First Detection of Hepatitis E Virus (Rocahepevirus ratti Genotype C1) in Synanthropic Norway Rats (Rattus norvegicus) in Romania
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
- -
- -
- Nine foxes (Vulpes vulpes) from Vrancea county (Figure 1);
- -
- Four ferrets (Mustela putorius), one from Iasi County and three from Tulcea County, one coypu (Myocastor coypus) and one muskrat (Ondatra zibethicus) from Tulcea County, one mole (Talpa europaea) from Suceava County and one mouse (Mus musculus) from Iasi County (Figure 1).
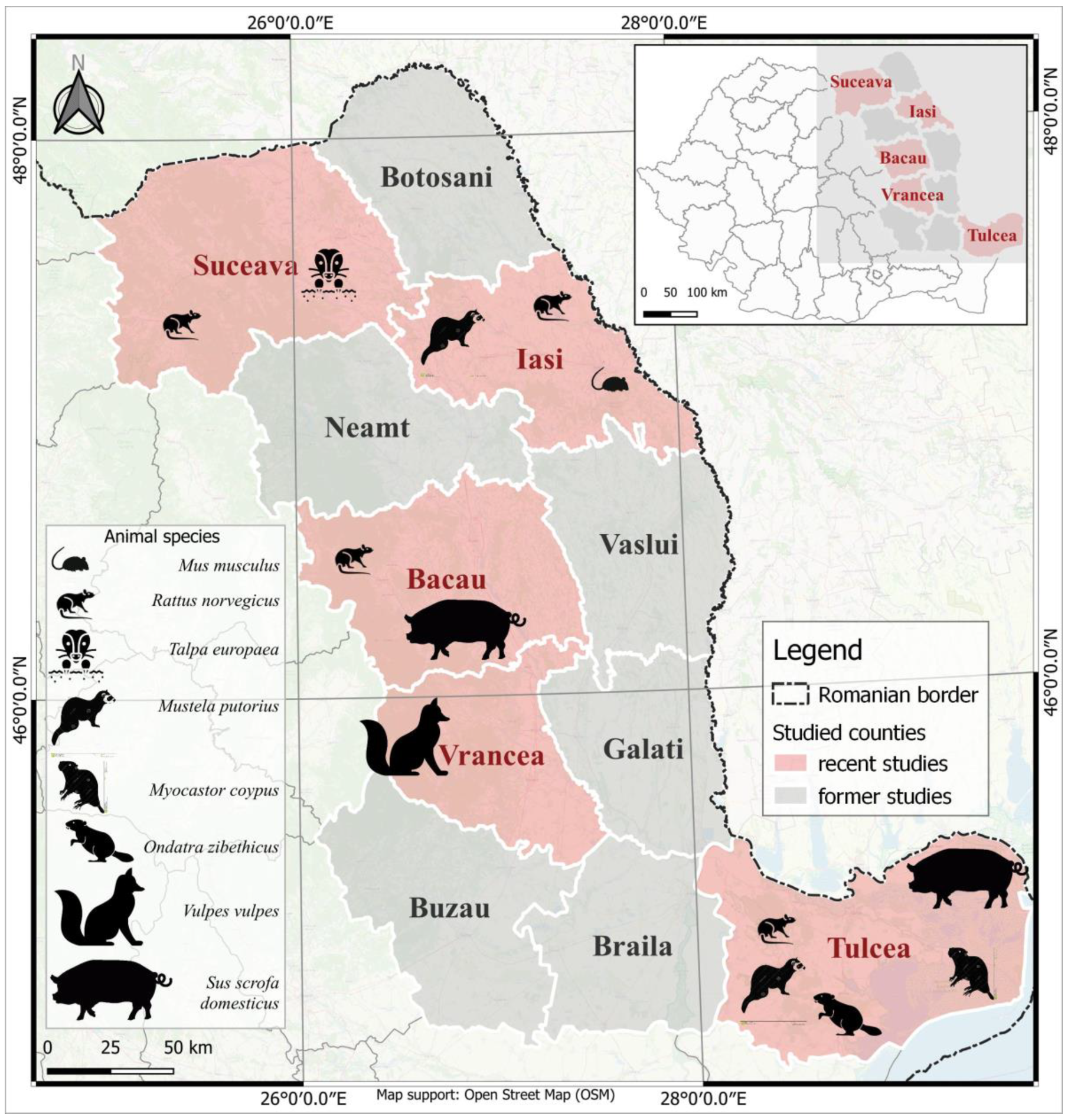
| No. | Location/Site | Number of Rats/Location/Counties | |||
|---|---|---|---|---|---|
| Suceava | Iasi | Bacau | Tulcea | ||
| 1 | Intensive system pig farm | - | - | - | 12 |
| 2 | Semi-intensive system pig farm | 5 | - | 11 | - |
| 3 | Extensive system pig farm | 1 | - | 5 | - |
| 4 | Farm with different animal species, forestry site | 10 | - | - | - |
| 5 | Cattle farm, metropolitan area | - | 1 | - | - |
| 6 | Urban settlements, near garbage cans | - | 5 | - | - |
| 7 | Rural settlements, population’s household | - | - | - | 2 |
| Total | 16 | 6 | 16 | 14 | |
2.2. RNA Extraction and Gene Amplification—PCR Assays
2.3. Nucleotide Sequencing and Phylogenetic Analysis
3. Results
3.1. Detection of HEV RNA
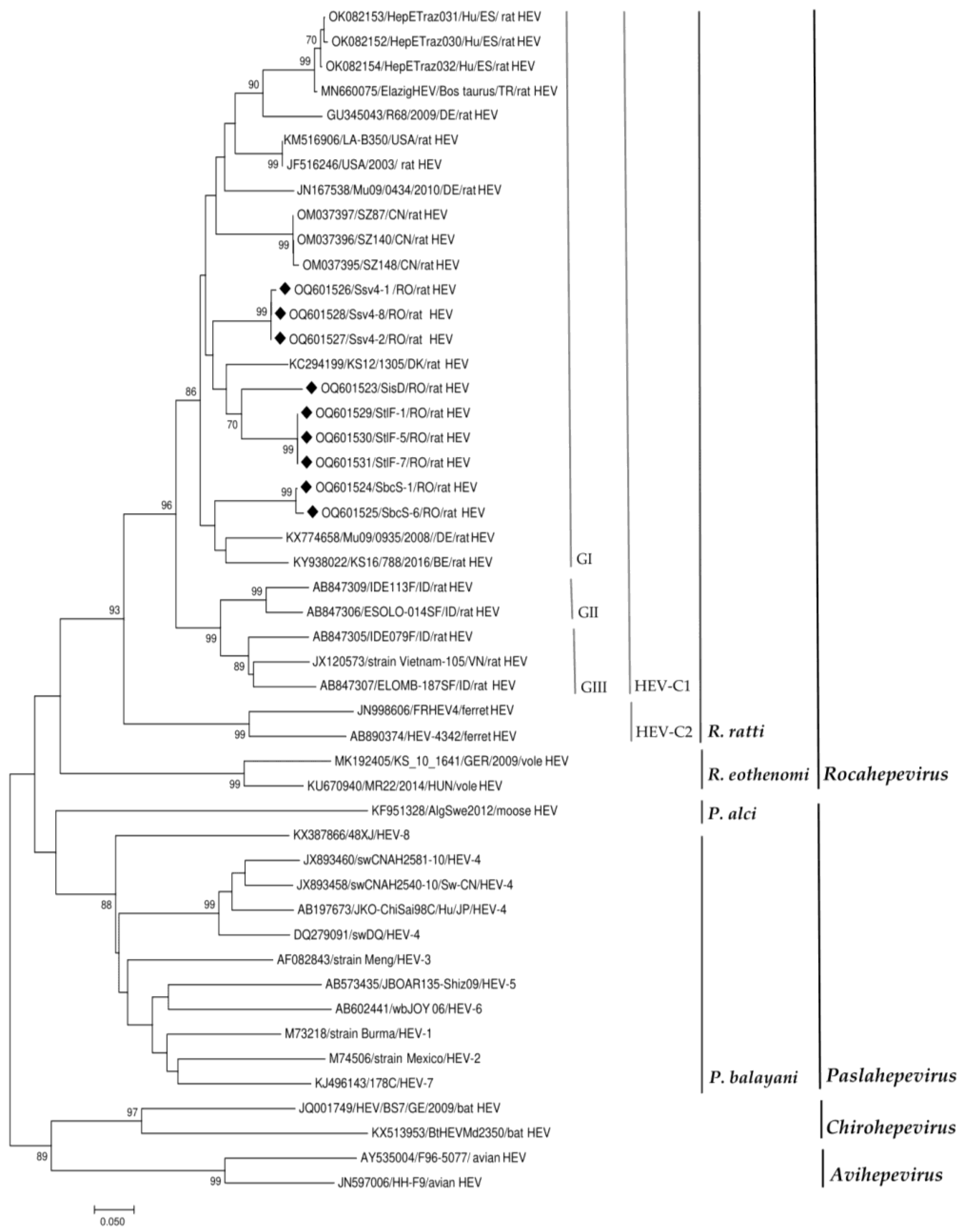
3.2. HEV Sequence Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Purdy, M.A.; Drexler, J.F.; Meng, X.-J.; Norder, H.; Okamoto, H.; Van der Poel, W.H.M.; Reuter, G.; de Souza, W.M.; Ulrich, R.G.; Smith, D.B. ICTV Virus Taxonomy Profile: Hepeviridae 2022. J. Gen. Virol. 2022, 103, 001778. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, X.-L. Chirohepevirus from Bats: Insights into Hepatitis E Virus Diversity and Evolution. Viruses 2022, 14, 905. [Google Scholar] [CrossRef] [PubMed]
