Abstract
Cats harbor many important viral pathogens, and the knowledge of their diversity has been greatly expanded thanks to increasingly popular molecular sequencing techniques. While the diversity is mostly described in numerous regionally defined studies, there lacks a global overview of the diversity for the majority of cat viruses, and therefore our understanding of the evolution and epidemiology of these viruses was generally inadequate. In this study, we analyzed 12,377 genetic sequences from 25 cat virus species and conducted comprehensive phylodynamic analyses. It revealed, for the first time, the global diversity for all cat viruses known to date, taking into account highly virulent strains and vaccine strains. From there, we further characterized and compared the geographic expansion patterns, temporal dynamics and recombination frequencies of these viruses. While respiratory pathogens such as feline calicivirus showed some degree of geographical panmixes, the other viral species are more geographically defined. Furthermore, recombination rates were much higher in feline parvovirus, feline coronavirus, feline calicivirus and feline foamy virus than the other feline virus species. Collectively, our findings deepen the understanding of the evolutionary and epidemiological features of cat viruses, which in turn provide important insight into the prevention and control of cat pathogens.
1. Introduction
Cats (species Felis catus, genus Felis of the family Felidae) harbor a wide range of viruses. Since the discovery of the first feline virus—feline parvovirus in 1928 [1]—more than 25 feline virus species have been identified, among which six have been extensively studied because of known disease association and/or worldwide prevalence. These viruses include feline immunodeficiency virus (FIV), feline coronavirus (FCoV), feline leukemia virus (FeLV), feline calicivirus (FCV), feline parvovirus (FPV) and felid alphaherpesvirus 1 (FHV-1). FCV and FHV-1 are important pathogens that cause feline upper respiratory tract disease (URTD) [2,3], FCoV and FPV are initially primary pathogens of the feline gastrointestinal tract disease [4,5], and FIV and FeLV can cause immunodeficiency disease in cats similar to acquired immunodeficiency syndrome (AIDS) in humans [6,7]. Furthermore, FeLV infection has been associated with quite a few unique diseases, such as acute myeloid leukemia (AML), multicentric lymphoma, aplastic anemia and other immunodeficiencies [8,9,10]. In addition to these well-known pathogens, a number of other feline viruses have been identified, but disease associations remained unclear; these include feline astrovirus (FAstV), feline rotavirus (FRV), feline foamy virus (FFV), Torque Teno feline virus (TTFV) and felis catus papillomavirus (FcaPV). Among these, FAstV and FRV are suggested to be part of the gastrointestinal virome of domestic cats [11], FcaPV is suspected to be associated with cutaneous squamous cell carcinomas (SCC) [12], whereas FFV and TTFV have been regarded as non-pathogenic in felines [13,14,15].
In recent years, with the development of PCR-based and next-generation sequencing techniques, a large number of novel viruses have been identified from cats, including feline morbillivirus (FeMV) [16], feline bocaparvovirus (FBoV) [17], felis catus gammaherpesvirus 1 (FcaGHV−1) [18,19], feline kobuvirus (FeKoV) [20], feline paramyxovirus (FePaV) [21], domestic cat hepadnavirus (DCH) [22], feline picornavirus (FePV) [23], feline norovirus (FNoV) [24], feline chaphamaparvovirus (FeChPV) [25], feline picobirnavirus (FePBV) [26], feline bufavirus (FeBuV) [27] and feline cyclovirus (FeCyCV) [28]. Nevertheless, the pathogenicity of these viruses is currently under-studied, although many have been identified in diseased animals. For example, FBoV, FeKoV, FNoV and FeChPV have been identified in cats with gastrointestinal disease [29,30,31,32,33], FcaGHV−1 from cats with immunosuppressive signs and ocular disorders [18,34,35] and FeMV from cats with renal diseases [16]. Nevertheless, confirming the disease association of these viruses requires more data from standard “case-and-control” studies.
While more viruses are being discovered in cats, our knowledge of the geographic range of cat viruses has also expanded, thanks to a huge number of sequence-based molecular epidemiological studies, each targeting a specific pathogen from a specific region. Indeed, for most well-known pathogens, namely, FCV, FIV, FPV, FeLV and FCoV, their geographic range has been shown to range from the place of discovery to worldwide. Global distribution has also been established for recently discovered viruses. For example, FeMV was first discovered in Hong Kong in 2012 [16], and by using PCR primers designed based on the first few genomes, it was then identified in Japan, the United Kingdom, Brazil, Turkey and Italy [36,37,38,39,40,41,42,43]. Similarly, FeChPV was first discovered in 2019 from a feline shelter in Canada [5] before its presence was expanded to Turkey [44], Italy [25] and China [45,46].
In general, recent studies based on molecular sequencing have witnessed a great expansion of virus species richness, intra-specific genomic diversity, and geographic range for feline viruses. While these diversities are mostly described in numerous regionally defined studies, there is a lack of an overview of global diversity for the majority of cat viruses. As a result, the general evolutionary and epidemiological characteristics of these viruses were never systematically compared. In this study, we downloaded the entire collection of genetic sequences for every feline virus identified thus far and conducted a comprehensive phylodynamic analysis. Our results revealed and compared the overall diversity, geographic and temporal dynamics, disease characteristics and recombination frequency of each cat virus, deepened our understanding of evolutionary and epidemiological features, and provided important insight into the prevention and control of cat pathogens.
2. Materials and Methods
2.1. Collection of Feline Virus Sequences and Information
The taxonomy of feline viruses was determined based on a comprehensive search of the NCBI PubMed database using the keyword “feline virus” or “cat virus” (Table S1). For each feline virus species, we downloaded all the nucleotide sequences, coding DNA sequences (CDS) and the associated sequence information, including GenBank number, virus strain, collection time, country of origin, PubMed (publication) ID and host information, from the NCBI GenBank database. For those with missing or incomplete information and for virulence, vaccine, and pathogenic information, an in-depth search was carried out to look for corresponding data from the original publications. All sequence and information collections were carried out in February 2022.
2.2. Data Processing and Sequence Alignment
For each virus, the CDS were first categorized based on encoded genes. Sequences with <300 bp in length and >20% of ambiguous nucleotides were treated as low-quality data and removed from the data set. Highly identical sequences (>99% nucleotide identity) were also removed by using Cluster Database at High Identity with Tolerance (CD-HIT) (version 4.8.1) and CD-HIT-EST (version 4.8.1) programs [47], given that they were from the same geographic regions. Based on the number of sequences available, sequence divergence level, and the representativeness of diversity and geographic distribution, it is subsequently determined which gene (and regions of genes) was used to represent the overall diversity of this virus. Corresponding sequences were then aligned using the program Mafft (version 7.480) [48] for later phylogenetic and evolutionary analyses.
2.3. Phylogenetic Analyses
The intra-specific phylogenetic trees for most of the viruses involved in this study were reconstructed based on gene or partial gene alignments representative of the virus diversity (see Section 2.2) using the maximum likelihood (ML) algorithm, General Time Reversible (GTR) nucleotide substitution model, and the subtree pruning and regrafting (SPR) branch-swapping algorithm implemented in software PhyML (version 3.0) [49]. For some of the newly discovered viruses, such as FAstV, FNoV and FRV, the inter-specific phylogenetic trees were reconstructed to demonstrate their diversity in the context of other related virus species. The inter-specific phylogenetic trees were reconstructed based on RNA dependent RNA polymerase (RdRp) (RNA viruses) and non-structural protein 1 (NS1) (FeChPV) protein alignments and utilizing a maximum likelihood algorithm, the Le Gascuel (LG) amino acid substitution model, and the SPR branch-swapping algorithm implemented in PhyML. All trees generated in this study were mid-point rooted and were labeled and demonstrated using the ggtree (version 3.2.1) [50] software package implemented in R (version 4.1.2) [51].
2.4. Analyses of Phylogeographic Structures
To assess whether the spread of a virus was confined by geographic distances or locations, we used the package adegenet (version 2.1.10) [52] implemented in R (version 4.1.2), which reveals the genetic structure present among geographic regions using discriminant analysis of principal components (DAPC). We also used Wilks’ lambda to evaluate the degree of geographic structure with the MANOVA method.
2.5. Analyses of Temporal Structure, Evolutionary Rate and Estimating Time to the Most Recent Common Ancestor (tMRCA)
Before time-scale analysis, sequences without sampling date information were removed from the alignment. The temporal structure of each cat virus species was examined using TempEst (v 1.5.3) [53], which carried out a regression of phylogenetic root-to-tip distances against the sampling date. In addition, time-scale trees were inferred using TreeTime (version 0.8.6) [54] and Least-squares dating (LSD) (version 0.3beta) [55], which transforms ML trees topology into time-scale trees with GTR nucleotide substitution model and autocorrelated molecular clock model.
2.6. Analyses of Genomic Recombination
To investigate the frequency of recombination for each cat virus species, phylogenies were reconstructed at the beginning (i.e., the first 2000 bp) and the end (i.e., the last 2000 bp) of the full genome alignments. The trees based on the two genomic regions were subsequently compared for (i) incongruences in topology or (ii) inconsistencies in pairwise genetic distance matrices as measures of recombination frequency. The incongruences in topology were visualized using the dendextend package (version 1.15.2) [56] implemented in R (version 4.1.2). The degree of inconsistencies was estimated based on pairwise comparisons of patristic genetic distance matrices and using the mantel test [57].
3. Results
3.1. Overview of Feline Viruses around the World
We collected a total of 12,377 sequences of feline viruses from NCBI GenBank database, among which the majority are associated with the five most common feline pathogens, namely, FIV (n = 3624), FCoV (n = 3066), FeLV (n = 1571), FCV (n = 1441) and FPV (n = 929) (Figure 1A), which also had worldwide distributions (Figure 1B). While FIV sequences are quite abundant across different continents, other viruses have a more uneven distribution. For example, FCoV is more frequently sampled in Asia and Europe than in North America, whereas FeLV is more frequently sampled in Asia and the Americas than in Europe (Figure 1B). Nevertheless, some of the unevenness in distribution might reflect sampling bias instead of true prevalence. Indeed, the sampling sizes in Africa and South America are much smaller than those of the other continents (Figure 1B). Additionally, within Europe, more sequences are obtained from Western than Eastern Europe. Collectively, these observations suggest wide distribution and potential gaps in the global sampling of feline viruses.
