The Influence of the Omicron Variant on RNA Extraction and RT-qPCR Detection of SARS-CoV-2 in a Laboratory in Brazil
Abstract
1. Introduction
2. Materials and Methods
2.1. Nucleic Acid Extraction
2.2. RT-qPCR Analysis
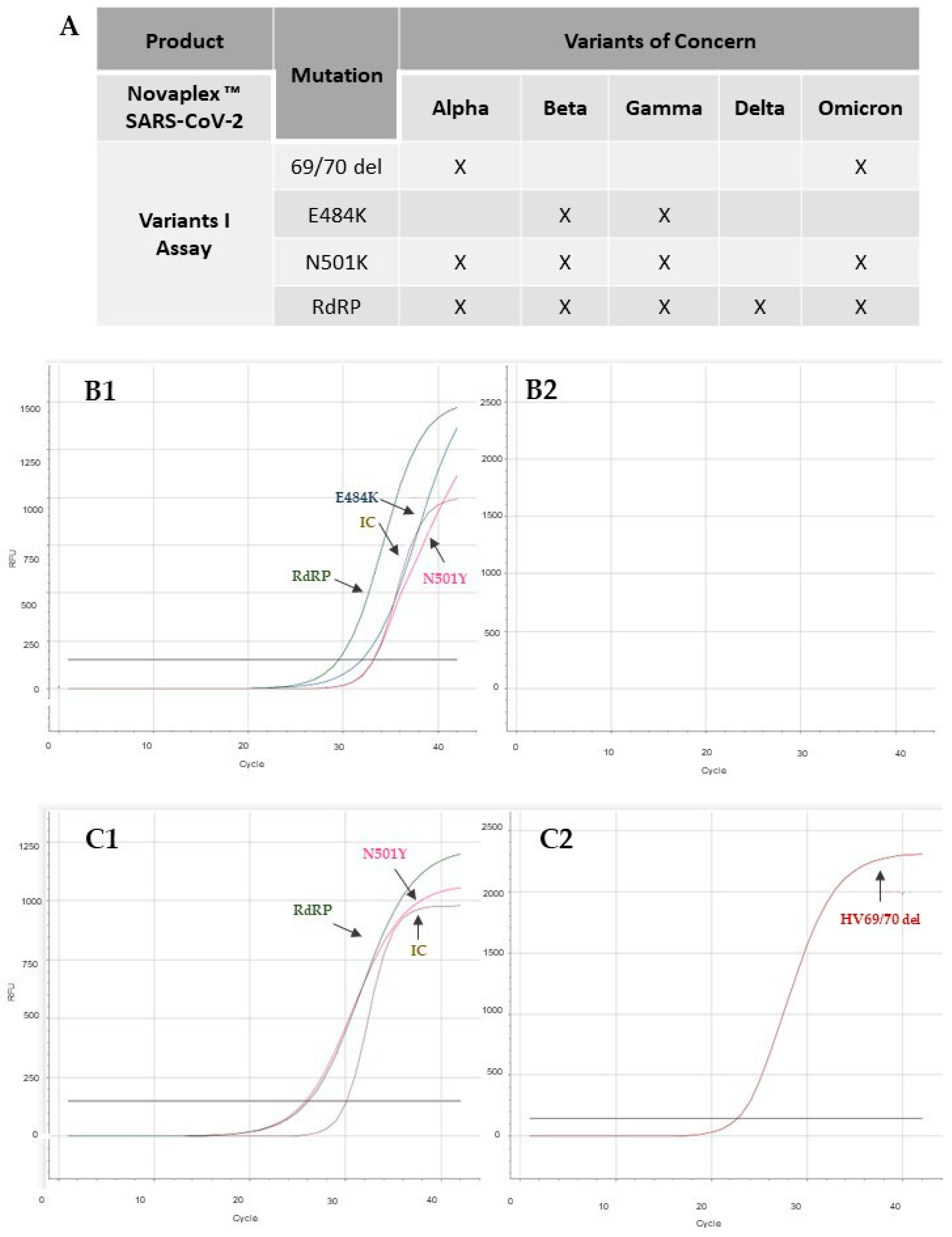
2.3. Analysis of SARS-CoV-2 Variant Using RT-qPCR
2.4. Ethics
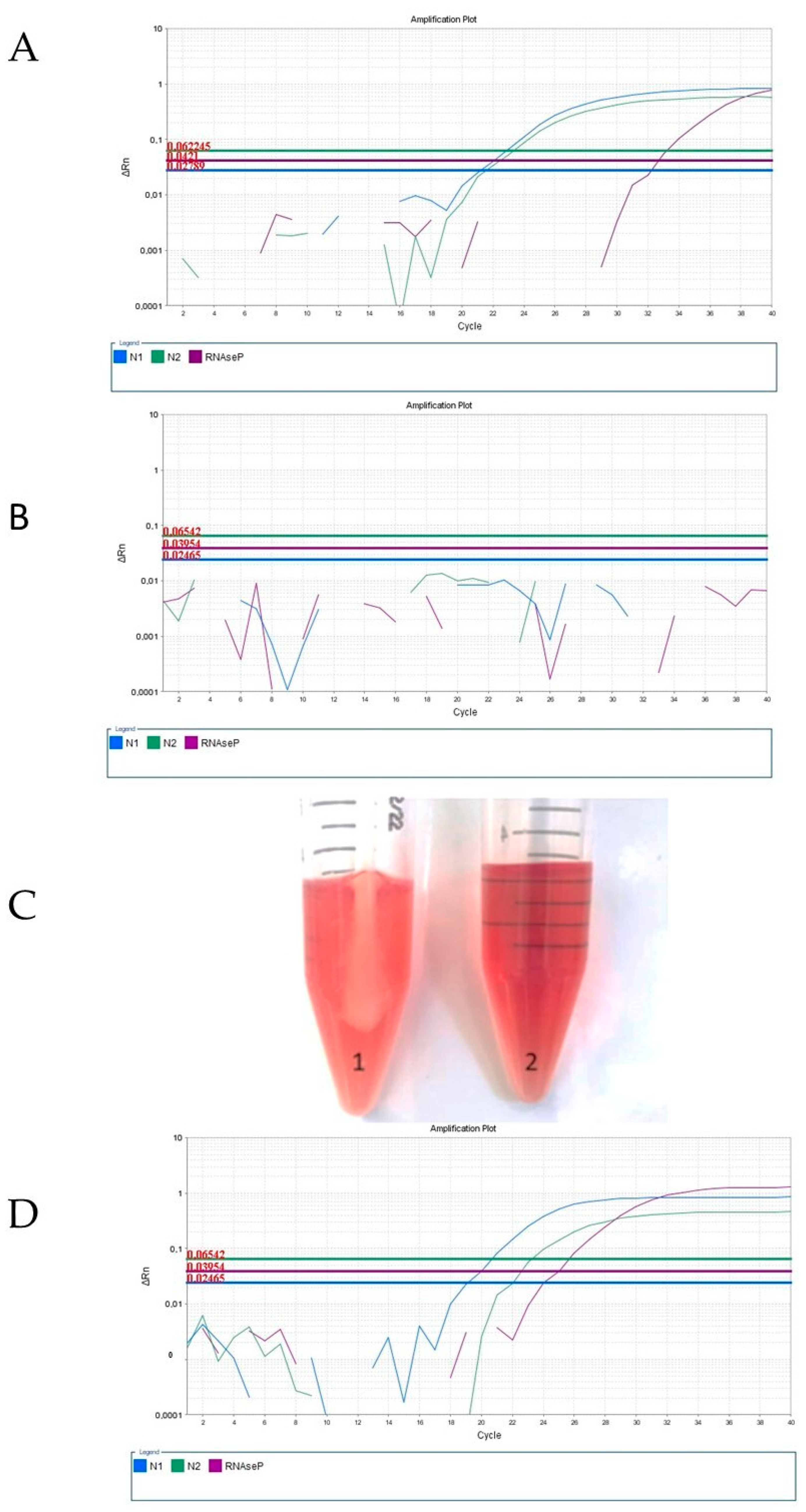
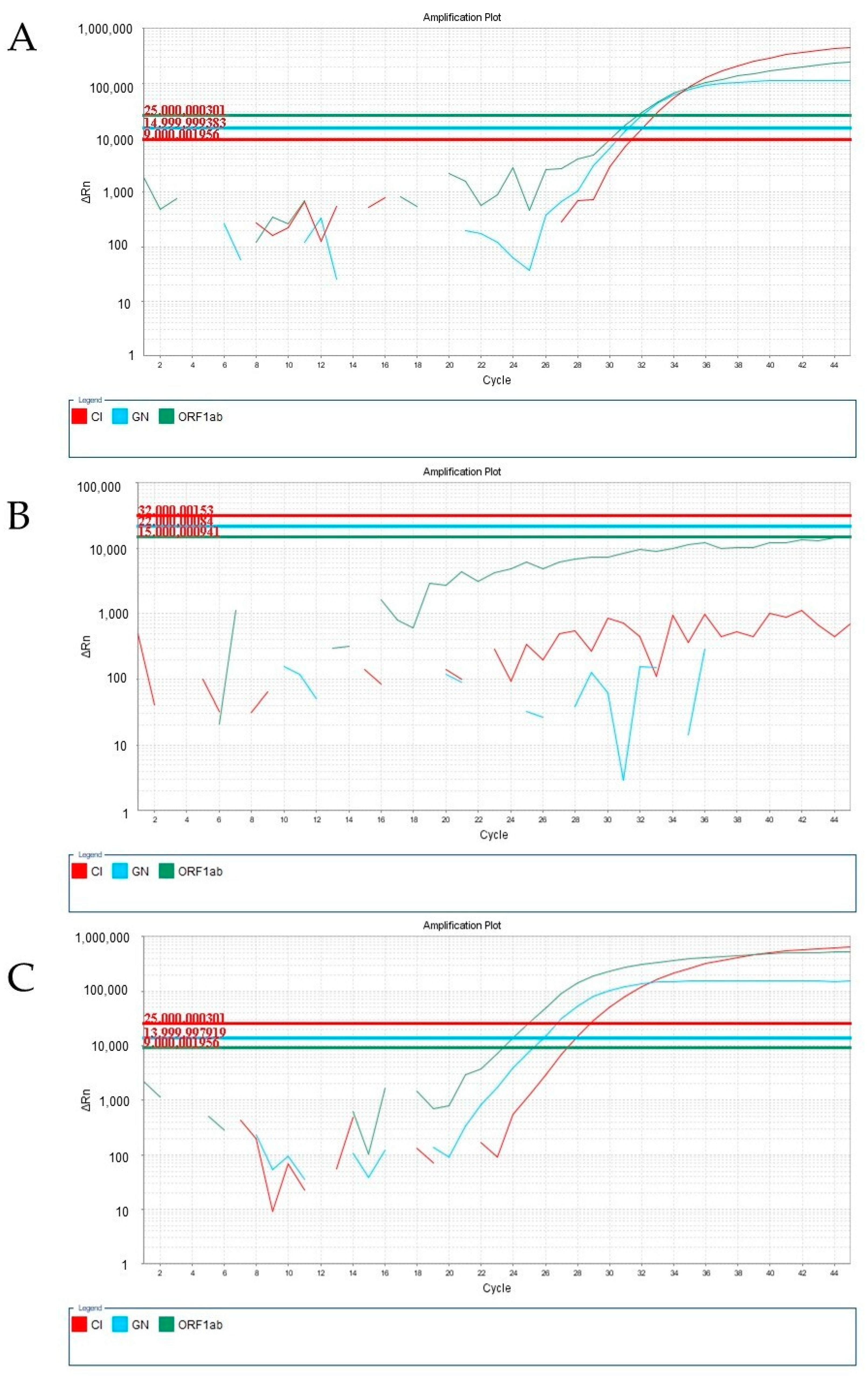
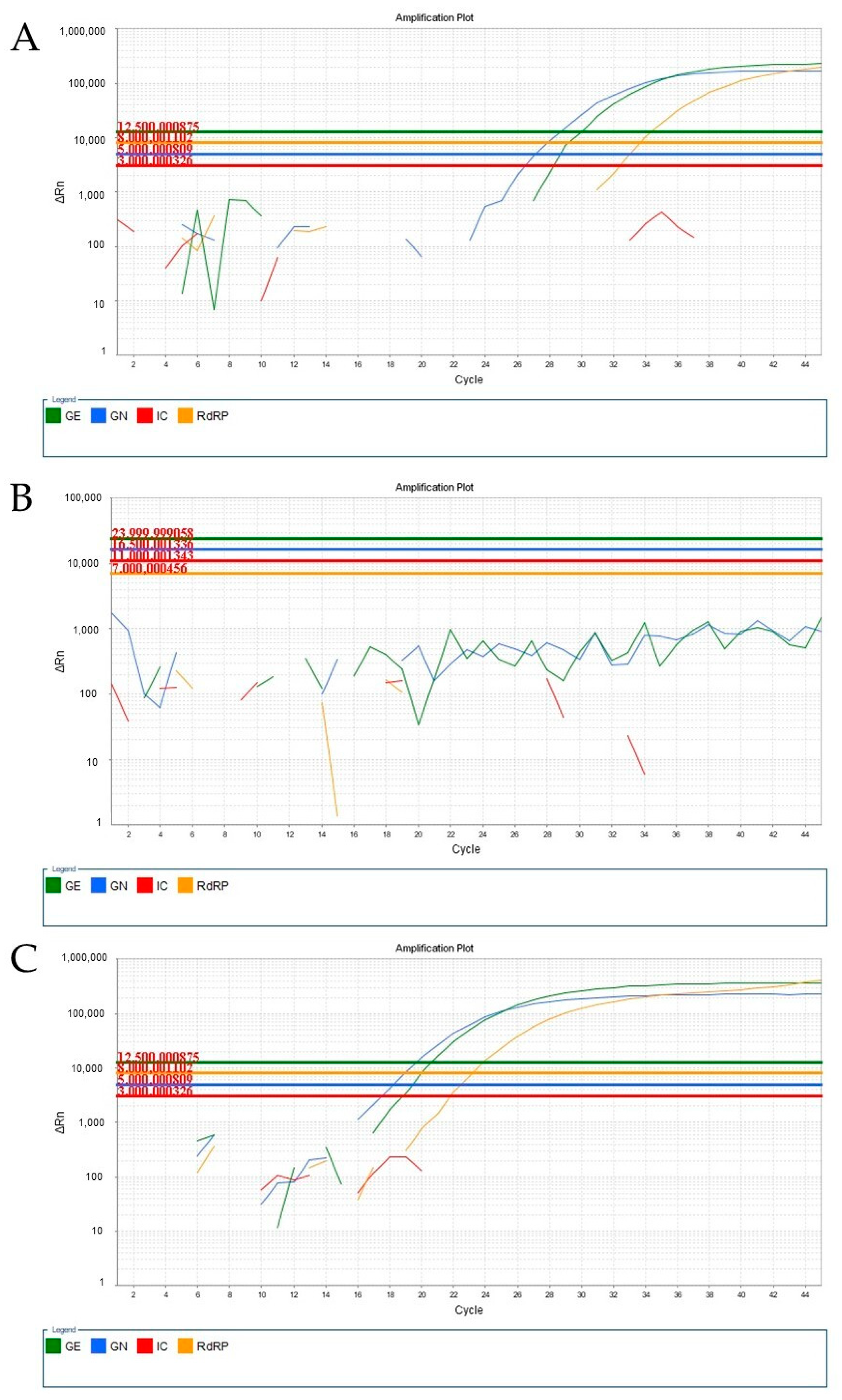
