Molecular and Metagenomic Analyses Reveal High Prevalence and Complexity of Viral Infections in French-American Hybrids and North American Grapes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Processing
2.2. Isolation of Total RNA
2.3. Primers and Multiplex RT-PCR
2.4. High-Throughput Sequencing
3. Results
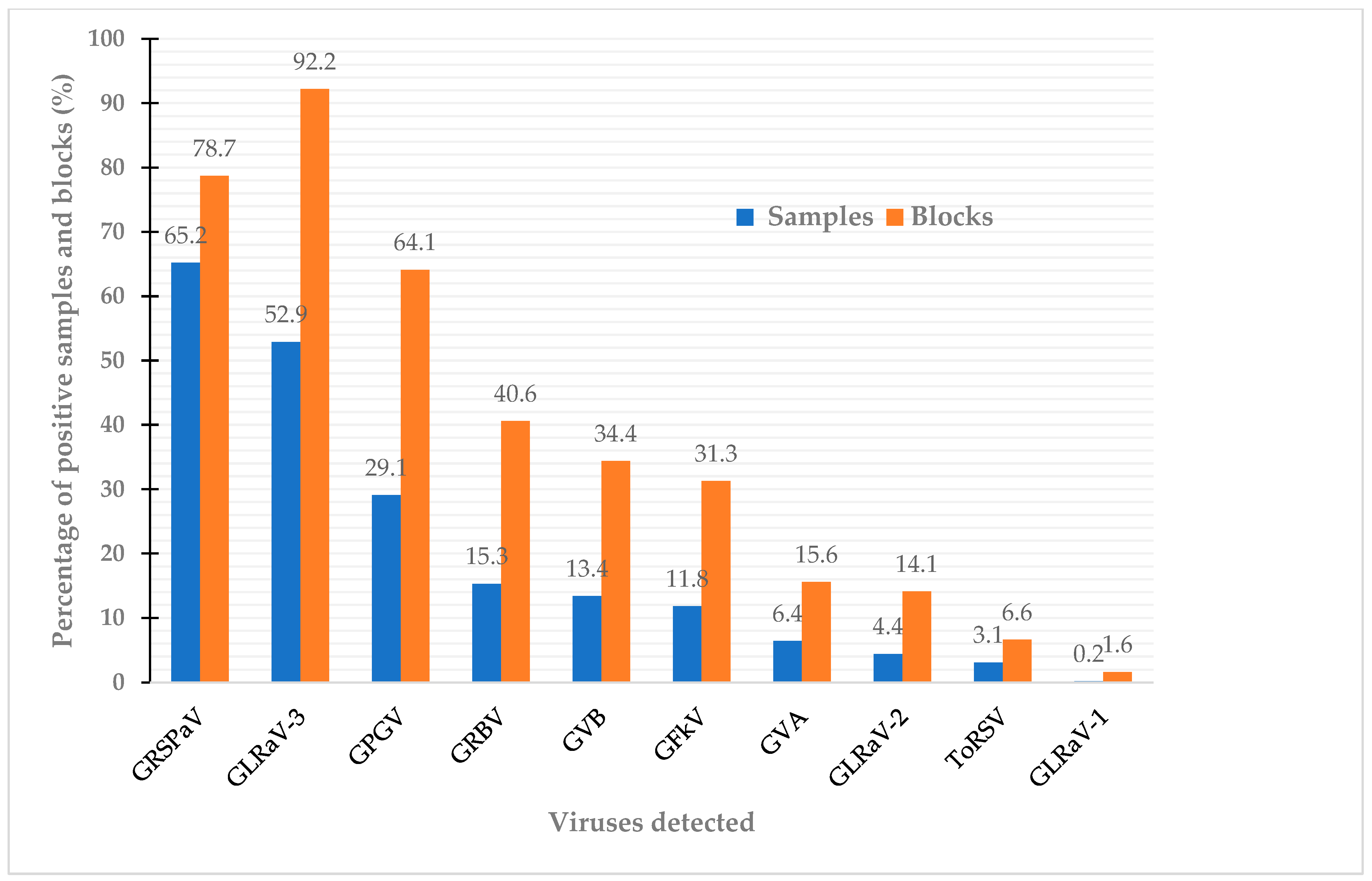
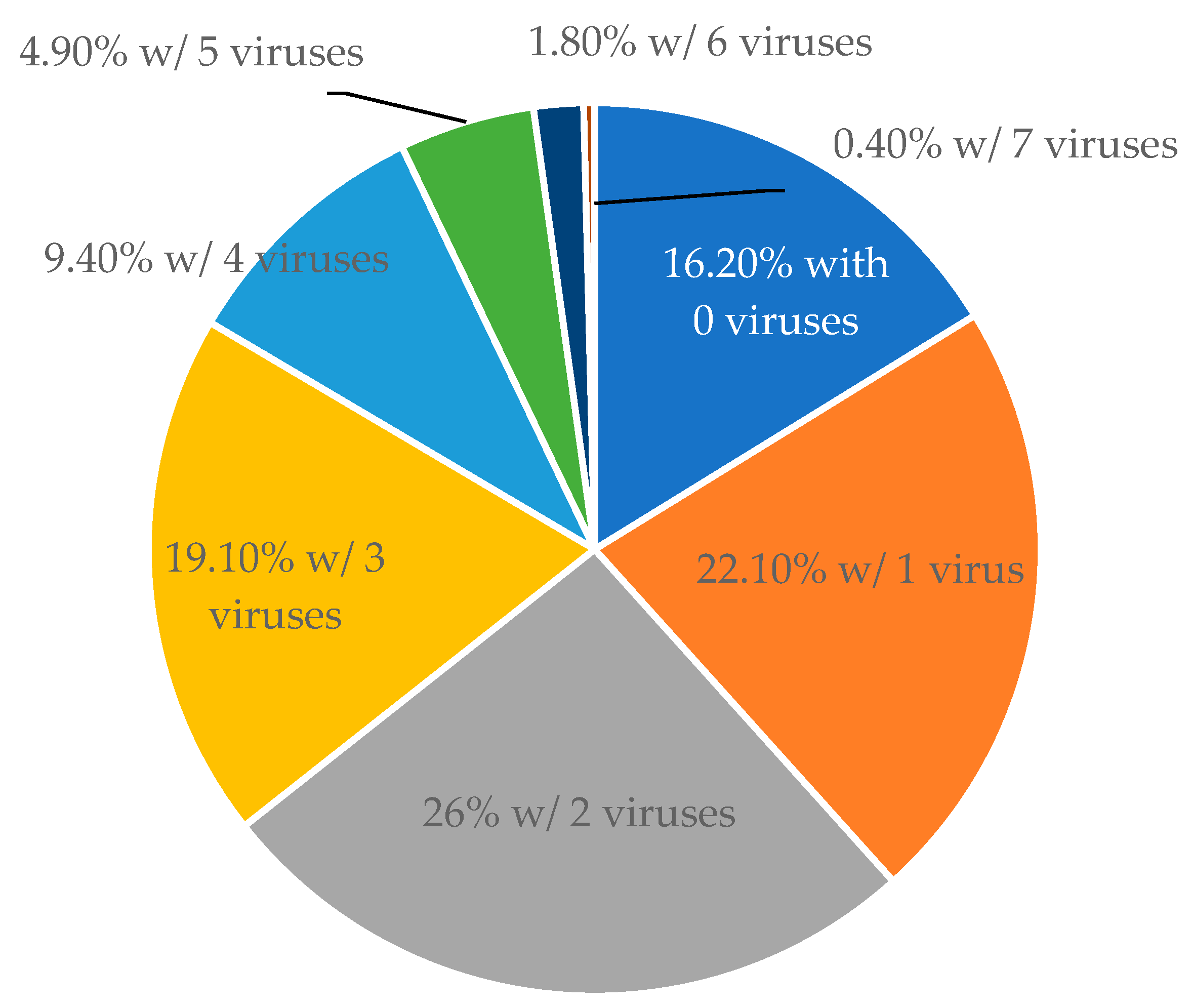
3.1. Status of Viral Infections among Non-Vinifera Grapes
3.2. Metagenomic Analysis Reveals Viromes of Varying Levels of Complexity among Hybrid Wine and Juice Grapes
3.2.1. Vidal
3.2.2. Baco
3.2.3. Niagara
3.2.4. Concord
3.2.5. Coronation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Reynolds, A.G. The Grapevine, Viticulture, and Winemaking: A Brief Introduction. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G., Golino, D., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 3–29. [Google Scholar]
- Bautista, J.; Dangl, G.S.; Yang, J.; Reisch, B.; Stover, E. Use of genetic markers to assess pedigrees of grape cultivars and breeding program. Am. J. Enol. Vitic. 2008, 59, 248–254. [Google Scholar] [CrossRef]
- Fuchs, M. Grapevine viruses: A multitude of diverse species with simple but overall poorly adopted management solutions in the vineyard. J. Plant Pathol. 2020, 102, 643–653. [Google Scholar] [CrossRef]
- Xiao, H.; Shabanian, M.; Moore, C.; Li, C.; Meng, B. Survey for major viruses in commercial Vitis vinifera wine grapes in Ontario. Virol. J. 2018, 15, 127. [Google Scholar] [CrossRef] [PubMed]
- Cieniewicz, E.; Perry, K.; Fuchs, M. Grapevine red blotch: Molecular biology of the virus and management of the disease. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G., Golino, D., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 303–314. [Google Scholar]
- Saldarelli, P.; Gualandri, V.; Malossini, U.; Glasa, M. Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G., Golino, D., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 351–364. [Google Scholar]
- Blouin, A.G.; Chooi, K.M.; Warren, B.; Napier, K.R.; Barrero, R.A.; MacDiarmid, R.M. Grapevine virus I, a putative new vitivirus detected in co-infection with grapevine virus G in New Zealand. Arch. Virol. 2018, 163, 1371–1374. [Google Scholar] [CrossRef] [PubMed]