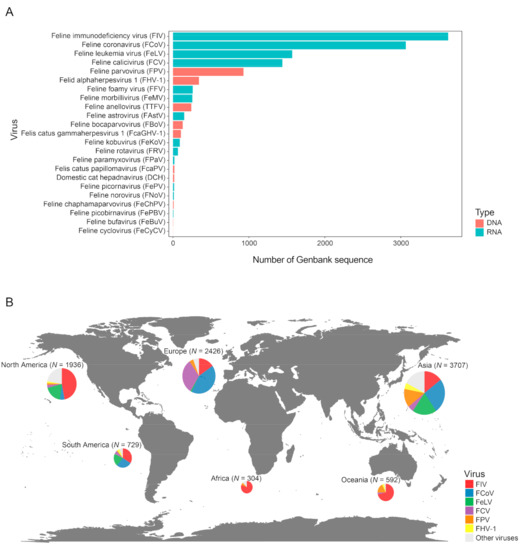
Figure 1.
Overview of the distribution of feline virus sequences. (A) Number of GenBank sequences available for each feline virus species. Red and blue bars represent DNA and RNA virus, respectively. (B) The global distribution of virus sequences collected for this study. The size of the circle reflects the number of sequences.
3.2. Phylogenetic Analysis
We performed phylogenetic analyses that revealed the diversity backbone for more than nine major cat virus species. Among these, FIV is an important cat pathogen whose infection usually leads to the depletion of CD4+ T cells and causes AIDS-like diseases in cats [58]. The virus has been previously divided into seven different clades (A–F and U), each associated with specific geographic locations [59,60,61]. We analyzed 831 representative sequences, which cover 501 nucleotides (nt) (501 bp out of 2571 bp complete length) in the env gene. The phylogenetic tree reveals a diversity comprised of at least six well-supported clades. The division is mostly consistent with the sub-typing system from previous studies, except for Clade E, which is merged into Clade B due to a lack of clear distinguishment between the two (Figure 2). In comparison with previously defined clades (A–F), the clade depicted here contains a much larger diversity and wider geographic ranges (Figure 2). Among these, Clades A–D harbor most of the sequences (816/831) identified from the field. Clade A and B are both worldwide clades with global distributions, although, within these clades, there is some degree of geographic structuring of virus diversity (Figure 2). In comparison, Clades C and D are more regionally defined: Clade D is only identified in Asia, whereas Clade C is mainly identified in both Oceania and Asia and contains a highly virulent strain, CABCpady00C, sampled from Canada, which causes high death rates from acute phase immunodeficiency disease [62,63] (Figure 2). Two vaccine strains are available for FIV, which belong to Clade A (i.e., strain Petaluma) and D (i.e., strain Shizuoka), and are used as inactivated dual-subtype FIV vaccines.
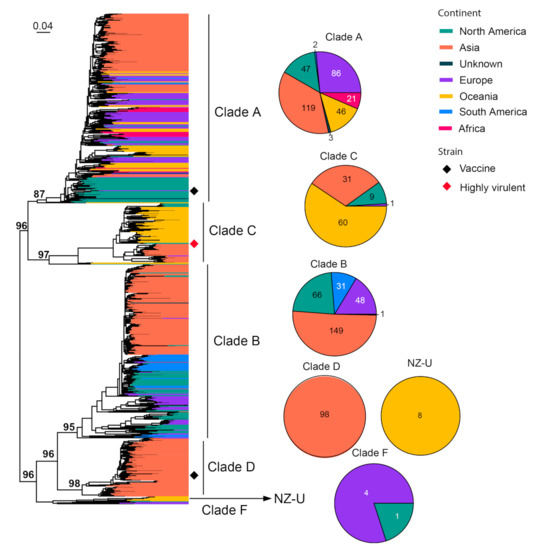
Figure 2.
Diversity and phylogeographic structure of FIV based on env gene. A maximum-likelihood tree reconstructed based on partial env gene alignments (501 nt) is shown on the left, which includes 831 virus sequences obtained from the GenBank. The tree is mid-point rooted. Geographic locations (continent) for these virus strains are marked with different colors. Highly virulent strains and vaccine strains are labeled using red and black diamonds, respectively. The detailed geographic distributions of each subtype are shown as pie charts on the right of the phylogenetic tree.
FCoV can cause two distinct types of diseases: some cause only mild (often subclinical) gastrointestinal illness, while others can lead to fatal multisystemic disease of feline infectious peritonitis (FIP) [64]. However, these diseases are not associated with specific FCoV genotypes or subtypes, and FIP arises as a result of viral mutations occurring following FCoV infection [65]. FCoVs are classified into two serotypes, Type I and Type II FCoVs [66]. We analyzed 107 complete S gene sequences (4494 nt) of FCoV in the context of 147 sequences under Alphacoronavirus 1, which includes FCoV, Canine coronavirus (CCoV), Transmissible gastroenteritis virus of swine (TGEV) and Porcine respiratory coronavirus (PRCoV) (Figure 3). Type I and II alphacoronaviruses are separated by an average of 58% nucleotide sequence divergence. FCoV is identified in both types, with the majority (92/107) of Type I and only a few (15/107) of Type II, and the latter show a relatively close relationship with Type II CCoV compared with TGEV and PRCoV (Figure 3). Highly virulent strains are identified in both Type I and II viruses, which include the HRB-17 strain from China [67], the 79–1146 strain from the USA [68] and the 26 M strain from the UK [69]. A vaccine strain is occasionally used to prevent FCoV infection, and it is derived from strain DF-2, a Type II FCoV.
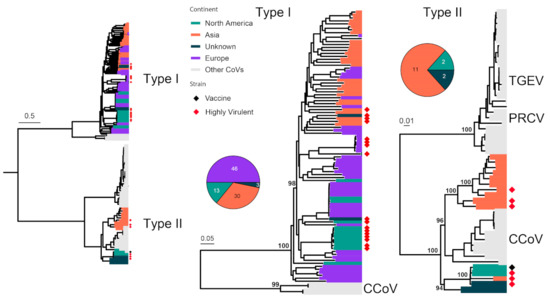
Figure 3.
Diversity and phylogeographic structure of FCoV based on S gene. A maximum-likelihood tree reconstructed based on S gene alignments (4494 nt) is shown on the left, which includes 147 virus sequences obtained from the GenBank. The tree is mid-point rooted, and detailed subtrees of each serotype (i.e., Type I and II) are shown on the right. For each serotype, the geographic distributions are shown as a pie chart and placed left next to the corresponding subtree. Geographic locations (continent) for these virus strains are marked with different colors. Highly virulent strains and vaccine strains are labeled using red and black diamonds, respectively.
FeLV infection in cats is associated with various health issues, including anemia, immune system-related diseases and cancer [70], and it has relatively high fatality rate [71]. We analyzed 278 representatives of the partial env gene (1756 bp out of 1929 bp complete length), which divided the FeLV diversity into two major clades, namely, Clade 1 and 2 (Figure 4). For each clade, a significant part of the diversity is associated with viruses sampled in Asia, whereas those from the Americas and Europe form geographically defined lineages that are nested within the diversity of Asian lineages. Furthermore, Asian viruses dominate the sequences (187/278). Two vaccine strains (i.e., FeLV Rickard and FeLV Glasgow-1) are identified in the phylogeny, and they all belong to Clade 1.
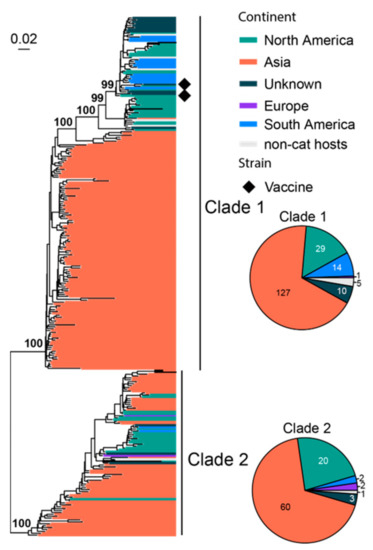
Figure 4.
Diversity and phylogeographic structure of FeLV based on partial env gene. A maximum-likelihood tree reconstructed based on partial env gene alignments (1756 nt) is shown on the left, which includes 278 virus sequences obtained from the GenBank. The tree is mid-point rooted. Geographic locations (continent) for these virus strains are marked with different colors. Vaccine strains are labeled using black diamonds. The detailed geographic distributions of each subtype are shown as pie charts on the right of the phylogenetic tree.
FCV commonly infects the respiratory tract of cats, and some strains with higher virulence can cause virulent systemic disease (VSD) [72,73,74,75]. A total of 801 representatives of partial viral protein 1 (VP1) genes (420 bp out of 2007 bp complete length) are used for phylogenetic analysis, which identifies seven well-supported clades (Clade 1–7) (Figure 5). Most of the diversity is dominated by sequences identified from Europe, a continent with much higher overall sequence numbers (644/801) and diversity than the rest of the continents. Interestingly, Clades 2, 3, 6 and 7 are all “cosmopolitan” with geographic expansion into more than three different continents. These clades are also ones that contain more strains with higher virulence and transmissibility than the rest of the clades (Figure 5). Specifically, Outbreak NSW 2015 [76], Outbreak ACT 2018 [76] and Outbreak Massachusetts 2001 [77] were from Clade 2, whereas Outbreak QLD 2017 [76], Outbreak Missouri 1995–1996 [77], Outbreak Sacramento 1998 [78], Outbreak Harrisburg 2003 [78] and Outbreak Florida 2003 [78] were from Clade 6 (Figure S1). Four vaccine strains are identified here, which belong to Clade 6 (FCV-F9) and Clade 7 (F4, FCV-255 and FCV-2024).
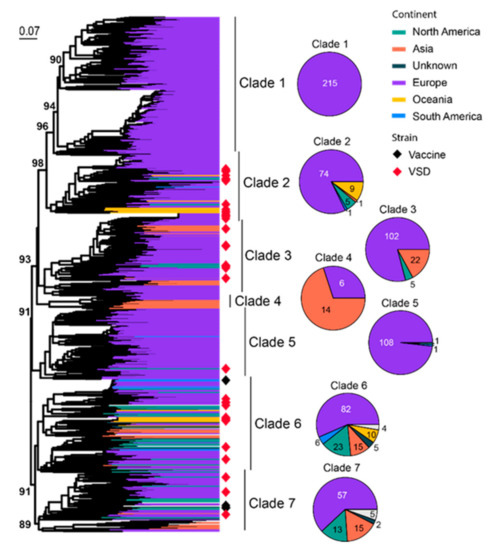
Figure 5.