- Kamar, N.; Izopet, J.; Pavio, N.; Aggarwal, R.; Labrique, A.; Wedemeyer, H.; Dalton, H.R. Hepatitis E Virus Infection. Nat. Rev. Dis. Prim. 2017, 3, 17086. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Teng, J.L.L.; Lau, S.K.P.; Sridhar, S.; Fu, H.; Gong, W.; Li, M.; Xu, Q.; He, Y.; Zhuang, H.; et al. Transmission of a Novel Genotype of Hepatitis E Virus from Bactrian Camels to Cynomolgus Macaques. J. Virol. 2019, 93, e02014-18. [Google Scholar] [CrossRef] [PubMed]
- Rivero-Juarez, A.; Frias, M.; Perez, A.B.; Pineda, J.A.; Reina, G.; Fuentes-Lopez, A.; Freyre-Carrillo, C.; Ramirez-Arellano, E.; Alados, J.C.; Rivero, A. Orthohepevirus C Infection as an Emerging Cause of Acute Hepatitis in Spain: First Report in Europe. J. Hepatol. 2022, 77, 326–331. [Google Scholar] [CrossRef]
- Wang, B.; Harms, D.; Yang, X.-L.; Bock, C.-T. Orthohepevirus C: An Expanding Species of Emerging Hepatitis E Virus Variants. Pathogens 2020, 9, 154. [Google Scholar] [CrossRef]
- Raj, V.S.; Smits, S.L.; Pas, S.D.; Provacia, L.B.V.; Moorman-Roest, H.; Osterhaus, A.D.M.E.; Haagmans, B.L. Novel Hepatitis E Virus in Ferrets, the Netherlands. Emerg. Infect. Dis. 2012, 18, 1369–1370. [Google Scholar] [CrossRef]
- Krog, J.S.; Breum, S.Ø.; Jensen, T.H.; Larsen, L.E. Hepatitis E Virus Variant in Farmed Mink, Denmark. Emerg. Infect. Dis. 2013, 19, 2028–2030. [Google Scholar] [CrossRef]
- Bodewes, R.; van der Giessen, J.; Haagmans, B.L.; Osterhaus, A.D.M.E.; Smits, S.L. Identification of Multiple Novel Viruses, Including a Parvovirus and a Hepevirus, in Feces of Red Foxes. J. Virol. 2013, 87, 7758–7764. [Google Scholar] [CrossRef]
- Eiden, M.; Dähnert, L.; Spoerel, S.; Vina-Rodriguez, A.; Schröder, R.; Conraths, F.J.; Groschup, M.H. Spatial-Temporal Dynamics of Hepatitis E Virus Infection in Foxes (Vulpes Vulpes) in Federal State of Brandenburg, Germany, 1993–2012. Front. Microbiol. 2020, 11, 115. [Google Scholar] [CrossRef]
