Novel RNA-Seq Signatures Post-Methamphetamine and Oxycodone Use in a Model of HIV-Associated Neurocognitive Disorders
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
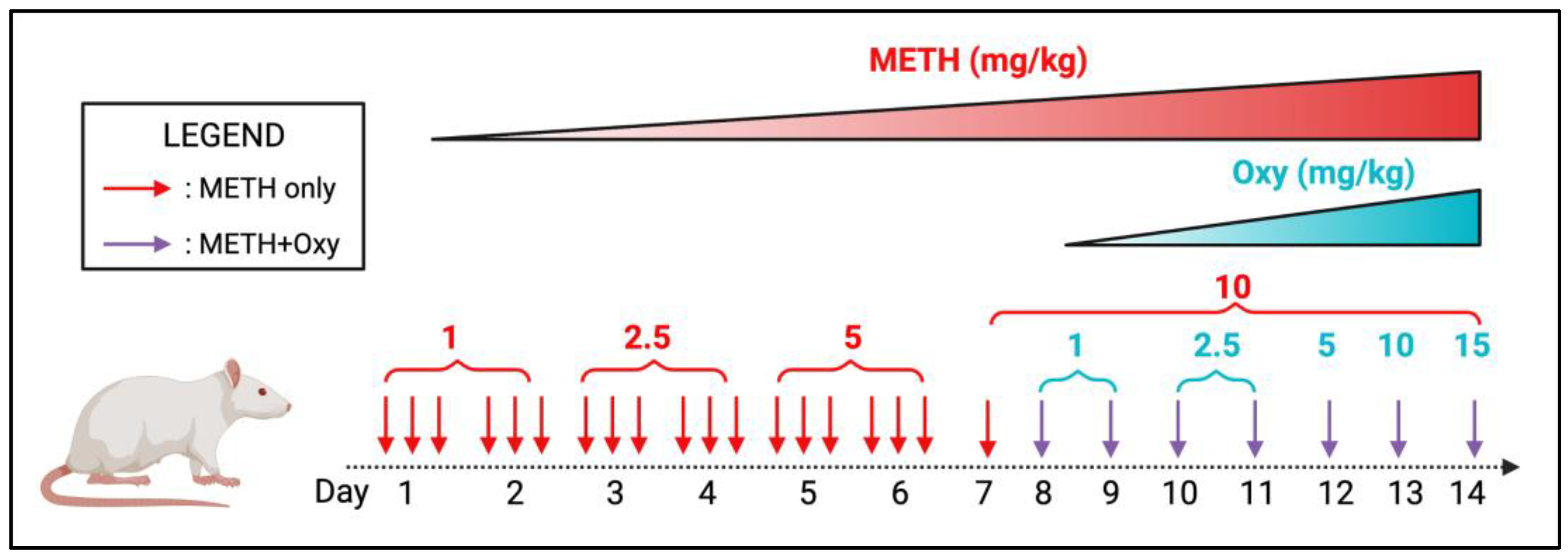
2.2. Polysubstance Administration
2.3. Total RNA Extraction, Quality Control, Library Preparation, and RNA Sequencing
2.4. Bioinformatic Data Analysis
3. Results
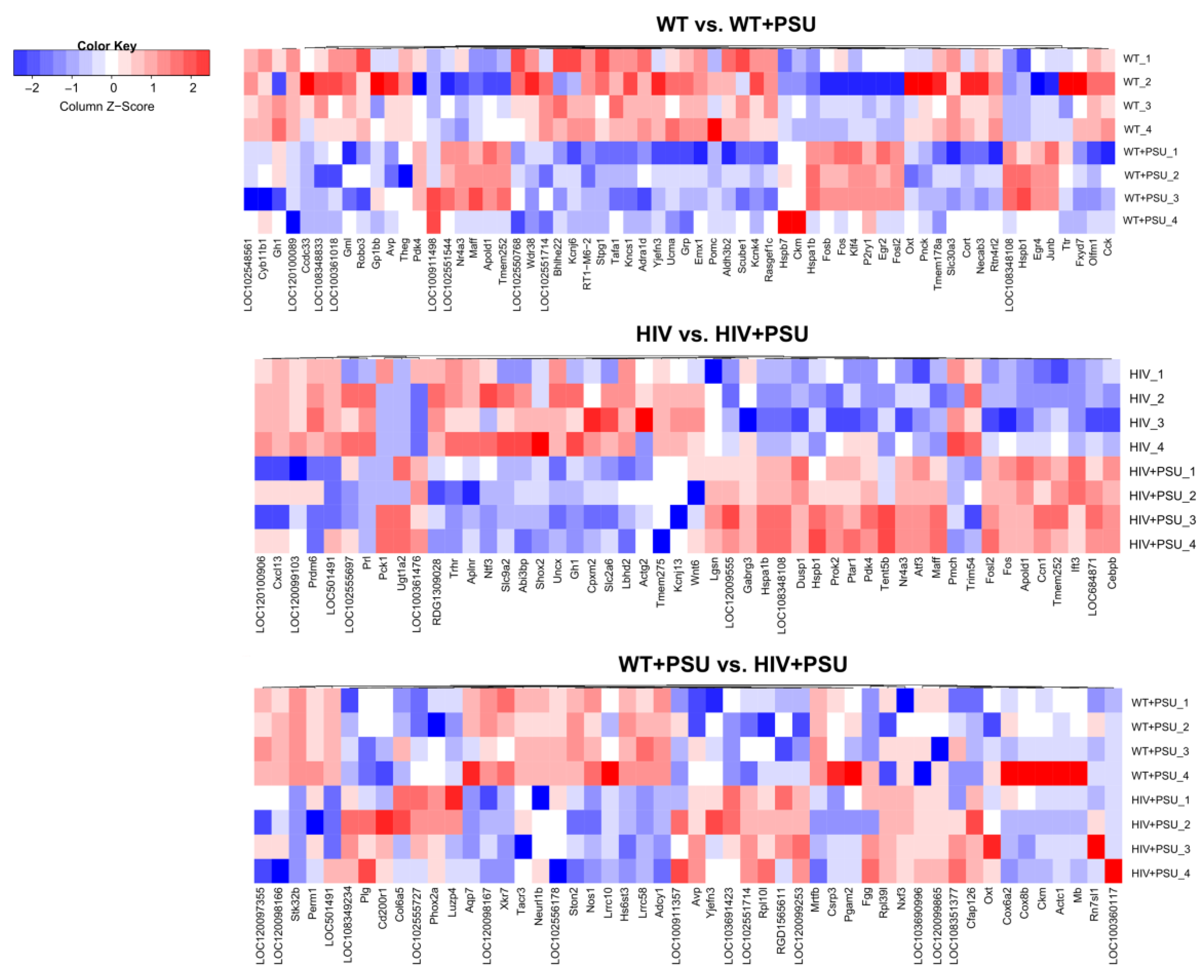
3.1. HIV and Chronic PSU Leads to Changes in the Striatum Transcriptome