Diversity and phylogeographic structure of FCV based on VP1 gene. A maximum-likelihood tree reconstructed based on partial VP1 gene alignments (420 nt) is shown on the left, which includes 801 representative virus sequences obtained from the GenBank. The tree is mid-point rooted. Geographic locations (continent) for these virus strains are marked with different colors. Highly virulent strains and vaccine strains are labeled using red and black diamonds, respectively. The detailed geographic distributions of each subtype are shown as pie charts on the right of the phylogenetic tree.
FPV is known as an important pathogen that causes gastrointestinal disease in cats [79]. This disease is also referred to as feline panleukopenia or feline infectious enteritis, which can result in severe or even fatal disease in kittens. We used 252 representatives of the complete minor capsid protein (VP2) gene (1755 bp) of FPV for phylogenetic analysis. All FPVs form a sister clade to canine parvovirus (CPV). Within FPV, the diversity can be divided, with low confidence, into two major clades and a number of small transmission chains close to a common ancestor of all FPVs (Figure 6). Asian strains dominate the entire phylogeny (144/252), followed by Europe (42/252) and Australia (19/252). Interestingly, FPV also contains a number of strains identified from non-cat feline hosts, such as the fox, lion, dog and raccoons, amongst others, resulting in gastroenteritis- and leukopenia-type diseases similar to those observed in cats [79,80]. Six vaccine strains (FPV-Felocell, FPV-Panocell, Philips Roxane, FPV-Purevax, PLI-IV and FPV-Dohyvac) are identified within Clade 2 (Figure 6). The outbreak strains, which were recorded in Outbreak Mildura 2015, Outbreak Sydney 2016–2017 and Outbreak New Zealand 2016 [81], are also located within Clade 2 (Figure S2).
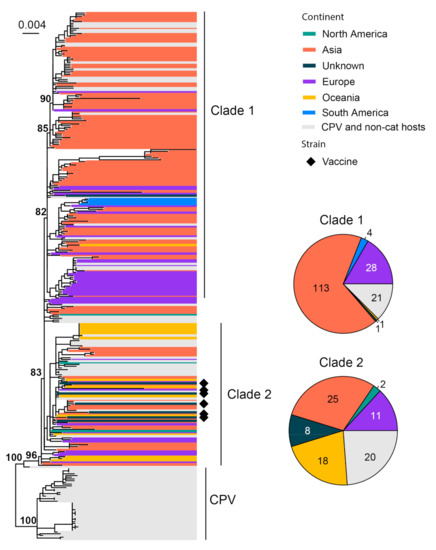
Figure 6.
Diversity and phylogeographic structure of FPV based on VP2 gene. A maximum-likelihood tree reconstructed based on partial VP2 gene alignments (1755 nt) is shown on the left, which includes 252 virus sequences of FPV obtained from the GenBank. The tree is mid-point rooted. Geographic locations (continent) for these virus strains are marked with different colors. Vaccine strains are labeled using black diamonds. The detailed geographic distributions of each subtype are shown as pie charts on the right of the phylogenetic tree.
FHV-1 normally infects the upper respiratory tract of cats and causes viral rhinotracheitis [82]. We collected 66 complete genomes for phylogenetic analysis, and it revealed three major clades with strong geographic structures, although there are a few genome sequences from continents other than North America and Oceania (Figure 7). Clade 1 is comprised of mainly North American strains, Clade 3 is comprised of only Australian strains and Clade 2 is comprised of strains sampled from both continents. Three vaccine strains (i.e., Companion, Merial Purevax MLV and Feligen) were identified, and they all belong to Clade 1.
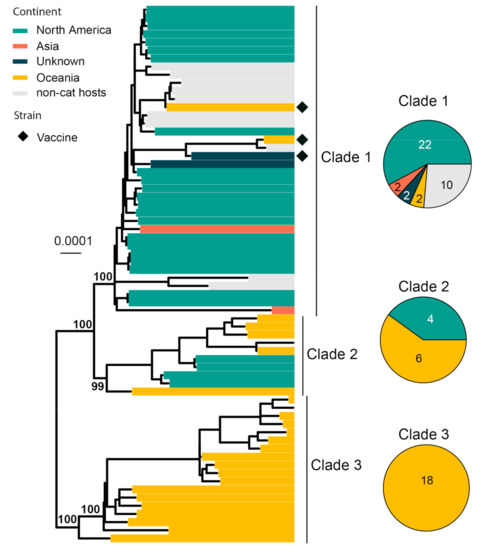
Figure 7.
Diversity and phylogeographic structure of FHV-1 based on complete genome alignment. A maximum-likelihood tree reconstructed based on complete genome alignments (134,883 nt) is shown on the left, which includes 66 virus sequences obtained from the GenBank. The tree is mid-point rooted. Geographic locations (continent) for these virus strains are marked with different colors. Vaccine strains are labeled using black diamonds. The detailed geographic distributions of each subtype are shown as pie charts on the right of the phylogenetic tree.
FFV is generally considered to be non-pathogenic in domestic and wild felids [15]. FFVs are classified into two subtypes, the F17/951-type and FUV-type [83]. We collected 104 env gene (2949 bp) sequences of FFV for phylogenetic analysis. FFV strains can be divided into two subtypes, including F17/951-type and FUV-type (Figure 8). Interestingly, the majority of the sequences obtained here were from cougars sampled from the United States [84]. Among the cat-associated FFV, the majority of the viral sequences are obtained from North America and Asia, although the sampling size is too small for reliable phylogeographical inference.
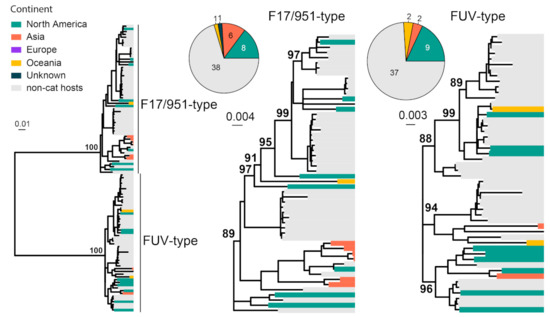
Figure 8.
Diversity and phylogeographic structure of FFV based on env gene. A maximum-likelihood tree reconstructed based on env gene alignments (2949 nt) is shown on the left, which includes 104 virus sequences obtained from the GenBank. The tree is mid-point rooted. Geographic locations (continent) for these virus strains are marked with different colors. For each serotype, the geographic distributions are shown as a pie chart and placed left next to the corresponding subtree.
FeMV is believed to be associated with renal disease, although more data from clinical studies are required to confirm this [38,85]. Previous analysis of FeMV genomes divided the diversity into two genotypic lineages, FeMV−1 and FeMV−2 [86]. In our study, a total of 186 sequences of RdRp gene sequences were analyzed, which divided the diversity into at least three clades (Figure 9). Among these, Clade 1 has cosmopolitan distribution except for Australia, Clade 2 is mainly found in Europe, South America and Asia, and Clade 3 had only four sequences which were all from Europe (Figure 9).
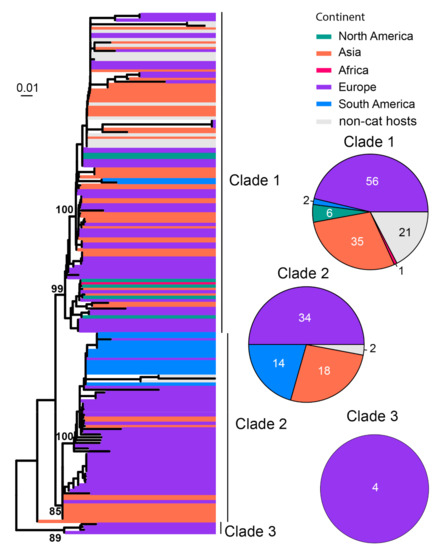
Figure 9.
Diversity and phylogeographic structure of FeMV based on L protein gene. A maximum-likelihood tree reconstructed based on complete L protein gene alignment (6618 nt) is shown on the left, which includes 186 virus sequences obtained from the GenBank. The tree is mid-point rooted. Geographic locations (continent) for these virus strains are marked with different colors. The detailed geographic distributions of each subtype are shown as pie charts on the right of the phylogenetic tree.
FBoV was often identified in the intestinal tract of cats and was suspected to be associated with gastrointestinal signs [29,30]. To date, three FBoV genotypes have been identified, namely, FBoV−1, FBoV−2 and FBoV−3. A total of 114 NS1 sequences were analyzed and compared. Three types of FBoVs have been identified, including FBOV−1 (n = 74), FBoV−2 (n = 33) and FBoV−3 (n = 7) (Figure 10). The majority of the sequences identified here were sampled in Asia (110/114), although strains from North America and Europe have also been identified (4/114).
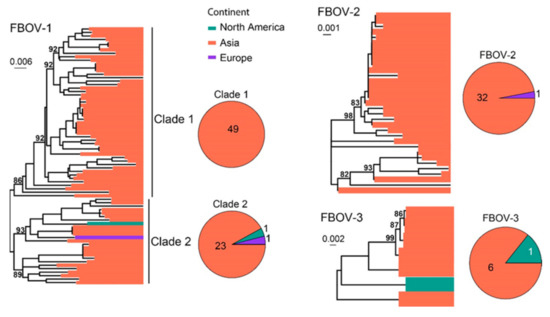
Figure 10.
Diversity and phylogeographic structure of FBoVs based on NS1 gene. Three maximum-likelihood trees reconstructed based on complete NS1 gene alignments (2415 nt, 2394 nt and 2391 nt) are shown, which include 74, 33 and 7 virus sequences obtained from the GenBank, respectively. The trees are mid-point rooted. Geographic locations (continent) for these virus strains are marked with different colors. The detailed geographic distributions of each subtype are shown as pie charts on the right of the phylogenetic trees.
In addition to the common feline viruses above, we also investigated the diversity and geographical distribution of less common or more recently discovered viruses. Interspecies phylogenetic analyses reveal that several viral families, namely, Anelloviridae, Astroviridae and Papillomaviridae, contain multiple virus species associated with cats, although no obvious disease association has been identified. Indeed, most of the feline viruses were identified from fecal samples, including FAstV [87], FNoV [24], FeCyCV [28], FeKoV [20], FeChPV [31] and FePBV [88]. Other virus species were identified from samples such as from blood (DCH [89] and FcaGHV−1 [19]) and the gut (FeBuV [27]). Furthermore, among these newly identified viruses, FcaGHV−1 has been identified from more than three continents despite their recent discovery, suggesting that latent or subclinical infections exist in a wide range of cat populations worldwide (Figure 11).