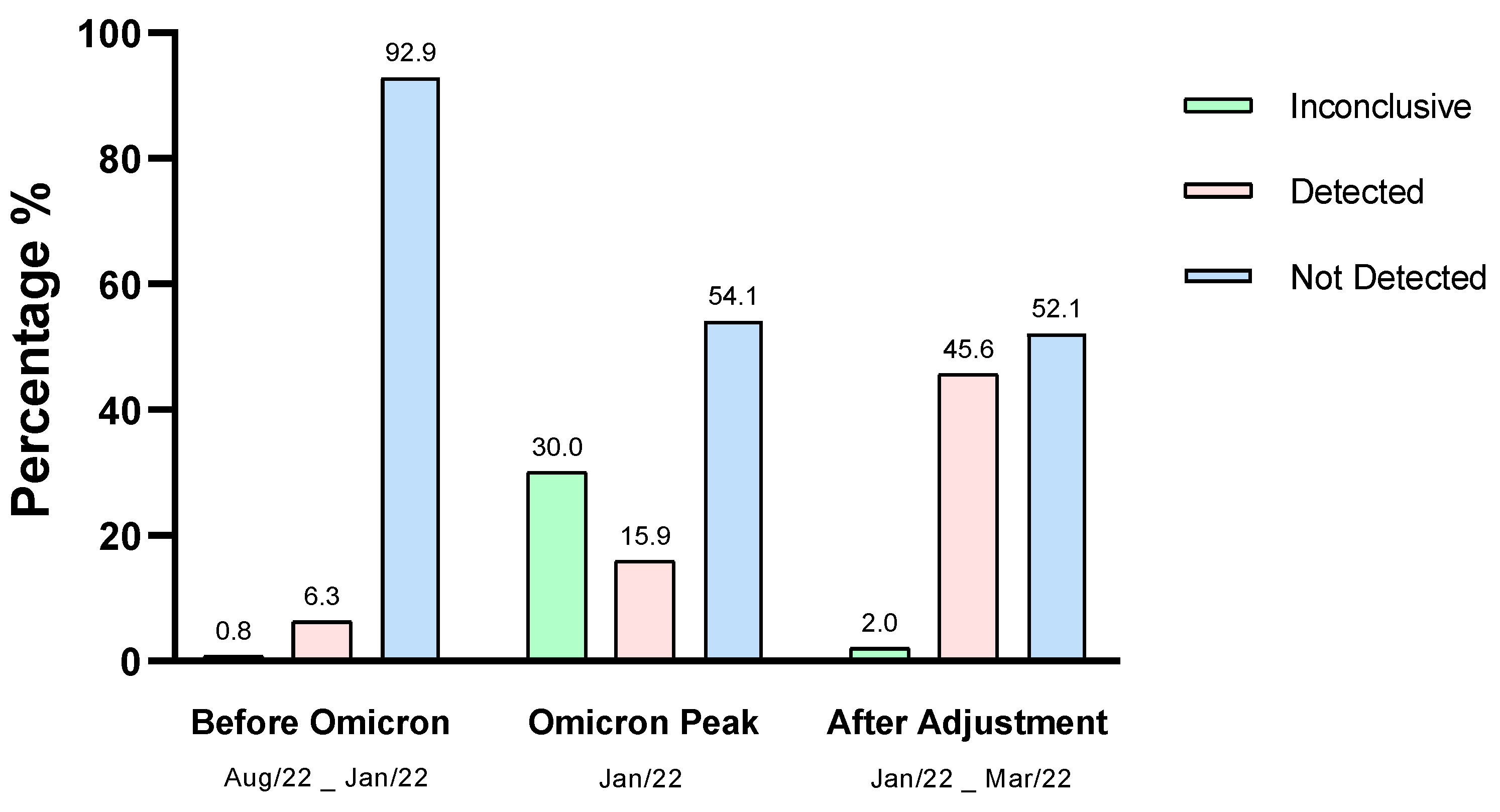
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. WHO Coronavirus (COVID-19) Dashboard. 2022. Available online: https://covid19.who.int/ (accessed on 27 June 2023).
- Koyama, T.; Platt, D.; Parida, L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020, 98, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Tzou, P.L.; Nouhin, J.; Gupta, R.K.; Oliveira, T.; Pond, S.L.K.; Fera, D.; Shafer, R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021, 22, 757–773. [Google Scholar] [CrossRef] [PubMed]
- WHO. Update on Omicron. 2021. Available online: https://www.who.int/news/item/28-11-2021-update-on-omicron (accessed on 15 February 2022).