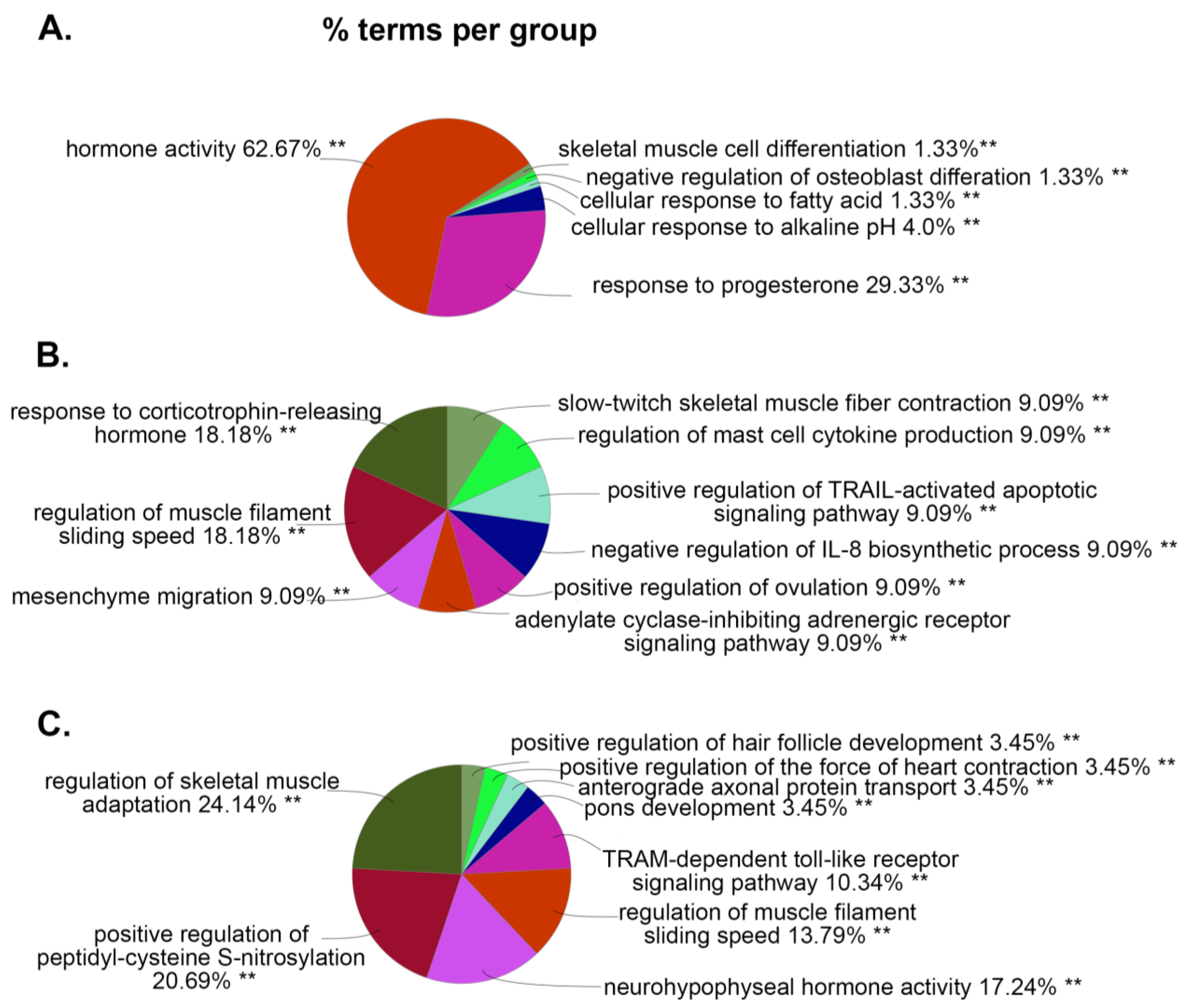
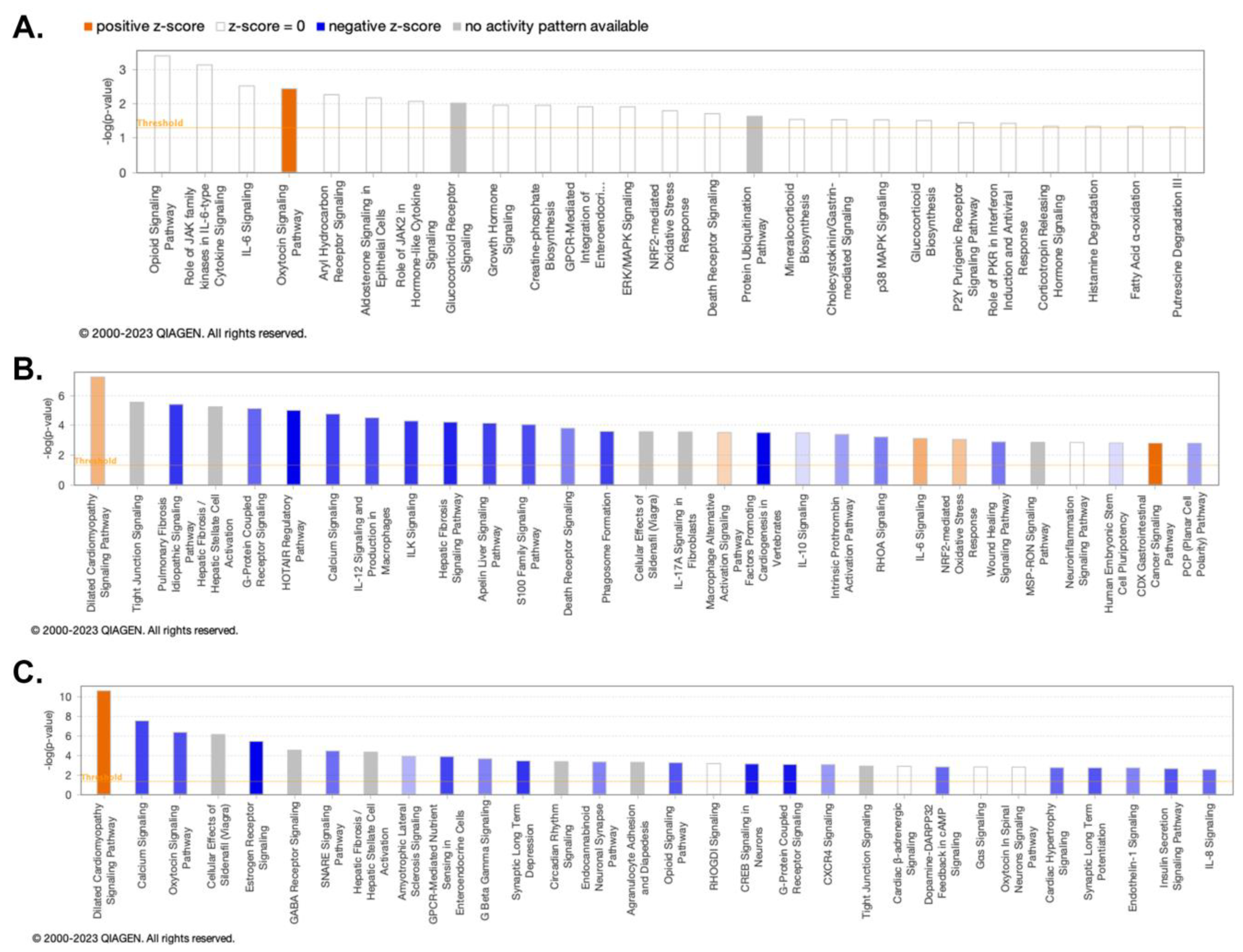
3.2. ClueGO and IPA Analysis Demonstrates the Molecular Activities and Pathways Associated with HIV and Chronic PSU
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cicero, T.J.; Ellis, M.S.; Kasper, Z.A. Polysubstance Use: A Broader Understanding of Substance Use during the Opioid Crisis. Am. J. Public Health 2020, 110, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Crummy, E.A.; O’Neal, T.J.; Baskin, B.M.; Ferguson, S.M. One Is Not Enough: Understanding and Modeling Polysubstance Use. Front. Neurosci. 2020, 14, 569. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, J.; Gladden, R.M.; Mattson, C.L.; Hunter, C.T.; Davis, N.L. Vital Signs: Characteristics of Drug Overdose Deaths Involving Opioids and Stimulants—24 States and the District of Columbia, January–June 2019. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 1189–1197. [Google Scholar] [CrossRef] [PubMed]
- Compton, W.M.; Valentino, R.J.; DuPont, R.L. Polysubstance use in the U.S. opioid crisis. Mol. Psychiatry 2021, 26, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, E.; Sivalingam, K.; Doke, M.; Samikkannu, T. Effects of Drugs of Abuse on the Blood-Brain Barrier: A Brief Overview. Front. Neurosci. 2020, 14, 513. [Google Scholar] [CrossRef]
- CDC. Substance Use and Sexual Risk Behaviors. Available online: https://www.cdc.gov/healthyyouth/substance-use/dash-substance-use-fact-sheet.htm (accessed on 26 January 2023).