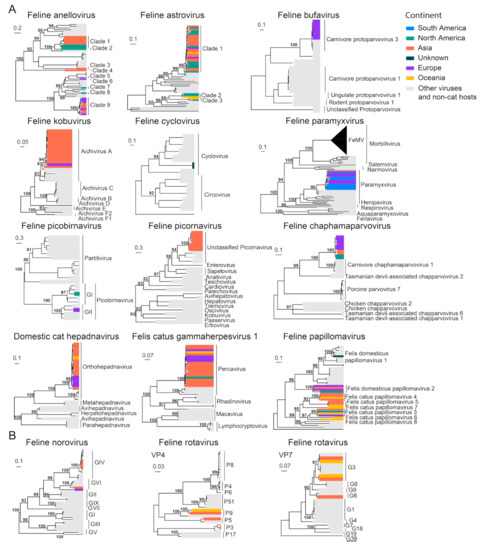
Figure 11.
Diversity and phylogeographic structure of other feline viruses based on their marker genes/proteins. (A) Interspecific phylogenetic trees and (B) intraspecific phylogenetics were reconstructed for less characterized cat virus species. The trees are mid-point rooted. The virus species’ name is labeled on the top of each tree. Geographic locations (continent) for these virus strains are marked with different colors.
3.3. Phylogeographic Structure Analysis
Phylogeographic structures were examined for a few feline virus species with relatively large sample sizes, namely, FCV, FIV, FPV, FCoV, FeLV, FHV-1 and FFV. Based on Wilk’s lambda, all seven viruses examined here had significant geographic structure (p < 0.001), although the degree of such structuring varied between different virus species (Table 1). Indeed, the differentiation of geographic groups is most obvious for FCoV, FeLV, FHV-1 and FFV but least obvious for FCV (Table 1). Similarly, DAPC scatter plots reveal clear geographic structure for all viruses examined except for FCV, which shows substantial mixing of virus diversity from different continents (Figure 12). In addition to FCV, there are also clear indications within other viral species where strains from different continents are indistinguishable (i.e., the mixing of North and South American strains in FIV and European and Asian strains in FPV) (Figure 12), suggesting cross-continent transmissions of the corresponding pathogens.

Table 1.
The degree of geographic structure evaluated using Wilks’ lambda value.
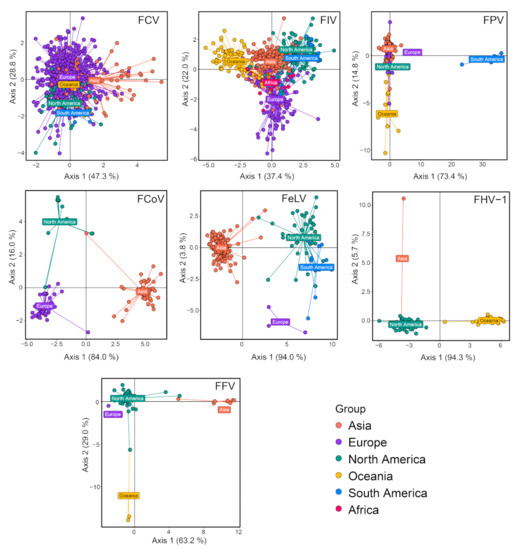
Figure 12.
Discriminant analysis of principal components (DAPC) scatter plot of main feline viruses, including FCV, FIV, FPV, FCoV, FeLV, FHV-1 and FFV. Different colors indicate virus strains of different geographic regions. The y-axis and x-axis represent two principal discriminant components.
3.4. Time-Scale Phylogenies and Analysis of Evolutionary Rates
To reveal temporal and epidemiological characteristics of the feline viruses, we first performed root-to-tip regression analysis, which shows that the temporal structures are poor for most of the viral species (Figure 13A), and therefore we only performed time-scaled evolutionary analyses for those with R > 0. Two approaches (i.e., TreeTime and LSD) were used for evolutionary rate and tMRCA estimations, and they generated consistent results for the five viruses examined here (Figure 13B,C and Table S2). The evolutionary rate significantly varied among different virus species. Among these, FCV and FCoV Type II have the fastest evolutionary rate, at a median of 6.71 × 10−4 (CI 4.57 × 10−4–7.89 × 10−4) and 2.778 × 10−4 (CI 9.33 × 10−5–6.50 × 10−4) substitutions/site/year based on LSD method, and 7.82 × 10−4 (CI 6.92 × 10−4–8.72 × 10−4) and 9.25 × 10−4 (CI 8.65 × 10−4–9.85 × 10−4) substitutions/site/year based on TreeTime method. Despite the fast evolutionary rate, the tMRCA for current FCV diversity was traced back to 1449 (CI 1381–1517) and 1428 (CI 1160–1516) using TreeTime and LSD methods, respectively, suggesting the early emergence of the FCV. Furthermore, the lowest evolutionary rate was estimated for FPV, an ssDNA virus, which has 4.11 × 10−5 (CI 2.11 × 10−5–6.11 × 10−5) substitutions/site/year and 5.29 × 10−5 (CI 2.95 × 10−5–6.29 × 10−5) using TreeTime and LSD approaches, respectively.
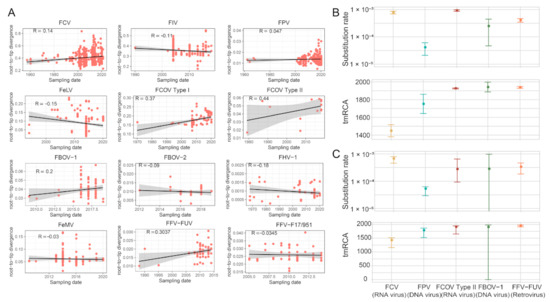
Figure 13.
Evolutionary analysis. (A) Regression of root-to-tip genetic distance against the year of sampling for several feline viruses. Black indicates a linear regression line. (B) Estimates of evolutionary parameters (substitution rate and tMRCA) for feline viruses via TreeTime software (version 0.8.6). (C) Estimates of evolutionary parameters (substitution rate and tMRCA) for feline viruses via LSD software (version 0.3beta).
3.5. Recombination Analysis
We assessed recombination frequencies for different viral species by comparing phylogenies reconstructed based on the first and last 2000 bp of the full-length genomes, which included FCV, FCoV, FFV, FPV, FeLV and FeMV. Correlation coefficiency estimations based on patristic distances (i.e., genetic distance derived from the maximum likelihood phylogenetic trees) revealed relatively greater phylogenetic incongruence for FPV, FCoV, FCV and FFV (Pearson’s coefficient < 0.8) and therefore higher recombination rates for these viruses (Figure 14). Strikingly, the coefficient for FPV was as low as 0.153 (p = 0.067), suggesting extremely frequent genomic exchange between diverse FPV genomes. Similarly, low recombination rates were inferred for FeMV and FeLV, which had highly congruent phylogenetic structures between the 5′ and 3′ ends of their genomes (i.e., coefficient > 0.8) (Figure 14).
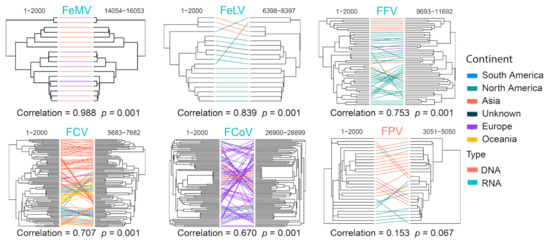
Figure 14.
Recombination analyses of representative cat virus species. For each species, phylogenies based on the 5′-end and 3′-end of genomes are reconstructed and compared for potential signs of phylogenetic incongruence. The incongruence is measured by Pearson’s correlation coefficient shown on the bottom of the trees, which reflects the recombination frequency for this specific virus.
4. Discussion
In this study, we obtained a thorough collection of sequence data associated with viruses infecting cats. We revealed, for each of the viruses, the most comprehensive diversity described to date. Specifically, our data set has greatly expanded the knowledge on the diversity of FCV [75,90,91,92,93], FPV [4,94,95,96], FCoV [69,97,98,99], FIV [60,61,100,101,102] and FeLV [8,103,104,105,106], such that in-depth analyses of feline viruses from across the globe could be performed. From there, we gained important insight into the global distribution, geographic spread pattern, evolutionary time-scale and recombination frequencies of these viruses. Generally, a larger diversity and wider distribution are described for the majority of viral species. Nevertheless, the diversity depicted here was by no means all-inclusive for these viruses. This is because the global sampling was largely uneven, with more diversity revealed for developed countries than for developing ones. Indeed, the number of sequences collected from African countries was substantially smaller than those from Asia (mainly China and Japan), European, and North American countries. Furthermore, viruses with low pathogenicity or less clinical impact, namely, TTFV, FFV, FePV, FePBV, FeCyCV and FePaV, contained fewer sequences and poorer geographical representation, most likely due to lack of surveillance from “targeted sequencing”. The global prevalence of these viruses is expected to be much higher, given that many cause subclinical or latent infections in cats.
Despite the global distribution of feline viruses, before this study, there was a lack of an overview of how they spread across the globe. This study revealed two distinct types of geographic structure: a few viruses or viral lineages showed geographical panmixes that features the dissemination of closely related virus over a long distance, whereas the others are more geographically contained. It is still unclear what causes the rapid transmission of cat viruses across the globe. One possibility is the long-distance transportation of pets [107,108,109,110]. Alternatively, the involvement of contaminated fomite or a secondary host could not be ruled out. Regardless, poor geographic structure and rapid long-distance distribution were mostly observed in FCV, which was a respiratory pathogen with frequent outbreaks of highly transmissible viral strains [76,111,112,113], implying that the mode of viral transmission might be a key contributor to the rapid spread of viral pathogens globally.
Our study revealed evolutionary rate and tMRCA estimations for a few RNA viruses (i.e., FCV, FCoV), retroviruses (i.e., FFV) and ssDNA viruses (i.e., FBoV and FPV). Our estimations of the median evolutionary rate of FCV were 7.82 × 10−4, which was much lower than a related virus, human norovirus, at 4.16 × 10−3 substitution/site/year [114] but similar to that of rabbit calicivirus at 7.7 × 10−4 substitution/site/year [115]. The rate estimation for FCoV and FPV was 9.25 × 10−4 and 4.11 × 10−5 substitution/site/year, which was similar to previous estimations for FPV [116] and coronavirus (e.g., SARS-CoV-2) [117] at 9.4 × 10−5 and 1.69 × 10−3 substitution/site/year, respectively. Accordingly, the estimation for tMRCA revealed that the FCV was circulating in the global cat population for at least 750 years, whereas FFV for only 90 years. Nevertheless, caution must be taken in interpreting tMRCA for FFV because the sample size obtained thus far is very limited, and it may not reflect the true diversity of the virus, and therefore the tMRCA might be massively under-estimated. Furthermore, our results also revealed poor temporal structure for a number of viruses, including FIV, FeLV and FHV-1. This is expected because these viruses, all of which are retroviruses or DNA viruses, have a slow evolutionary rate such that their time-scales are unlikely to be inferred from contemporary sampled sequences [117].