- Poudel, S.; Ishak, A.; Perez-Fernandez, J.; Garcia, E.; Leon-Figueroa, D.A.; Romaní, L.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. Highly mutated SARS-CoV-2 Omicron variant sparks significant concern among global experts—What is known so far? Travel Med. Infect. Dis. 2022, 45, e102234. [Google Scholar] [CrossRef] [PubMed]
- Mistry, P.; Barmania, F.; Mellet, J.; Peta, K.; Strydom, A.; Viljoen, I.M.; James, W.; Gordon, S.; Pepper, M.S. SARS-CoV-2 Variants, Vaccines, and Host Immunity. Front. Immunol. 2022, 12, 809244. [Google Scholar] [CrossRef] [PubMed]
- Bei, Y.; Vrtis, K.B.; Borgaro, J.G.; Langhorst, B.W.; Nichols, N.M. The Omicron variant mutation at position 28,311 in the SARS-CoV-2 N gene does not perturb CDC N1 target detection. medRxiv 2021, 12, 1–8. [Google Scholar] [CrossRef]
- Agency USFD. SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests. 2021. Available online: https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests#omicronvariantimpact (accessed on 15 February 2022).
- Jaeger, L.H.; Nascimento, T.C.; Rocha, F.D.; Vilela, F.M.P.; Duque, A.P.N.; Silva, L.M.; Riani, L.R.; Moreira, J.P.; Chagas, J.M.D.A.; Pereira, T.V.; et al. Adjusting RT-qPCR conditions to avoid unspecific amplification in SARS-CoV-2 diagnosis. Int. J. Infect. Dis. 2021, 102, 437–439. [Google Scholar] [CrossRef] [PubMed]
- Bekliz, M.; Perez-Rodriguez, F.; Puhach, O.; Adea, K.; Melancia, S.M.; Baggio, S. Sensitivity of SARS-CoV-2 antigen-detecting rapid tests for Omicron variant. medRxiv 2021, 18, 1–17. [Google Scholar] [CrossRef]
- Wagenhäuser, I.; Knies, K.; Hofmann, D.; Rauschenberger, V.; Eisenmann, M.; Reusch, J.; Gabel, A.; Flemming, S.; Andres, O.; Petri, N.; et al. Virus variant-specific clinical performance of SARS coronavirus two rapid antigen tests in point-of-care use, from November 2020 to January 2022. Clin. Microbiol. Infect. 2023, 29, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Ambrosi, C.; Prezioso, C.; Checconi, P.; Scribano, D.; Sarshar, M.; Capannari, M.; Tomino, C.; Fini, M.; Garaci, E.; Palamara, A.T.; et al. SARS-CoV-2: Comparative analysis of different RNA extraction methods. J. Virol. Methods 2021, 287, 114008. [Google Scholar] [CrossRef] [PubMed]
- SES-MG. Informe Epidemiológico Coronavírus—Painel de Monitoramento de Casos. Secretaria de Estado de Saúde de Minas Gerais; 2022. Available online: https://coronavirus.saude.mg.gov.br (accessed on 10 August 2022).
- ANVISA. Anvisa Informa Sobre Identificação Preliminar de Dois Casos da Variante Ômicron em Território Nacional. 2021. Available online: https://www.gov.br/anvisa/pt-br/assuntos/noticias-anvisa/2021/anvisa-informa-sobre-a-identificacao-preliminar-de-dois-casos-da-variante-omicron-em-territorio-nacional (accessed on 22 February 2022).
- Silva, L.M.; Riani, L.R.; Silvério, M.S.; Pereira-Júnior, O.S.; Pittella, F. Comparison of Rapid Nucleic Acid Extraction Methods for SARS-CoV-2 Detection by RT-qPCR. Diagnostics 2022, 12, 601. [Google Scholar] [CrossRef] [PubMed]
- Seegene. Novaplex SARS-CoV-2 Variants II Assay. Available online: https://seegenetech.com/novaplex-sars-cov-2-variants-ii-assay/ (accessed on 12 December 2022).