- Baskin-Sommers, A.; Sommers, I. The co-occurrence of substance use and high-risk behaviors. J. Adolesc. Health 2006, 38, 609–611. [Google Scholar] [CrossRef]
- Wang, S.-C.; Maher, B. Substance Use Disorder, Intravenous Injection, and HIV Infection: A Review. Cell Transplant. 2019, 28, 1465–1471. [Google Scholar] [CrossRef]
- Scutari, R.; Alteri, C.; Perno, C.F.; Svicher, V.; Aquaro, S. The Role of HIV Infection in Neurologic Injury. Brain Sci. 2017, 7, 38. [Google Scholar] [CrossRef]
- Lashomb, A.L.; Vigorito, M.; Chang, S.L. Further characterization of the spatial learning deficit in the human immunodeficiency virus-1 transgenic rat. J. Neurovirol. 2009, 15, 14–24. [Google Scholar] [CrossRef]
- Kass, M.D.; Liu, X.; Vigorito, M.; Chang, L.; Chang, S.L. Methamphetamine-induced behavioral and physiological effects in adolescent and adult HIV-1 transgenic rats. J. Neuroimmune Pharmacol. Off. J. Soc. NeuroImmune Pharmacol. 2010, 5, 566–573. [Google Scholar] [CrossRef]
- Vigorito, M.; Cao, J.; Li, M.D.; Chang, S.L. Acquisition and long-term retention of spatial learning in the human immunodeficiency virus-1 transgenic rat: Effects of repeated nicotine treatment. J. Neurovirol. 2013, 19, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Reid, W.; Sadowska, M.; Denaro, F.; Rao, S.; Foulke, J., Jr.; Hayes, N.; Jones, O.; Doodnauth, D.; Davis, H.; Sill, A.; et al. An HIV-1 transgenic rat that develops HIV-related pathology and immunologic dysfunction. Proc. Natl. Acad. Sci. USA 2001, 98, 9271–9276. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Schrott, L.M.; Arnold, T.; Snelling, S.; Rao, M.; Graham, D.; Cornelius, A.; Korneeva, N.L. Chronic oxycodone induces axonal degeneration in rat brain. BMC Neurosci. 2018, 19, 15. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 14 July 2023).
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2013, 30, 523–530. [Google Scholar] [CrossRef]
- Honda, S.; Sasaki, Y.; Ohsawa, K.; Imai, Y.; Nakamura, Y.; Inoue, K.; Kohsaka, S. Extracellular ATP or ADP Induce Chemotaxis of Cultured Microglia through Gi/o-Coupled P2Y Receptors. J. Neurosci. 2001, 21, 1975–1982. [Google Scholar] [CrossRef]
- Davalos, D.; Grutzendler, J.; Yang, G.; Kim, J.V.; Zuo, Y.; Jung, S.; Littman, D.R.; Dustin, M.L.; Gan, W.-B. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 2005, 8, 752–758. [Google Scholar] [CrossRef]
- Berríos-Cárcamo, P.; Quezada, M.; Quintanilla, M.E.; Morales, P.; Ezquer, M.; Herrera-Marschitz, M.; Israel, Y.; Ezquer, F. Oxidative Stress and Neuroinflammation as a Pivot in Drug Abuse. A Focus on the Therapeutic Potential of Antioxidant and Anti-Inflammatory Agents and Biomolecules. Antioxidants 2020, 9, 830. [Google Scholar] [CrossRef]
- Cuzzo, B.; Padala, S.A.; Lappin, S.L. Physiology, Vasopressin. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Caldwell, H.K.; Lee, H.J.; Macbeth, A.H.; Young, W.S., 3rd. Vasopressin: Behavioral roles of an “original” neuropeptide. Prog. Neurobiol. 2008, 84, 1–24. [Google Scholar] [CrossRef]
- Logrip, M.L.; Koob, G.F.; Zorrilla, E.P. Role of corticotropin-releasing factor in drug addiction: Potential for pharmacological intervention. CNS Drugs 2011, 25, 271–287. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Macbeth, A.H.; Pagani, J.H.; Young, W.S., 3rd. Oxytocin: The great facilitator of life. Prog. Neurobiol. 2009, 88, 127–151. [Google Scholar] [CrossRef] [PubMed]