- Kurucz, K.; Hederics, D.; Bali, D.; Kemenesi, G.; Horváth, G.; Jakab, F. Hepatitis E Virus in Common Voles (Microtus Arvalis) from an Urban Environment, Hungary: Discovery of a Cricetidae-Specific Genotype of Orthohepevirus C. Zoonoses Public Health 2019, 66, 259–263. [Google Scholar] [CrossRef]
- Wang, B.; Li, W.; Zhou, J.-H.; Li, B.; Zhang, W.; Yang, W.-H.; Pan, H.; Wang, L.-X.; Bock, C.T.; Shi, Z.-L.; et al. Chevrier’s Field Mouse (Apodemus Chevrieri) and Père David’s Vole (Eothenomys Melanogaster) in China Carry Orthohepeviruses That Form Two Putative Novel Genotypes Within the Species Orthohepevirus C. Virol. Sin. 2018, 33, 44–58. [Google Scholar] [CrossRef]
- Ryll, R.; Heckel, G.; Corman, V.M.; Drexler, J.F.; Ulrich, R.G. Genomic and Spatial Variability of a European Common Vole Hepevirus. Arch. Virol. 2019, 164, 2671–2682. [Google Scholar] [CrossRef]
- Doceul, V.; Bagdassarian, E.; Demange, A.; Pavio, N. Zoonotic Hepatitis E Virus: Classification, Animal Reservoirs and Transmission Routes. Viruses 2016, 8, 270. [Google Scholar] [CrossRef]
- Sridhar, S.; Yip, C.C.Y.; Lo, K.H.Y.; Wu, S.; Situ, J.; Chew, N.F.S.; Leung, K.H.; Chan, H.S.Y.; Wong, S.C.Y.; Leung, A.W.S.; et al. Hepatitis E Virus Species C Infection in Humans, Hong Kong. Clin. Infect. Dis. 2022, 75, 288–296. [Google Scholar] [CrossRef]
- Andonov, A.; Robbins, M.; Borlang, J.; Cao, J.; Hatchette, T.; Stueck, A.; Deschambault, Y.; Murnaghan, K.; Varga, J.; Johnston, L. Rat Hepatitis E Virus Linked to Severe Acute Hepatitis in an Immunocompetent Patient. J. Infect. Dis. 2019, 220, 951–955. [Google Scholar] [CrossRef]
- Rodriguez, C.; Marchand, S.; Sessa, A.; Cappy, P.; Pawlotsky, J.-M. Orthohepevirus C Hepatitis, an Underdiagnosed Disease? J. Hepatol. 2023, 77, S016882782300096X. [Google Scholar] [CrossRef]