Recombination is an important driving force of evolutionary diversity and has been frequently reported for positive sense RNA viruses and DNA viruses [118]. In our study, the highest recombination rate was observed for FPV, which is a ssDNA virus. Although FPV has a low evolutionary rate, its recombination rate is high and is the main driving force of its genetic diversity, as expected for other ssDNA viruses [116]. For RNA viruses, the highest recombination rates were observed in FCoV and FCV, belonging to Coronaviridae and Caliciviridae, respectively, both of which were among the RNA viruses families with the highest recombination rates [118]. Indeed, the recombination hotspot of FCV is located at the breakpoint between the nonstructural protein coding region (Open reading frame 1(ORF 1)) and the structural protein coding region (ORF 2) [119]. Whereas that of FCoV is also located between the structural (Spike) and nonstructural (ORF1b) proteins [120]. Furthermore, it has also been reported that the Type II FCoV sequence originated from recombination of the Type II CCoV sequence and the Type I FCoV sequence [121], suggesting recombination as a common mechanism that drives the evolutionary diversity of positive sense RNA viruses in general.
5. Conclusions
In summary, our study performed an all-inclusive sequence collection from the GenBank and comprehensive phylodynamic analyses for 25 feline virus species. Our results revealed, for the first time, a global diversity much larger than previously depicted, based on which we further characterized the geographic expansion patterns, temporal dynamics and recombination frequencies of these viruses. Importantly, the representative sequence data set obtained here forms a knowledge basis for cat virus diversity, evolution and epidemiology, and they could be used as reference data sets or diversity backbones for future surveillance work.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v15061338/s1, Figures S1 and S2: Detailed phylogenetic subclades for FCV and FPV; Table S1: Information of feline viruses; Table S2: Phylogenetic estimates of evolutionary parameters Information of feline viruses; Alignments and trees.zip: all sequences alignments and phylogenetic trees.
Author Contributions
Conceptualization, M.S.; methodology, S.-J.L., G.-Y.X., W.-C.W. and M.S.; formal analysis, S.-J.L., G.-Y.X. and W.-C.W.; writing—original draft, S.-J.L.; writing—review and editing, W.-C.W. and M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Shenzhen Science and Technology Program (KQTD20200820145822023), National Natural Science Foundation of China (32270160), Natural Science Foundation of Guangdong Province (2022A1515011854) and Guangdong Province “Pearl River Talent Plan” Innovation and Entrepreneurship Team Project (2019ZT08Y464).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All alignments and phylogenetic trees in Newick format were included in the Supplementary File “Alignments and trees.zip”.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Verge, J.; Christoforoni, N. La Gastroenterite Infectieuse des Chats; Est-Elle Due à un Virus Filtrable. CR Seances Soc. Biol. Fil. 1928, 99, 312. [Google Scholar]
- Zhou, L.; Fu, N.; Ding, L.; Li, Y.; Huang, J.; Sha, X.; Zhou, Q.; Song, X.; Zhang, B. Molecular Characterization and Cross-Reactivity of Feline Calicivirus Circulating in Southwestern China. Viruses 2021, 13, 1812. [Google Scholar] [CrossRef]
- Sun, H.; Li, Y.; Jiao, W.; Liu, C.; Liu, X.; Wang, H.; Hua, F.; Dong, J.; Fan, S.; Yu, Z.; et al. Isolation and Identification of Feline Herpesvirus Type 1 from a South China Tiger in China. Viruses 2014, 6, 1004–1014. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.-C.; Kim, S.-J.; Nguyen, V.G.; Shin, S.; Kim, J.Y.; Lim, S.-K.; Park, Y.H.; Park, B. New Genotype Classification and Molecular Characterization of Canine and Feline Parvoviruses. J. Vet. Sci. 2020, 21, e43. [Google Scholar] [CrossRef] [PubMed]
- Palombieri, A.; Di Profio, F.; Fruci, P.; Sarchese, V.; Martella, V.; Marsilio, F.; Di Martino, B. Emerging Respiratory Viruses of Cats. Viruses 2022, 14, 663. [Google Scholar] [CrossRef] [PubMed]
- Westman, M.; Coggins, S.; Dorsselaer, M.; Norris, J.; Squires, R.; Thompson, M.; Malik, R. Feline Immunodeficiency Virus (FIV) Infection in Domestic Pet Cats in Australia and New Zealand: Guidelines for Diagnosis, Prevention and Management. Aust. Vet. J. 2022, 100, 345–359. [Google Scholar] [CrossRef]
- Stickney, A. Lack of Protection against Feline Immunodeficiency Virus Infection among Domestic Cats in New Zealand Vaccinated with the Fel-O-Vax® FIV Vaccine. Vet. Microbiol. 2020, 250, 108865. [Google Scholar] [CrossRef]
- Watanabe, S.; Kawamura, M.; Odahara, Y.; Anai, Y.; Ochi, H.; Nakagawa, S.; Endo, Y.; Tsujimoto, H.; Nishigaki, K. Phylogenetic and Structural Diversity in the Feline Leukemia Virus Env Gene. PLoS ONE 2013, 8, e61009. [Google Scholar] [CrossRef]
- Muz, D.; Can, H.; Karakavuk, M.; Döşkaya, M.; Özdemir, H.G.; Döşkaya, A.D.; Şahar, E.A.; Pektaş, B.; Karakuş, M.; Töz, S.; et al. The Molecular and Serological Investigation of Feline Immunodeficiency Virus and Feline Leukemia Virus in Stray Cats of Western Turkey. Comp. Immunol. Microbiol. Infect. Dis. 2021, 78, 101688. [Google Scholar] [CrossRef]
- Kraberger, S.; Serieys, L.E.K.; Richet, C.; Fountain-Jones, N.M.; Baele, G.; Bishop, J.M.; Nehring, M.; Ivan, J.S.; Newkirk, E.S.; Squires, J.R.; et al. Complex Evolutionary History of Felid Anelloviruses. Virology 2021, 562, 176–189. [Google Scholar] [CrossRef]
- Brussel, K.V.; Wang, X.; Shi, M.; Carrai, M.; Li, J.; Martella, V.; Beatty, J.A.; Holmes, E.C.; Barrs, V.R. Astrovirus_Identification of Novel Astroviruses in the Gastrointestinal Tract of Domestic Cats. Viruses 2020, 12, 1301. [Google Scholar] [CrossRef]
- Carrai, M.; Van Brussel, K.; Shi, M.; Li, C.-X.; Chang, W.-S.; Munday, J.S.; Voss, K.; McLuckie, A.; Taylor, D.; Laws, A.; et al. Identification of a Novel Papillomavirus Associated with Squamous Cell Carcinoma in a Domestic Cat. Viruses 2020, 12, 124. [Google Scholar] [CrossRef]
- Koç, B.T. First Report on the Prevalence and Genetic Relatedness of Feline Foamy Virus (FFV) from Turkish Domestic Cats. Virus Res. 2019, 274, 197768. [Google Scholar] [CrossRef]
- Roy, J.; Rudolph, W.; Juretzek, T.; Gärtner, K.; Bock, M.; Herchenröder, O.; Lindemann, D.; Heinkelein, M.; Rethwilm, A. Feline Foamy Virus Genome and Replication Strategy. J. Virol. 2003, 77, 11324–11331. [Google Scholar] [CrossRef] [PubMed]
- Winkler, I.G.; Löchelt, M.; Flower, R.L.P. Epidemiology of Feline Foamy Virus and Feline Immunodeficiency Virus Infections in Domestic and Feral Cats: A Seroepidemiological Study. J. Clin. Microbiol. 1999, 37, 2848–2851. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Wong, B.H.L.; Fan, R.Y.Y.; Wong, A.Y.P.; Zhang, A.J.X.; Wu, Y.; Choi, G.K.Y.; Li, K.S.M.; Hui, J.; et al. Feline Morbillivirus, a Previously Undescribed Paramyxovirus Associated with Tubulointerstitial Nephritis in Domestic Cats. Proc. Natl. Acad. Sci. USA 2012, 109, 5435–5440. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Woo, P.C.Y.; Yeung, H.C.; Teng, J.L.L.; Wu, Y.; Bai, R.; Fan, R.Y.Y.; Chan, K.-H.; Yuen, K.-Y. Identification and Characterization of Bocaviruses in Cats and Dogs Reveals a Novel Feline Bocavirus and a Novel Genetic Group of Canine Bocavirus. J. Gen. Virol. 2012, 93, 1573–1582. [Google Scholar] [CrossRef]
- Beatty, J.A.; Troyer, R.M.; Carver, S.; Barrs, V.R.; Espinasse, F.; Conradi, O.; Stutzman-Rodriguez, K.; Chan, C.C.; Tasker, S.; Lappin, M.R.; et al. Felis Catus Gammaherpesvirus 1; A Widely Endemic Potential Pathogen of Domestic Cats. Virology 2014, 460–461, 100–107. [Google Scholar] [CrossRef]
- Troyer, R.M.; Beatty, J.A.; Stutzman-Rodriguez, K.R.; Carver, S.; Lozano, C.C.; Lee, J.S.; Lappin, M.R.; Riley, S.P.D.; Serieys, L.E.K.; Logan, K.A.; et al. Novel Gammaherpesviruses in North American Domestic Cats, Bobcats, and Pumas: Identification, Prevalence, and Risk Factors. J. Virol. 2014, 88, 3914–3924. [Google Scholar] [CrossRef]
