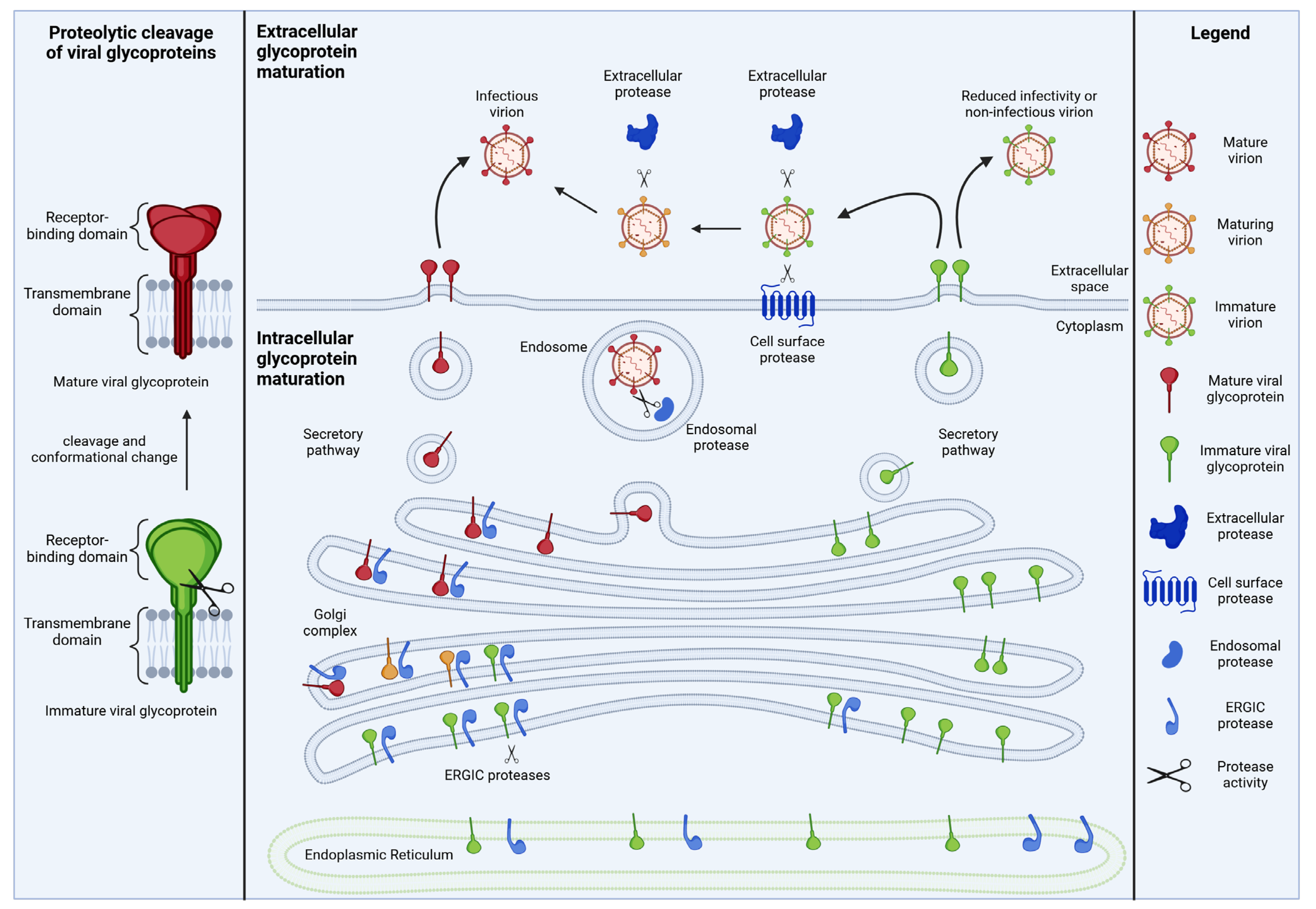
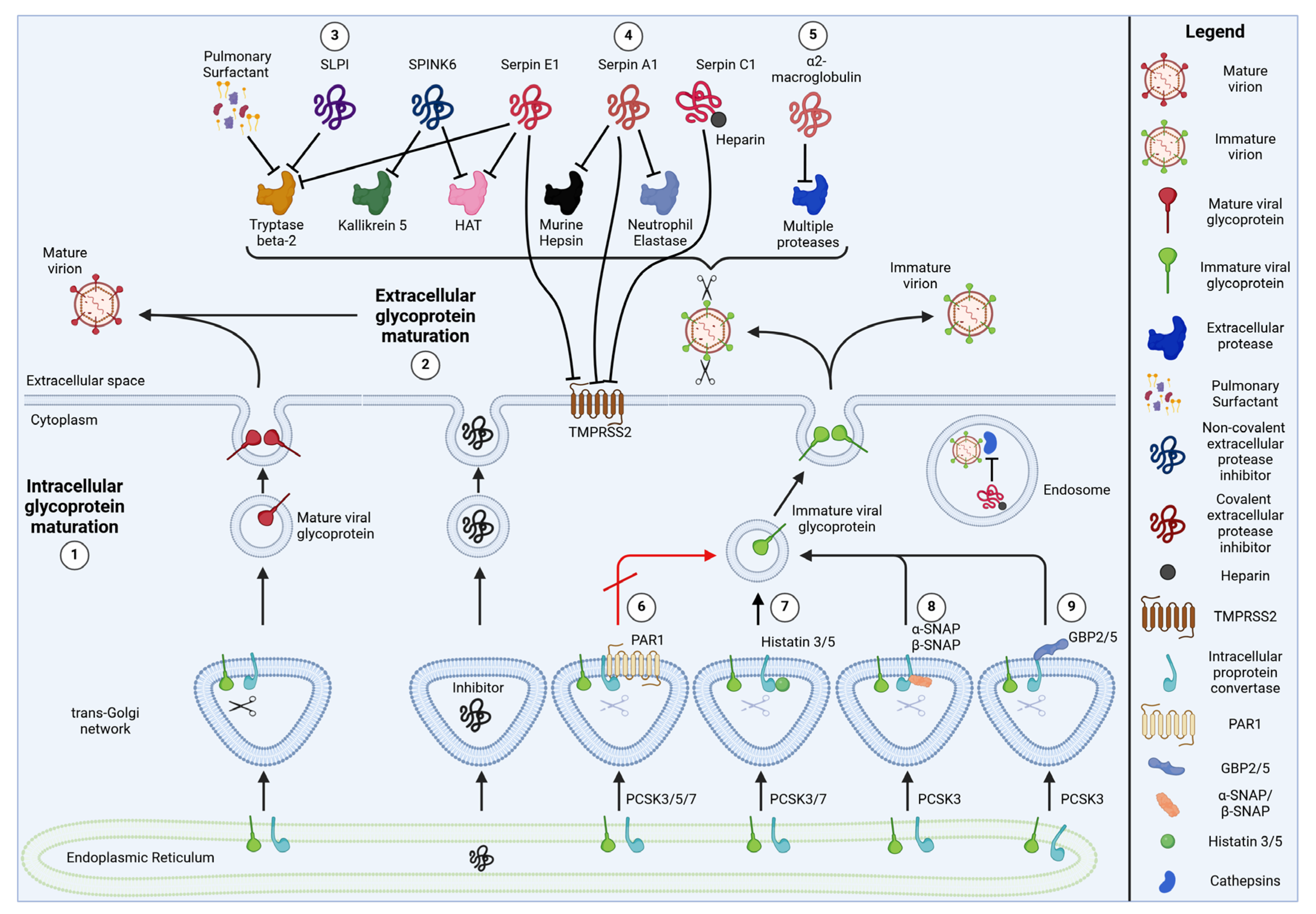

| Cellular Protease Inhibitor | Target Proteases | Modes of Inhibition | Target Viruses (Viral Glycoprotein) | References |
|---|---|---|---|---|
| Serpin E1 | HAT Tryptase beta-2 TMPRSS2 | The formation of covalently linked inactive serpin-protease dimers | Influenza A virus (HA) Sendai virus (F) SARS-CoV-2 (Spike) | [35,39] |
| Serpin A1 | TMPRSS2 Neutrophil elastase Murine Hepsin | Most likely the formation of covalently linked inactive serpin–protease dimers | SARS-CoV-2 (Spike) Influenza A virus (HA) Hepatitis C virus (unknown) | [36,37,39,40,41] |
| Serpin C1 | TMPRSS2 Cathepsin L | Most likely the formation of covalently linked inactive serpin–protease dimers | SARS-CoV-2 (Spike) MERS-CoV (Spike) SARS-CoV (Spike) hCoV-229E (Spike) | [39,42] |
| α2-macroglobulin | Many | The cleavage of a bait region induces a conformational change in the inhibitor; in a Venus flytrap-like mechanism, the inhibitor collapses and masks the active site of the protease | Influenza A virus (HA) (?) | [43] |