- Blouin, A.G.; Keenan, S.; Napier, K.R.; Barrero, R.A.; MacDiarmid, R.M. Identification of a novel vitivirus from grapevines in New Zealand. Arch. Virol. 2018, 163, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Candresse, T.; Theil, S.; Faure, C.; Marais, A. Determination of the complete genomic sequence of grapevine virus H, a novel vitivirus infecting grapevine. Arch. Virol. 2018, 163, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Lara, A.; Golino, D.; Al Rwahnih, M. Genomic characterization of grapevine virus J, a novel virus identified in grapevine. Arch. Virol. 2018, 163, 1965–1967. [Google Scholar] [CrossRef] [PubMed]
- Alabi, O.J.; McBride, S.; Appel, D.N.; Al Rwahnih, M.; Pontasch, F.M. Grapevine virus M, a novel vitivirus discovered in the American hybrid bunch grape cultivar Blanc du Bois in Texas. Arch. Virol. 2019, 164, 1739–1741. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Kim, W.S.; Meng, B. A highly effective and versatile technology for the isolation of RNAs from grapevines and other woody perennials for use in virus diagnostics. Virol. J. 2015, 12, 171. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Rowhani, A. Grapevine rupestris stem pitting-associated virus. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G., Golino, D., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 257–288. [Google Scholar]
- Diaz-Lara, A.; Klaassen, V.; Stevens, K.; Sudarshana, M.R.; Rowhani, A.; Maree, H.J.; Chooi, K.M.; Blouin, A.G.; Habili, N.; Song, Y.; et al. Characterization of grapevine leafroll-associated virus 3 genetic variants and application towards RT-qPCR assay design. PLoS ONE 2018, 13, e0208862. [Google Scholar] [CrossRef] [PubMed]
- Alabi, O.J.; Poojari, S.; Sarver, K.; Martin, R.R.; Naidu, R.A. Complete genome sequence analysis of an American isolate of grapevine virus E. Virus Genes 2013, 46, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Poojari, S.; Boulé, J.; DeLury, N.; Lowery, D.T.; Rott, M.; Schmidt, A.M.; Úrbez-Torres, J.R. Epidemiology and genetic diversity of grapevine leafroll-associated viruses in British Columbia. Plant Dis. 2017, 101, 2088–2097. [Google Scholar] [CrossRef] [PubMed]