- Chung, J.-Y.; Kim, S.-H.; Kim, Y.-H.; Lee, M.-H.; Lee, K.-K.; Oem, J.-K. Detection and Genetic Characterization of Feline Kobuviruses. Virus Genes 2013, 47, 559–562. [Google Scholar] [CrossRef]
- Balbo, L.C.; Fritzen, J.T.T.; Lorenzetti, E.; Medeiros, T.N.S.; Jardim, A.M.; Alfieri, A.A.; Alfieri, A.F. Molecular Characterization of Feline Paramyxovirus and Feline Morbillivirus in Cats from Brazil. Braz. J. Microbiol. 2021, 52, 961–965. [Google Scholar] [CrossRef] [PubMed]
- Capozza, P.; Lanave, G.; Diakoudi, G.; Stasi, F.; Ghergo, P.; Ricci, D.; Santo, G.; Arena, G.; Grillo, I.; Delle Donne, E.; et al. A Longitudinal Observational Study in Two Cats Naturally-Infected with Hepadnavirus. Vet. Microbiol. 2021, 254, 108999. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Woo, P.C.Y.; Yip, C.C.Y.; Choi, G.K.Y.; Wu, Y.; Bai, R.; Fan, R.Y.Y.; Lai, K.K.Y.; Chan, K.-H.; Yuen, K.-Y. Identification of a Novel Feline Picornavirus from the Domestic Cat. J. Virol. 2012, 86, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Pinto, P.; Wang, Q.; Chen, N.; Dubovi, E.J.; Daniels, J.B.; Millward, L.M.; Buonavoglia, C.; Martella, V.; Saif, L.J. Discovery and Genomic Characterization of Noroviruses from a Gastroenteritis Outbreak in Domestic Cats in the US. PLoS ONE 2012, 7, e32739. [Google Scholar] [CrossRef] [PubMed]
- Di Profio, F.; Sarchese, V.; Palombieri, A.; Fruci, P.; Massirio, I.; Martella, V.; Fulvio, M.; Di Martino, B. Feline Chaphamaparvovirus in Cats with Enteritis and Upper Respiratory Tract Disease. Transbound. Emerg. Dis. 2022, 69, 660–668. [Google Scholar] [CrossRef]
- Ng, T.F.F.; Mesquita, J.R.; Nascimento, M.S.J.; Kondov, N.O.; Wong, W.; Reuter, G.; Knowles, N.J.; Vega, E.; Esona, M.D.; Deng, X.; et al. Feline Fecal Virome Reveals Novel and Prevalent Enteric Viruses. Vet. Microbiol. 2014, 171, 102–111. [Google Scholar] [CrossRef]
- Diakoudi, G.; Lanave, G.; Capozza, P.; Di Profio, F.; Melegari, I.; Di Martino, B.; Pennisi, M.G.; Elia, G.; Cavalli, A.; Tempesta, M.; et al. Identification of a Novel Parvovirus in Domestic Cats. Vet. Microbiol. 2019, 228, 246–251. [Google Scholar] [CrossRef]
- Zhang, W.; Li, L.; Deng, X.; Kapusinszky, B.; Pesavento, P.A.; Delwart, E. Faecal Virome of Cats in an Animal Shelter. J. Gen. Virol. 2014, 95, 2553–2564. [Google Scholar] [CrossRef]
- Garigliany, M.; Gilliaux, G.; Jolly, S.; Casanova, T.; Bayrou, C.; Gommeren, K.; Fett, T.; Mauroy, A.; Lévy, E.; Cassart, D.; et al. Feline Panleukopenia Virus in Cerebral Neurons of Young and Adult Cats. BMC Vet. Res. 2016, 12, 28. [Google Scholar] [CrossRef]
- Liu, C.; Liu, F.; Li, Z.; Qu, L.; Liu, D. First Report of Feline Bocavirus Associated with Severe Enteritis of Cat in Northeast China, 2015. J. Vet. Med. Sci. 2018, 80, 731–735. [Google Scholar] [CrossRef]
- Li, Y.; Gordon, E.; Idle, A.; Altan, E.; Seguin, M.A.; Estrada, M.; Deng, X.; Delwart, E. Virome of a Feline Outbreak of Diarrhea and Vomiting Includes Bocaviruses and a Novel Chapparvovirus. Viruses 2020, 12, 506. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, B.; Di Profio, F.; Melegari, I.; Marsilio, F. Feline Virome—A Review of Novel Enteric Viruses Detected in Cats. Viruses 2019, 11, 908. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, B.; Di Profio, F.; Melegari, I.; Sarchese, V.; Cafiero, M.A.; Robetto, S.; Aste, G.; Lanave, G.; Marsilio, F.; Martella, V. A Novel Feline Norovirus in Diarrheic Cats. Infect. Genet. Evol. 2016, 38, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Beatty, J.A.; Sharp, C.R.; Duprex, W.P.; Munday, J.S. Novel Feline Viruses: Emerging Significance of Gammaherpesvirus and Morbillivirus Infections. J. Feline Med. Surg. 2019, 21, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Koç, B.T.; BiRcan, M. Molecular Presence of Felis Catus Gammaherpesvirus -1 in Cats with Ocular Disorders in Turkey. Ank. Üniv. Vet. Fak. Derg. 2020, 68, 53–59. [Google Scholar] [CrossRef]
- Marcacci, M.; De Luca, E.; Zaccaria, G.; Di Tommaso, M.; Mangone, I.; Aste, G.; Savini, G.; Boari, A.; Lorusso, A. Genome Characterization of Feline Morbillivirus from Italy. J. Virol. Methods 2016, 234, 160–163. [Google Scholar] [CrossRef]
- Furuya, T.; Sassa, Y.; Omatsu, T.; Nagai, M.; Fukushima, R.; Shibutani, M.; Yamaguchi, T.; Uematsu, Y.; Shirota, K.; Mizutani, T. Existence of Feline Morbillivirus Infection in Japanese Cat Populations. Arch. Virol. 2014, 159, 371–373. [Google Scholar] [CrossRef]
- Darold, G.M.; Alfieri, A.A.; Muraro, L.S.; Amude, A.M.; Zanatta, R.; Yamauchi, K.C.I.; Alfieri, A.F.; Lunardi, M. First Report of Feline Morbillivirus in South America. Arch. Virol. 2017, 162, 469–475. [Google Scholar] [CrossRef]
- McCallum, K.E.; Stubbs, S.; Hope, N.; Mickleburgh, I.; Dight, D.; Tiley, L.; Williams, T.L. Detection and Seroprevalence of Morbillivirus and Other Paramyxoviruses in Geriatric Cats with and without Evidence of Azotemic Chronic Kidney Disease. J. Vet. Intern. Med. 2018, 32, 1100–1108. [Google Scholar] [CrossRef]
- Yilmaz, H.; Tekelioglu, B.K.; Gurel, A.; Bamac, O.E.; Ozturk, G.Y.; Cizmecigil, U.Y.; Altan, E.; Aydin, O.; Yilmaz, A.; Berriatua, E.; et al. Frequency, Clinicopathological Features and Phylogenetic Analysis of Feline Morbillivirus in Cats in Istanbul, Turkey. J. Feline Med. Surg. 2017, 19, 1206–1214. [Google Scholar] [CrossRef]
- Darold, G.M.; Alfieri, A.A.; Araújo, J.P.; Cruz, T.F.; de Bertti, K.M.L.B.; Silva, G.C.P.; Amude, A.M.; Muraro, L.S.; Lavorente, F.L.P.; Lunardi, M. High Genetic Diversity of Paramyxoviruses Infecting Domestic Cats in Western Brazil. Transbound. Emerg. Dis. 2021, 68, 3453–3462. [Google Scholar] [CrossRef] [PubMed]
- Chaiyasak, S.; Techangamsuwan, S. First Evidence of Feline Morbillivirus Detected in Sheltered Cats in Thailand. Thai. J. Vet. Med. Suppl. 2017, 47, 127–128. [Google Scholar]
- Sharp, C.R.; Nambulli, S.; Acciardo, A.S.; Rennick, L.J.; Drexler, J.F.; Rima, B.K.; Williams, T.; Duprex, W.P. Chronic Infection of Domestic Cats with Feline Morbillivirus, United States. Emerg. Infect. Dis. 2016, 22, 760–762. [Google Scholar] [CrossRef]
- Abayli, H.; Can-Sahna, K. First Detection of Feline Bocaparvovirus 2 and Feline Chaphamaparvovirus in Healthy Cats in Turkey. Vet. Res. Commun. 2022, 46, 127–136. [Google Scholar] [CrossRef]
- Ji, J.; Hu, W.; Liu, Q.; Zuo, K.; Zhi, G.; Xu, X.; Kan, Y.; Yao, L.; Xie, Q. Genetic Analysis of Cachavirus-Related Parvoviruses Detected in Pet Cats: The First Report from China. Front. Vet. Sci. 2020, 7, 580836. [Google Scholar] [CrossRef] [PubMed]
- Shao, R.; Ye, C.; Zhang, Y.; Sun, X.; Cheng, J.; Zheng, F.; Cai, S.; Ji, J.; Ren, Z.; Zhong, L.; et al. Novel Parvovirus in Cats, China. Virus Res. 2021, 304, 198529. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T. GGTREE: An R Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Paradis, E. Analysis of Phylogenetics and Evolution with R; Use R! Springer: New York, NY, USA, 2006; ISBN 978-1-4614-1743-9. [Google Scholar]
- Jombart, T.; Ahmed, I. Adegenet 1.3-1: New Tools for the Analysis of Genome-Wide SNP Data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Lam, T.T.; Max Carvalho, L.; Pybus, O.G. Exploring the Temporal Structure of Heterochronous Sequences Using TempEst (Formerly Path-O-Gen). Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef]
- Sagulenko, P.; Puller, V.; Neher, R.A. TreeTime: Maximum-Likelihood Phylodynamic Analysis. Virus Evol. 2018, 4, vex042. [Google Scholar] [CrossRef] [PubMed]
- To, T.-H.; Jung, M.; Lycett, S.; Gascuel, O. Fast Dating Using Least-Squares Criteria and Algorithms. Syst. Biol. 2016, 65, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Galili, T. Dendextend: An R Package for Visualizing, Adjusting, and Comparing Trees of Hierarchical Clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef]
- Legendre, P.; Borcard, D.; Peres-Neto, P.R. Analyzing Beta Diversity: Partitioning The Spatial Variation of Community Composition Data. Ecol. Monogr. 2005, 75, 435–450. [Google Scholar] [CrossRef]