- Sridhar, S.; Situ, J.; Cai, J.-P.; Yip, C.C.-Y.; Wu, S.; Zhang, A.J.-X.; Wen, L.; Chew, N.F.-S.; Chan, W.-M.; Poon, R.W.-S.; et al. Multimodal Investigation of Rat Hepatitis E Virus Antigenicity: Implications for Infection, Diagnostics, and Vaccine Efficacy. J. Hepatol. 2021, 74, 1315–1324. [Google Scholar] [CrossRef]
- Johne, R.; Heckel, G.; Plenge-Bönig, A.; Kindler, E.; Maresch, C.; Reetz, J.; Schielke, A.; Ulrich, R.G. Novel Hepatitis E Virus Genotype in Norway Rats, Germany. Emerg. Infect. Dis. 2010, 16, 1452–1455. [Google Scholar] [CrossRef]
- Reuter, G.; Boros, Á.; Pankovics, P. Review of Hepatitis E Virus in Rats: Evident Risk of Species Orthohepevirus C to Human Zoonotic Infection and Disease. Viruses 2020, 12, 1148. [Google Scholar] [CrossRef]
- Widén, F.; Ayral, F.; Artois, M.; Olofson, A.-S.; Lin, J. PCR Detection and Analyzis of Potentially Zoonotic Hepatitis E Virus in French Rats. Virol. J. 2014, 11, 90. [Google Scholar] [CrossRef] [PubMed]
- Ryll, R.; Bernstein, S.; Heuser, E.; Schlegel, M.; Dremsek, P.; Zumpe, M.; Wolf, S.; Pépin, M.; Bajomi, D.; Müller, G.; et al. Detection of Rat Hepatitis E Virus in Wild Norway Rats (Rattus Norvegicus) and Black Rats (Rattus Rattus) from 11 European Countries. Vet. Microbiol. 2017, 208, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Simanavicius, M.; Juskaite, K.; Verbickaite, A.; Jasiulionis, M.; Tamosiunas, P.L.; Petraityte-Burneikiene, R.; Zvirbliene, A.; Ulrich, R.G.; Kucinskaite-Kodze, I. Detection of Rat Hepatitis E Virus, but Not Human Pathogenic Hepatitis E Virus Genotype 1–4 Infections in Wild Rats from Lithuania. Vet. Microbiol. 2018, 221, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.G.; Williams, N.J.; Jennings, D.; Chantrey, J.; Verin, R.; Grierson, S.; McElhinney, L.M.; Bennett, M. First Detection of Hepatitis E Virus (Orthohepevirus C) in Wild Brown Rats (Rattus Norvegicus) from Great Britain. Zoonoses Public Health 2019, 66, 686–694. [Google Scholar] [CrossRef] [PubMed]