- Poojari, S.; Lowery, D.T.; Rott, M.; Schmidt, A.M.; Úrbez-Torres, J.R. Incidence, distribution and genetic diversity of grapevine red blotch virus in British Columbia. Can. J. Plant Pathol. 2017, 39, 201–211. [Google Scholar] [CrossRef]
- Milkus, B.N.; Goodman, R.N. A survey of Missouri vineyards for the presence of five grape viruses. Am. J. Enol. Vitic. 1999, 50, 133–134. [Google Scholar] [CrossRef]
- Soule, M.J.; Eastwell, K.C.; Naidu, R. First report of grapevine leafroll-associated virus 3 in American Vitis spp. grapevines in Washington State. Plant Dis. 2006, 90, 1461. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, M.; Martinson, T.E.; Loeb, G.M.; Hoch, H.C. Survey for the three major leafroll-associated viruses in Finger Lakes vineyards in New York. Plant Dis. 2009, 93, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Bahder, B.W.; Poojari, S.; Walsh, D.B.; Naidu, R.A. A survey for grapevine viruses in Washington State ‘Concord’ (Vitis x labruscana L.) vineyards. Plant Health Progress. 2013, 14. [Google Scholar] [CrossRef]
- MacKenzie, D.J.; Johnson, R.C.; Warner, C. Incidence of four important viral pathogens in Canadian vineyards. Plant Dis. 1996, 80, 955–958. [Google Scholar] [CrossRef]
- Poojari, S.; Moreau, D.L.; Kahl, D.; Ritchie, M.; Ali, S.; Úrbez-Torres, J.R. Disease incidence and genetic variability of economically important grapevine viruses in Nova Scotia. Can. J. Plant Pathol. 2020, 42, 584–594. [Google Scholar] [CrossRef]
- Fall, M.L.; Xu, D.; Lemoyne, P.; Moussa, I.E.B.; Beaulieu, C.; Carisse, O. A Diverse Virome of Leafroll-Infected Grapevine Unveiled by dsRNA Sequencing. Viruses 2020, 12, 1142. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Harding, J.; Vouillamoz, J. Wine Grapes—A Complete Guide to 1368 Vine Varieties, Including Their Origins and Flavours; Penguin: London, UK; Harper Collins: Broadway, NY, USA, 2012; pp. 851, 875 & 1136–1135. [Google Scholar]