- Miyazawa, T.; Tomonaga, K.; Kawaguchi, Y.; Mikami, T. The Genome of Feline Immunodeficiency Virus. Arch. Virol. 1994, 134, 221–234. [Google Scholar] [CrossRef]
- Frankenfeld, J.; Meili, T.; Meli, M.; Riond, B.; Helfer-Hungerbuehler, A.; Bönzli, E.; Pineroli, B.; Hofmann-Lehmann, R. Decreased Sensitivity of the Serological Detection of Feline Immunodeficiency Virus Infection Potentially due to Imported Genetic Variants. Viruses 2019, 11, 697. [Google Scholar] [CrossRef]
- Duarte, A.; Tavares, L. Phylogenetic Analysis of Portuguese Feline Immunodeficiency Virus Sequences Reveals High Genetic Diversity. Vet. Microbiol. 2006, 114, 25–33. [Google Scholar] [CrossRef]
- Hayward, J.J.; Rodrigo, A.G. Molecular Epidemiology of Feline Immunodeficiency Virus in the Domestic Cat (Felis Catus). Vet. Immunol. Immunopathol. 2010, 134, 68–74. [Google Scholar] [CrossRef]
- De Rozières, S.; Mathiason, C.K.; Rolston, M.R.; Chatterji, U.; Hoover, E.A.; Elder, J.H. Characterization of a Highly Pathogenic Molecular Clone of Feline Immunodeficiency Virus Clade C. J. Virol. 2004, 78, 8971–8982. [Google Scholar] [CrossRef] [PubMed]
- De Rozières, S.; Swan, C.H.; Sheeter, D.A.; Clingerman, K.J.; Lin, Y.-C.; Huitron-Resendiz, S.; Henriksen, S.; Torbett, B.E.; Elder, J.H. Assessment of FIV-C Infection of Cats as a Function of Treatment with the Protease Inhibitor, TL-3. Retrovirology 2004, 1, 38. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C.; Evermann, J.F.; McKeirnan, A.J.; Ott, R.L. Pathogenicity Studies of Feline Coronavirus Isolates 79-1146 and 79-1683. Am. J. Vet. Res. 1984, 45, 2580–2585. [Google Scholar] [PubMed]
- Felten, S.; Hartmann, K. Diagnosis of Feline Infectious Peritonitis: A Review of the Current Literature. Viruses 2019, 11, 1068. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C.; Black, J.W.; Boyle, J.F.; Evermann, J.F.; McKeirnan, A.J.; Ott, R.L. Pathogenic Differences between Various Feline Coronavirus Isolates. Adv. Exp. Med. Biol. 1984, 173, 365–380. [Google Scholar]
- Guan, X.; Li, H.; Han, M.; Jia, S.; Feng, B.; Gao, X.; Wang, Z.; Jiang, Y.; Cui, W.; Wang, L.; et al. Epidemiological Investigation of Feline Infectious Peritonitis in Cats Living in Harbin, Northeast China from 2017 to 2019 Using a Combination of an EvaGreen-Based Real-Time RT-PCR and Serum Chemistry Assays. Mol. Cell. Probes 2020, 49, 101495. [Google Scholar] [CrossRef]
- Herrewegh, A.A.P.M.; Smeenk, I.; Horzinek, M.C.; Rottier, P.J.M.; de Groot, R.J. Feline Coronavirus Type II Strains 79-1683 and 79-1146 Originate from a Double Recombination between Feline Coronavirus Type I and Canine Coronavirus. J. Virol. 1998, 72, 4508–4514. [Google Scholar] [CrossRef]
- Lewis, C.S.; Porter, E.; Matthews, D.; Kipar, A.; Tasker, S.; Helps, C.R.; Siddell, S.G. Genotyping Coronaviruses Associated with Feline Infectious Peritonitis. J. Gen. Virol. 2015, 96, 1358–1368. [Google Scholar] [CrossRef]
- Compans, R.W.; Cooper, M.; Koprowski, H.; McConnell, I.; Melchers, F.; Nussenzweig, V.; Oldstone, M.; Olsnes, S.; Potter, M.; Saedler, H.; et al. Feline Leukaemia Virus: Generation of Pathogenic and Oncogenic Variants. Curr. Top. Microbiol. Immunol. 1991, 171, 67–93. [Google Scholar]
- Addie, D.D.; Toth, S.; Reid, S.; Jarrett, O.; Dennis, J.M.; Callanan, J.J. Long-Term Impact on a Closed Household of Pet Cats of Natural Infection with Feline Coronavirus, Feline Leukaemia Virus and Feline Immunodeficiency Virus. Vet. Rec. 2000, 146, 419–424. [Google Scholar] [CrossRef]
- Hurley, K.F.; Pesavento, P.A.; Pedersen, N.C.; Poland, A.M.; Wilson, E.; Foley, J.E. An Outbreak of Virulent Systemic Feline Calicivirus Disease. J. Am. Vet. Med. Assoc. 2004, 224, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Hurley, K.F.; Sykes, J.E. Update on Feline Calicivirus: New Trends. Vet. Clin. N. Am. Small Anim. Pract. 2003, 33, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C.; Elliott, J.B.; Glasgow, A.; Poland, A.; Keel, K. An Isolated Epizootic of Hemorrhagic-like Fever in Cats Caused by a Novel and Highly Virulent Strain of Feline Calicivirus. Vet. Microbiol. 2000, 73, 281–300. [Google Scholar] [CrossRef]
- Schorr-Evans, E.M.; Poland, A.; Johnson, W.; Pedersen, N.C. An Epizootic of Highly Virulent Feline Calicivirus Disease in a Hospital Setting in New England. J. Feline Med. Surg. 2003, 5, 217–226. [Google Scholar] [CrossRef]
- Bordicchia, M.; Fumian, T.M.; Van Brussel, K.; Russo, A.G.; Carrai, M.; Le, S.-J.; Pesavento, P.A.; Holmes, E.C.; Martella, V.; White, P.; et al. Feline Calicivirus Virulent Systemic Disease: Clinical Epidemiology, Analysis of Viral Isolates and In Vitro Efficacy of Novel Antivirals in Australian Outbreaks. Viruses 2021, 13, 2040. [Google Scholar] [CrossRef] [PubMed]
- Prikhodko, V.G.; Sandoval-Jaime, C.; Abente, E.J.; Bok, K.; Parra, G.I.; Rogozin, I.B.; Ostlund, E.N.; Green, K.Y.; Sosnovtsev, S.V. Genetic Characterization of Feline Calicivirus Strains Associated with Varying Disease Manifestations during an Outbreak Season in Missouri (1995–1996). Virus Genes 2014, 48, 96–110. [Google Scholar] [CrossRef]
- Ossiboff, R.J.; Sheh, A.; Shotton, J.; Pesavento, P.A.; Parker, J.S.L. Feline Caliciviruses (FCVs) Isolated from Cats with Virulent Systemic Disease Possess In Vitro Phenotypes Distinct from Those of Other FCV Isolates. J. Gen. Virol. 2007, 88, 506–517. [Google Scholar] [CrossRef]
- Stuetzer, B.; Hartmann, K. Feline Parvovirus Infection and Associated Diseases. Vet. J. 2014, 201, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Inthong, N.; Sutacha, K.; Kaewmongkol, S.; Sinsiri, R.; Sribuarod, K.; Sirinarumitr, K.; Sirinarumitr, T. Feline Panleukopenia Virus as the Cause of Diarrhea in a Banded Linsang (Prionodon linsang) in Thailand. J. Vet. Med. Sci. 2019, 81, 1763–1768. [Google Scholar] [CrossRef]
- Van Brussel, K.; Carrai, M.; Lin, C.; Kelman, M.; Setyo, L.; Aberdein, D.; Brailey, J.; Lawler, M.; Maher, S.; Plaganyi, I.; et al. Distinct Lineages of Feline Parvovirus Associated with Epizootic Outbreaks in Australia, New Zealand and the United Arab Emirates. Viruses 2019, 11, 1155. [Google Scholar] [CrossRef]
- Willemse, M.J.; Chalmers, W.S.K.; Cronenberg, A.M.; Pfundt, R.; Strijdveen, I.G.L.; Sondermeijer, P.J.A. The Gene Downstream of the GC Homologue in Feline Herpes Virus Type 1 Is Involved in the Expression of Virulence. J. Gen. Virol. 1994, 75, 3107–3116. [Google Scholar] [CrossRef]
- Winkler, I.G.; Flügel, R.M.; Löchelt, M.; Flower, R.L.P. Detection and Molecular Characterisation of Feline Foamy Virus Serotypes in Naturally Infected Cats. Virology 1998, 247, 144–151. [Google Scholar] [CrossRef]
- Kechejian, S.R.; Dannemiller, N.; Kraberger, S.; Ledesma-Feliciano, C.; Malmberg, J.; Roelke Parker, M.; Cunningham, M.; McBride, R.; Riley, S.P.D.; Vickers, W.T.; et al. Feline Foamy Virus Is Highly Prevalent in Free-Ranging Puma Concolor from Colorado, Florida and Southern California. Viruses 2019, 11, 359. [Google Scholar] [CrossRef]
- Piewbang, C.; Chaiyasak, S.; Kongmakee, P.; Sanannu, S.; Khotapat, P.; Ratthanophart, J.; Banlunara, W.; Somporn, L. Feline Morbillivirus Infection Associated with Tubulointerstitial Nephritis in Black Leopards (Panthera pardus). Vet. Pathol. 2020, 57, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Sieg, M.; Busch, J.; Eschke, M.; Böttcher, D.; Heenemann, K.; Vahlenkamp, A.; Reinert, A.; Seeger, J.; Heilmann, R.; Scheffler, K.; et al. A New Genotype of Feline Morbillivirus Infects Primary Cells of the Lung, Kidney, Brain and Peripheral Blood. Viruses 2019, 11, 146. [Google Scholar] [CrossRef] [PubMed]
- HosI, Y.; ZrMa, J.F.; MoIs, N.S.; Scott, F.W. Detection of astroviruses in feces of a cat with diarrhea. Brief report. Arch. Virol. 1981, 70, 373–376. [Google Scholar]
- Navarro, R.; Yibin, C.; Nair, R.; Peda, A.; Aung, M.S.; Ketzis, J.; Malik, Y.S.; Kobayashi, N.; Ghosh, S. Molecular Characterization of Complete Genomic Segment-2 of Picobirnavirus Strains Detected in a Cat and a Dog. Infect. Genet. Evol. 2017, 54, 200–204. [Google Scholar] [CrossRef]