- Yu, F.; Qiu, T.; Zeng, Y.; Wang, Y.; Zheng, S.; Chen, X.; Chen, Y. Comparative Evaluation of Three Preprocessing Methods for Extraction and Detection of Influenza a Virus Nucleic Acids from Sputum. Front. Med. 2018, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Khan, Z.A.; Charles, M.; Pratap, P.; Naeem, A.; Siddiqui, Z.; Naqvi, N.; Srivastava, S. Cytokine Storm and Mucus Hypersecretion in COVID-19: Review of Mechanisms. J. Inflamm. Res. 2021, 14, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Strand, D.A.; Holst-Jensen, A.; Viljugrein, H.; Edvardsen, B.; Klaveness, D.; Jussila, J.; Vrålstad, T. Detection and quantification of the crayfish plague agent in natural waters: Direct monitoring approach for aquatic environments. Dis. Aquat. Org. 2011, 95, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, C.; Sepulveda, A.; Ray, A.; Baumgardt, J.A. Waits Environmental DNA as a new method for early detection of New Zealand mudsnails (Potamopyrgus antipodarum). Freshwater Sci. 2013, 32, 792–800. [Google Scholar] [CrossRef]
- Umunnakwe, C.N.; Makatini, Z.N.; Maphanga, M.; Mdunyelwa, A.; Mlambo, K.M.; Manyaka, P.; Nijhuis, M.; Wensing, A.; Tempelman, H.A. Evaluation of a commercial SARS-CoV-2 multiplex PCR genotyping assay for variant identification in resource-scarce settings. PLoS ONE 2022, 17, e0269071. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, P.J.; Iida, S.; Iwatsuki-Horimoto, K.; Maemura, T.; Kiso, M. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 2022, 603, 687–692. [Google Scholar] [CrossRef] [PubMed]
- Kimura, Y.; Hirabayashi, E.; Yano, M.; Fujitani, S.; Shioiri, S. COVID-19 Omicron variant-induced laryngitis. Auris Nasus Larynx 2022, 23, 637–640. [Google Scholar] [CrossRef] [PubMed]
- Dulfano, M.J.; Kenneth, A.; Philippoff, W. Sputum viscoelasticity in chronic bronchitis. Am. Rev. Respir. Dis. 1971, 104, 88–98. [Google Scholar] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, L.M.; Riani, L.R.; Leite, J.B.; de Assis Chagas, J.M.; Fernandes, L.S.; Fochat, R.C.; Perches, C.G.P.; Nascimento, T.C.; Jaeger, L.H.; Silvério, M.S.; et al. The Influence of the Omicron Variant on RNA Extraction and RT-qPCR Detection of SARS-CoV-2 in a Laboratory in Brazil. Viruses 2023, 15, 1690. https://doi.org/10.3390/v15081690
Silva LM, Riani LR, Leite JB, de Assis Chagas JM, Fernandes LS, Fochat RC, Perches CGP, Nascimento TC, Jaeger LH, Silvério MS, et al. The Influence of the Omicron Variant on RNA Extraction and RT-qPCR Detection of SARS-CoV-2 in a Laboratory in Brazil. Viruses. 2023; 15(8):1690. https://doi.org/10.3390/v15081690
Chicago/Turabian StyleSilva, Lívia Mara, Lorena Rodrigues Riani, Juliana Brovini Leite, Jessica Mara de Assis Chagas, Laura Silva Fernandes, Romário Costa Fochat, Carmen Gomide Pinto Perches, Thiago César Nascimento, Lauren Hubert Jaeger, Marcelo Silva Silvério, and et al. 2023. "The Influence of the Omicron Variant on RNA Extraction and RT-qPCR Detection of SARS-CoV-2 in a Laboratory in Brazil" Viruses 15, no. 8: 1690. https://doi.org/10.3390/v15081690
APA StyleSilva, L. M., Riani, L. R., Leite, J. B., de Assis Chagas, J. M., Fernandes, L. S., Fochat, R. C., Perches, C. G. P., Nascimento, T. C., Jaeger, L. H., Silvério, M. S., dos Santos Pereira-Júnior, O., & Pittella, F. (2023). The Influence of the Omicron Variant on RNA Extraction and RT-qPCR Detection of SARS-CoV-2 in a Laboratory in Brazil. Viruses, 15(8), 1690. https://doi.org/10.3390/v15081690