- Appellation America “Vidal Blanc”. Available online: https://www.casalarga.com/vidal-blanc/ (accessed on 8 April 2013.).
- Burger, J.T.; Maree, H.J.; Gouveia, P.; Naidu, R.A. Grapevine leafroll-associated virus 3. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G., Golino, D., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 167–195. [Google Scholar]
- Digiaro, M.; Elbeaino, T.; Martelli, G.P. Grapevine fanleaf virus and other old world nepoviruses. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G., Golino, D., Fuchs, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 47–82. [Google Scholar]
| Type of Grapes | Cultivar | No. of Vineyard Blocks | No. of Samples |
|---|---|---|---|
| French-American hybrid wine grapes | Vidal | 10 | 78 |
| Baco | 7 | 80 | |
| De Chaunac | 7 | 44 | |
| Chambourcin | 4 | 24 | |
| Marquette | 3 | 44 | |
| Marechal Foch | 5 | 31 | |
| Frontenac 1 | 5 | 49 | |
| North American grapes | Concord | 7 | 59 |
| Niagara | 8 | 70 | |
| Table grapes | Sovereign coronation | 7 | 54 |
| Total | 10 | 63 | 533 |
| Cultivars | GRBV | GLRaV-1 | GLRaV-2 | GLRaV-3 | GRSPaV | GPGV | GVA | GVB | GFkV | ToRSV |
|---|---|---|---|---|---|---|---|---|---|---|
| North American juice grapes: | ||||||||||
| Niagara | 12.9 | 0 | 0 | 67.1 | 73.3 | 25.7 | 18.6 | 0 | 10 | 0 |
| Concord | 23.3 | 0 | 0 | 64.4 | 40.8 | 13.6 | 1.7 | 0 | 0 | 0 |
| North American table grapes: | ||||||||||
| Coronation | 13 | 0 | 0 | 44.4 | 63 | 7.4 | 0 | 5.6 | 37 | 0 |
| French-American hybrid wine grapes: | ||||||||||
| Vidal | 11.5 | 0 | 30.8 | 65.4 | 90.5 | 56.4 | 7.7 | 38.5 | 39.7 | 4.8 |
| Baco | 45 | 1.25 | 0 | 36.3 | 55.4 | 26.3 | 5.0 | 37.5 | 2.5 | 0 |
| De Chaunac | 2.3 | 0 | 0 | 50 | 72.7 | 13.6 | 0 | 0 | 6.8 | 22.8 |
| Chambourcin | 29.1 | 0 | 0 | 17 | 83.3 | 58.3 | 0 | 0 | 4.2 | 0 |
| Marquette | 0 | 0 | 0 | 40.9 | 52.2 | 15.9 | 25.0 | 0 | 2.3 | 0 |
| Marechal Foch | 0 | 0 | 0 | 83.9 | 87.1 | 32.3 | 0 | 9.7 | 22.6 | 0 |
| Frontenac | 0 | 0 | 0 | 4.1 | 32.7 | 8.2 | 0 | 0 | 8.2 | 4.1 |
| Names of Viruses and Viroids | Read Counts (% of Total Viral Reads) | ||||
|---|---|---|---|---|---|
| Vidal | Baco | Niagara | Concord | Coronation | |