- Aghazadeh, M.; Shi, M.; Barrs, V.; McLuckie, A.; Lindsay, S.; Jameson, B.; Hampson, B.; Holmes, E.; Beatty, J. A Novel Hepadnavirus Identified in an Immunocompromised Domestic Cat in Australia. Viruses 2018, 10, 269. [Google Scholar] [CrossRef]
- Henzel, A.; Sá e Silva, M.; Luo, S.; Lovato, L.T.; Weiblen, R. Genetic and Phylogenetic Analyses of Capsid Protein Gene in Feline Calicivirus Isolates from Rio Grande Do Sul in Southern Brazil. Virus Res. 2012, 163, 667–671. [Google Scholar] [CrossRef]
- Radford, A.D.; Dawson, S.; Ryvar, R.; Coyne, K.; Johnson, D.R.; Cox, M.B.; Acke, E.F.J.; Addie, D.D.; Gaskell, R.M. High Genetic Diversity of the Immunodominant Region of the Feline Calicivirus Capsid Gene in Endemically Infected Cat Colonies. Virus Genes 2003, 27, 145–155. [Google Scholar] [CrossRef]
- Hou, J.; Sánchez-Vizcaíno, F.; McGahie, D.; Lesbros, C.; Almeras, T.; Howarth, D.; O’Hara, V.; Dawson, S.; Radford, A.D. European Molecular Epidemiology and Strain Diversity of Feline Calicivirus. Vet. Rec. 2016, 178, 114–115. [Google Scholar] [CrossRef] [PubMed]
- Willi, B.; Spiri, A.M.; Meli, M.L.; Samman, A.; Hoffmann, K.; Sydler, T.; Cattori, V.; Graf, F.; Diserens, K.A.; Padrutt, I.; et al. Molecular Characterization and Virus Neutralization Patterns of Severe, Non-Epizootic Forms of Feline Calicivirus Infections Resembling Virulent Systemic Disease in Cats in Switzerland and in Liechtenstein. Vet. Microbiol. 2016, 182, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Koç, B.T.; Oğuzoğlu, T.Ç. The Investigation of Feline Parvoviruses (FPVs) into Two Different Phylogenetic Lineages in Turkey. J. Appl. Biol. Sci. 2016, 10, 4–7. [Google Scholar]
- Li, X.; Wu, H.; Wang, L.; Spibey, N.; Liu, C.; Ding, H.; Liu, W.; Liu, Y.; Tian, K. Genetic Characterization of Parvoviruses in Domestic Cats in Henan Province, China. Transbound. Emerg. Dis. 2018, 65, 1429–1435. [Google Scholar] [CrossRef]
- Tucciarone, C.M.; Franzo, G.; Legnardi, M.; Lazzaro, E.; Zoia, A.; Petini, M.; Furlanello, T.; Caldin, M.; Cecchinato, M.; Drigo, M. Genetic Insights into Feline Parvovirus: Evaluation of Viral Evolutionary Patterns and Association between Phylogeny and Clinical Variables. Viruses 2021, 13, 1033. [Google Scholar] [CrossRef]
- Dye, C.; Siddell, S.G. Genomic RNA Sequence of Feline Coronavirus Strain FCoV C1Je. J. Feline Med. Surg. 2007, 9, 202–213. [Google Scholar] [CrossRef]
- Li, C.; Liu, Q.; Kong, F.; Guo, D.; Zhai, J.; Su, M.; Sun, D. Circulation and Genetic Diversity of Feline Coronavirus Type I and II from Clinically Healthy and FIP-Suspected Cats in China. Transbound. Emerg. Dis. 2019, 66, 763–775. [Google Scholar] [CrossRef]
- An, D.-J.; Jeong, W.; Jeoung, H.-Y.; Yoon, S.H.; Kim, H.-J.; Park, J.-Y.; Park, B.-K. Phylogenetic Analysis of Feline Panleukopenia Virus (FPLV) Strains in Korean Cats. Res. Vet. Sci. 2011, 90, 163–167. [Google Scholar] [CrossRef]
- Teixeira, B.M.; Logan, N.; Cruz, J.C.M.; Reis, J.K.P.; Brandão, P.E.; Richtzenhain, L.J.; Hagiwara, M.K.; Willett, B.J.; Hosie, M.J. Genetic Diversity of Brazilian Isolates of Feline Immunodeficiency Virus. Arch. Virol. 2010, 155, 379–384. [Google Scholar] [CrossRef]
- Ravi, M.; Wobeser, G.A.; Taylor, S.M.; Jackson, M.L. Naturally Acquired Feline Immunodeficiency Virus (FIV) Infection in Cats from Western Canada: Prevalence, Disease Associations, and Survival Analysis. Can. Vet. J. 2010, 51, 271–276. [Google Scholar]
- Roukaerts, I.D.M. Phylogenetic Analysis of Feline Immunodeficiency Virus Strains from Naturally Infected Cats in Belgium and The Netherlands. Virus Res. 2015, 196, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Ortega, C.; Valencia, A.C.; Duque-Valencia, J.; Ruiz-Saenz, J. Prevalence and Genomic Diversity of Feline Leukemia Virus in Privately Owned and Shelter Cats in Aburrá Valley, Colombia. Viruses 2020, 12, 464. [Google Scholar] [CrossRef] [PubMed]
- Miyake, A.; Watanabe, S.; Hiratsuka, T.; Ito, J.; Ngo, M.H.; Makundi, I.; Kawasaki, J.; Endo, Y.; Tsujimoto, H.; Nishigaki, K. Novel Feline Leukemia Virus Interference Group Based on the Env Gene. J. Virol. 2016, 90, 4832–4837. [Google Scholar] [CrossRef] [PubMed]
- Makundi, I.; Koshida, Y.; Kuse, K.; Hiratsuka, T.; Ito, J.; Baba, T.; Watanabe, S.; Kawamura, M.; Odahara, Y.; Miyake, A.; et al. Epidemiologic Survey of Feline Leukemia Virus in Domestic Cats on Tsushima Island, Japan: Management Strategy for Tsushima Leopard Cats. J. Vet. Diagn. Investig. 2017, 29, 889–895. [Google Scholar] [CrossRef]
- Coelho, F.M.; Bomfim, M.R.Q.; de Andrade Caxito, F.; Ribeiro, N.A.; Luppi, M.M.; Costa, É.A.; Oliveira, M.E.; Da Fonseca, F.G.; Resende, M. Naturally Occurring Feline Leukemia Virus Subgroup A and B Infections in Urban Domestic Cats. J. Gen. Virol. 2008, 89, 2799–2805. [Google Scholar] [CrossRef]
- Lauzi, S.; Stranieri, A.; Giordano, A.; Luzzago, C.; Zehender, G.; Paltrinieri, S.; Ebranati, E. Origin and Transmission of Feline Coronavirus Type I in Domestic Cats from Northern Italy: A Phylogeographic Approach. Vet. Microbiol. 2020, 244, 108667. [Google Scholar] [CrossRef]
- Myrrha, L.W.; Silva, F.M.F.; Vidigal, P.M.P.; Resende, M.; Bressan, G.C.; Fietto, J.L.R.; Santos, M.R.; Silva, L.M.N.; Assao, V.S.; Nior, A.S.-J.; et al. Feline Coronavirus Isolates from a Part of Brazil: Insights into Molecular Epidemiology and Phylogeny Inferred from the 7b Gene. J. Vet. Med. Sci. 2019, 81, 1455–1460. [Google Scholar] [CrossRef]
- Cano-Ortiz, L.; Junqueira, D.M.; Comerlato, J.; Costa, C.S.; Zani, A.; Duda, N.B.; Tochetto, C.; dos Santos, R.N.; da Costa, F.V.A.; Roehe, P.M.; et al. Phylodynamics of the Brazilian Feline Immunodeficiency Virus. Infect. Genet. Evol. 2017, 55, 166–171. [Google Scholar] [CrossRef]
- Pedersen, N.C. A Review of Feline Infectious Peritonitis Virus Infection: 1963–2008. J. Feline Med. Surg. 2009, 11, 225–258. [Google Scholar] [CrossRef]
- Drummond, A.; Pybus, O.G.; Rambaut, A. Inference of Viral Evolutionary Rates from Molecular Sequences. Adv. Parasitol. 2003, 54, 331–358. [Google Scholar]
- Abd-Eldaim, M.; Potgieter, L.; Kennedy, M. Genetic Analysis of Feline Caliciviruses Associated with a Hemorrhagic-Like Disease. J. Vet. Diagn. Investig. 2005, 17, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Foley, J.; Hurley, K.; Pesavento, P.A.; Poland, A.; Pedersen, N.C. Virulent Systemic Feline Calicivirus Infection: Local Cytokine Modulation and Contribution of Viral Mutants. J. Feline Med. Surg. 2006, 8, 55–61. [Google Scholar] [CrossRef] [PubMed]
- De Graaf, M.; Van Beek, J.; Koopmans, M.P.G. Human Norovirus Transmission and Evolution in a Changing World. Nat. Rev. Microbiol. 2016, 14, 421–433. [Google Scholar] [CrossRef]
- Jahnke, M.; Holmes, E.C.; Kerr, P.J.; Wright, J.D.; Strive, T. Evolution and Phylogeography of the Nonpathogenic Calicivirus RCV-A1 in Wild Rabbits in Australia. J. Virol. 2010, 84, 12397–12404. [Google Scholar] [CrossRef] [PubMed]
- Shackelton, L.A.; Parrish, C.R.; Truyen, U.; Holmes, E.C. High Rate of Viral Evolution Associated with the Emergence of Carnivore Parvovirus. Proc. Natl. Acad. Sci. USA 2005, 102, 379–384. [Google Scholar] [CrossRef]
- Boni, M.F.; Lemey, P.; Jiang, X.; Lam, T.T.-Y.; Perry, B.W.; Castoe, T.A.; Rambaut, A.; Robertson, D.L. Evolutionary Origins of the SARS-CoV-2 Sarbecovirus Lineage Responsible for the COVID-19 Pandemic. Nat. Microbiol. 2020, 5, 1408–1417. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of Evolutionary Change in Viruses: Patterns and Determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef]
- Symes, S.J.; Job, N.; Ficorilli, N.; Hartley, C.A.; Browning, G.F.; Gilkerson, J.R. Novel Assay to Quantify Recombination in a Calicivirus. Vet. Microbiol. 2015, 177, 25–31. [Google Scholar] [CrossRef]
- Lai, M.M.C. RNA Recombination in Animal and Plant Viruses. Microbiol. Rev. 1992, 56, 61–79. [Google Scholar] [CrossRef]
- Terada, Y.; Matsui, N.; Noguchi, K.; Kuwata, R.; Shimoda, H.; Soma, T.; Mochizuki, M.; Maeda, K. Emergence of Pathogenic Coronaviruses in Cats by Homologous Recombination between Feline and Canine Coronaviruses. PLoS ONE 2014, 9, e106534. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).