- Neumann, I.D.; Slattery, D.A. Oxytocin in General Anxiety and Social Fear: A Translational Approach. Biol. Psychiatry 2016, 79, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Lindenberg, A.; Domes, G.; Kirsch, P.; Heinrichs, M. Oxytocin and vasopressin in the human brain: Social neuropeptides for translational medicine. Nat. Rev. Neurosci. 2011, 12, 524–538. [Google Scholar] [CrossRef] [PubMed]
- Uhrig, S.; Hirth, N.; Broccoli, L.; von Wilmsdorff, M.; Bauer, M.; Sommer, C.; Zink, M.; Steiner, J.; Frodl, T.; Malchow, B.; et al. Reduced oxytocin receptor gene expression and binding sites in different brain regions in schizophrenia: A post-mortem study. Schizophr. Res. 2016, 177, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Bartholomeusz, C.F.; Ganella, E.P.; Labuschagne, I.; Bousman, C.; Pantelis, C. Effects of oxytocin and genetic variants on brain and behaviour: Implications for treatment in schizophrenia. Schizophr. Res. 2015, 168, 614–627. [Google Scholar] [CrossRef]
- Sundar, M.; Patel, D.; Young, Z.; Leong, K.C. Oxytocin and Addiction: Potential Glutamatergic Mechanisms. Int. J. Mol. Sci. 2021, 22, 2405. [Google Scholar] [CrossRef]
- Leong, K.C.; Zhou, L.; Ghee, S.M.; See, R.E.; Reichel, C.M. Oxytocin decreases cocaine taking, cocaine seeking, and locomotor activity in female rats. Exp. Clin. Psychopharmacol. 2016, 24, 55–64. [Google Scholar] [CrossRef]
- Edinoff, A.N.; Thompson, E.; Merriman, C.E.; Alvarez, M.R.; Alpaugh, E.S.; Cornett, E.M.; Murnane, K.S.; Kozinn, R.L.; Shah-Bruce, M.; Kaye, A.M.; et al. Oxytocin, a Novel Treatment for Methamphetamine Use Disorder. Neurol. Int. 2022, 14, 186–198. [Google Scholar] [CrossRef]
- King, C.E.; Gano, A.; Becker, H.C. The role of oxytocin in alcohol and drug abuse. Brain Res. 2020, 1736, 146761. [Google Scholar] [CrossRef]
- Berridge, M.J. Dysregulation of neural calcium signaling in Alzheimer disease, bipolar disorder and schizophrenia. Prion 2013, 7, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.T. HIV-1 Tat-Mediated Calcium Dysregulation and Neuronal Dysfunction in Vulnerable Brain Regions. Curr. Drug Targets 2016, 17, 4–14. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Athota, P.; Nguyen, N.M.; Schaal, V.L.; Jagadesan, S.; Guda, C.; Yelamanchili, S.V.; Pendyala, G. Novel RNA-Seq Signatures Post-Methamphetamine and Oxycodone Use in a Model of HIV-Associated Neurocognitive Disorders. Viruses 2023, 15, 1948. https://doi.org/10.3390/v15091948
Athota P, Nguyen NM, Schaal VL, Jagadesan S, Guda C, Yelamanchili SV, Pendyala G. Novel RNA-Seq Signatures Post-Methamphetamine and Oxycodone Use in a Model of HIV-Associated Neurocognitive Disorders. Viruses. 2023; 15(9):1948. https://doi.org/10.3390/v15091948
Chicago/Turabian StyleAthota, Pranavi, Nghi M. Nguyen, Victoria L. Schaal, Sankarasubramanian Jagadesan, Chittibabu Guda, Sowmya V. Yelamanchili, and Gurudutt Pendyala. 2023. "Novel RNA-Seq Signatures Post-Methamphetamine and Oxycodone Use in a Model of HIV-Associated Neurocognitive Disorders" Viruses 15, no. 9: 1948. https://doi.org/10.3390/v15091948
APA StyleAthota, P., Nguyen, N. M., Schaal, V. L., Jagadesan, S., Guda, C., Yelamanchili, S. V., & Pendyala, G. (2023). Novel RNA-Seq Signatures Post-Methamphetamine and Oxycodone Use in a Model of HIV-Associated Neurocognitive Disorders. Viruses, 15(9), 1948. https://doi.org/10.3390/v15091948





_Guda.png)