| Total reads | 88,054,982 | 80,463,420 | 92,320,068 | 100,548,656 | 94,379,964 |
| Reads not matching grapevine sequence | 3,379,815 | 1,780,046 | 5,425,877 | 2,615,202 | 2,163,617 |
| GRBV | - | 5881 | 6798 (2.5) | 1551 | - |
| GLRaV-2 | 186,764 (24.6) | - | - | - | - |
| GLRaV-3 | 45,254 (6.0) | - | 105,966 (39) | 1044 | - |
| GRSPaV | 187,726 (24.7) | 539 | 120,285 (44.3) | 46,423 | 223 |
| GPGV | 8913 (1.2) | 3248 | 17,264 (6.4) | - | 21,178 |
| GVA | 3963 (0.52) | - | 3141 (1.2) | - | - |
| GVB | 127,982 (16.9) | 110 | - | - | - |
| GVE | 53,004 (7.0) | - | 17,323 (6.4) | - | - |
| GVQ | 2595 (0.34) | - | - | - | - |
| ArMV | 102,627 (13.5) | - | - | - | - |
| GFkV | 34,892 (4.6) | - | - | - | - |
| GSyV-1 | 2138 (0.28) | - | - | - | - |
| GAMaV | 1764 (0.23) | - | - | - | - |
| GRVFV | 542 (0.07) | - | - | - | - |
| GRGV | 720 (0.095) | - | - | - | - |
| TSV | - | - | 744 (0.27) | - | 793 |
| Total viral reads | 758,856 | 9778 | 271,321 | 49,018 | 21,401 |
| Viroids (HSVd, GYSVd1 and GYSVd2) | 6626 | 4362 | 17,483 | 7003 | 26,455 |
| Viruses | GenBank Accession No. | Reference Isolates | Sequence Identity (%) | No. of Contigs | Genome Coverage (%) |
|---|---|---|---|---|---|
| GLRaV-2 | KX774192.1 | ISA-BR | 99 | 1 | 100 |
| FJ436234.1 | OR1 | 99–100 | 3 | 88 | |
| GLRaV-3 | GU983863.1 | WA-MR | 97–100 | 3 | 99 |
| MK032068.1 | Vdl | 100 | 1 | 99 | |
| GRSPaV | KX925556.1 | TEMP-BR | 98 | 1 | 88 |
| KX925556.1 | TEMP-BR | 94 | 1 | 73 | |
| KX274275.1 | SK704-B | 94 | 1 | 96 | |
| KX274275.1 | SK704-B | 96–98 | 11 | 94 | |
| AY881627.1 | BS | 97 | 1 | 58 | |
| KT948710.1 | VF1 | 94 | 2 | 99 | |
| HE591388.1 | PG | 97–98 | 7 | 99 | |
| FR691076.1 | MG | 98 | 1 | 93 | |
| KX958435.1 | CS-BR | 97–98 | 2 | 62 | |
| AY368590.1 | Syrah | 93–96 | 2 | 31 | |
| GPGV | KR528581.1 | Tannat-GvPGV | 97–99 | 9 | 100 |
| GVA | DQ855084.2 | GTG11-1 | 82 | 1 | 99 |
| GVB | KY426923,1 | 8415 | 92 | 13 | 80 |
| GVE | JX402759.1 | WAHH2 | 98 | 1 | 100 |
| JX402759.1 | WAHH2 | 73–83 | 5 | 94 | |
| GVG | MF405923.1 | VID561 | 68–78 | 3 | 28 |
| ArMV | AY303786.1 | NW (RNA1) | 84 | 1 | 97 |