| PAR1 | PCSK3/furin PCSK5/PC5B PCSK7/PC7 | The sequestration of inactive protease–PAR1 complexes in the trans-Golgi network; PACS1-mediated sequestration | HIV-1 (Env) hMPV (F) | [44,45] |
| Histatins 3 and 5 | PCSK3/furin PCSK7/PC7 | Reversible and competitive inhibition; sequence similarity with inhibitory PCSK prodomains (?) | HIV (Env) (?) | [46,47] |
| α-SNAP β-SNAP | PCSK3/furin | Interaction with the P domain of furin; substrate binding not affected | SARS-CoV-2 (Spike) MERS-CoV (Spike) Ebola virus (Gp) Marburg virus (Gp) | [22] |
| GBP2/GBP5 | PCSK3/furin PCSK8/S1P (?) | The binding of the C-terminal cytosolic domain of furin; the inhibition of furin shedding; inhibition requires the isoprenylation of GBP2/GBP5 but not their GTPase activity; reduced shedding of furin | HIV-1 (Env) MLV (Env) HERV-K (Env) SARS-CoV-2 (Spike) Influenza A virus (HA) | [20,48,49] |
| MARCH8 | PCSK3/furin (?) | Interaction with furin; the sequestration of furin/glycoprotein complexes in the Golgi | Ebola virus (Gp) Influenza A virus (HA) HIV-1 (Env) | [50,51,52] |
| SPINK6 | HAT KLK5 | Unknown | Influenza A virus (HA) | [53,54] |
| SLPI | Tryptase beta-2 | Inhibition requires the C-terminal domain of SLPI (Leu72/Met73) | Sendai virus (F) Influenza A virus (HA) | |
| Surfactant | Tryptase beta-2 | Non-competitive inhibition; multiple surfactant components required | Sendai virus Influenza A virus | [55] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lotke, R.; Petersen, M.; Sauter, D. Restriction of Viral Glycoprotein Maturation by Cellular Protease Inhibitors. Viruses 2024, 16, 332. https://doi.org/10.3390/v16030332
Lotke R, Petersen M, Sauter D. Restriction of Viral Glycoprotein Maturation by Cellular Protease Inhibitors. Viruses. 2024; 16(3):332. https://doi.org/10.3390/v16030332
Chicago/Turabian StyleLotke, Rishikesh, Moritz Petersen, and Daniel Sauter. 2024. "Restriction of Viral Glycoprotein Maturation by Cellular Protease Inhibitors" Viruses 16, no. 3: 332. https://doi.org/10.3390/v16030332
APA StyleLotke, R., Petersen, M., & Sauter, D. (2024). Restriction of Viral Glycoprotein Maturation by Cellular Protease Inhibitors. Viruses, 16(3), 332. https://doi.org/10.3390/v16030332