| AY017339.1 | NW (RNA2) | 89 | 1 | 78 | |
| GFkV | AJ309022.1 | M48 | 89 | 10 | 78 |
| GSyV-1 | KT037017.1 | MH | 95–97 | 2 | 94 |
| KX130754.1 | TRAJ-BR | 96 | 1 | 61 | |
| GAMaV | KY123917.1 | CS | 86–91 | 3 | 28 |
| GRVFV | KY513701.1 | Mauzae | 82–86 | 5 | 94 |
| GRGV | KX171167.1 | Graciano-T53 | 87–90 | 2 | 29 |
| Viruses | GenBank Accession Numbers | Reference Isolates | Sequence Identity (%) | No. of Contigs | Genome Coverage (%) |
|---|---|---|---|---|---|
| GLRaV-3 | GQ352632.1 | 623 | 99–100 | 2 | 98.8 |
| GU983863.1 | WA-MR | 100 | 1 | 98.6 | |
| GU983863.1 | WA-MR | 93–96 | 6 | 86.5 | |
| KY073324.1 | 8415B | 100 | 1 | 99.7 | |
| JQ655295.1 | Vdl | 91.9–93.5 | 3 | 97.6 | |
| GRSPaV | KR054734.1 | JF | 98 | 1 | 99.1 |
| FR691076.1 | MG | 98 | 1 | 100 | |
| KX035004.1 | SGM5 clone 1 | 98 | 2 | 99.3 | |
| GVA | KC962564.1 | I327-5 | 84 | 1 | 99.9 |
| GPGV | KM491305.1 | MER | 99 | 1 | 98 |
| GVE | GU903012.1 | SA94 | 76 | 1 | 97 |
| TSV | FJ403375.1 | Illinois, RNA1 | 99 | 2 | 66.9 |
| FJ403376.1 | Illinois, RNA2 | 100 | 2 | 59.4 | |
| FJ403377.1 | Illinois, RNA3 | 91 | 1 | 97.0 | |
| GRBV | KY426922.1 | 93–26 | 100 | 1 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, H.; Meng, B. Molecular and Metagenomic Analyses Reveal High Prevalence and Complexity of Viral Infections in French-American Hybrids and North American Grapes. Viruses 2023, 15, 1949. https://doi.org/10.3390/v15091949
Xiao H, Meng B. Molecular and Metagenomic Analyses Reveal High Prevalence and Complexity of Viral Infections in French-American Hybrids and North American Grapes. Viruses. 2023; 15(9):1949. https://doi.org/10.3390/v15091949
Chicago/Turabian StyleXiao, Huogen, and Baozhong Meng. 2023. "Molecular and Metagenomic Analyses Reveal High Prevalence and Complexity of Viral Infections in French-American Hybrids and North American Grapes" Viruses 15, no. 9: 1949. https://doi.org/10.3390/v15091949
APA StyleXiao, H., & Meng, B. (2023). Molecular and Metagenomic Analyses Reveal High Prevalence and Complexity of Viral Infections in French-American Hybrids and North American Grapes. Viruses, 15(9), 1949. https://doi.org/10.3390/v15091949