- De Sabato, L.; Ianiro, G.; Monini, M.; De Lucia, A.; Ostanello, F.; Di Bartolo, I. Detection of Hepatitis E Virus RNA in Rats Caught in Pig Farms from Northern Italy. Zoonoses Public Health 2020, 67, 62–69. [Google Scholar] [CrossRef]
- Niendorf, S.; Harms, D.; Hellendahl, K.F.; Heuser, E.; Böttcher, S.; Jacobsen, S.; Bock, C.-T.; Ulrich, R.G. Presence and Diversity of Different Enteric Viruses in Wild Norway Rats (Rattus Norvegicus). Viruses 2021, 13, 992. [Google Scholar] [CrossRef]
- Kanai, Y.; Miyasaka, S.; Uyama, S.; Kawami, S.; Kato-Mori, Y.; Tsujikawa, M.; Yunoki, M.; Nishiyama, S.; Ikuta, K.; Hagiwara, K. Hepatitis E Virus in Norway Rats (Rattus Norvegicus) Captured around a Pig Farm. BMC Res. Notes 2012, 5, 4. [Google Scholar] [CrossRef]
- Lack, J.B.; Volk, K.; Van Den Bussche, R.A. Hepatitis E Virus Genotype 3 in Wild Rats, United States. Emerg. Infect. Dis. 2012, 18, 1268. [Google Scholar] [CrossRef]
- Prpić, J.; Keros, T.; Vucelja, M.; Bjedov, L.; Đaković Rode, O.; Margaletić, J.; Habrun, B.; Jemeršić, L. First Evidence of Hepatitis E Virus Infection in a Small Mammal (Yellow-Necked Mouse) from Croatia. PLoS ONE 2019, 14, e0225583. [Google Scholar] [CrossRef]
- Cunha, L.; Luchs, A.; Azevedo, L.S.; Silva, V.C.M.; Lemos, M.F.; Costa, A.C.; Compri, A.P.; França, Y.; Viana, E.; Malta, F.; et al. Detection of Hepatitis E Virus Genotype 3 in Feces of Capybaras (Hydrochoeris Hydrochaeris) in Brazil. Viruses 2023, 15, 335. [Google Scholar] [CrossRef]
- Pavio, N.; Kooh, P.; Cadavez, V.; Gonzales-Barron, U.; Thébault, A. Risk Factors for Sporadic Hepatitis E Infection: A Systematic Review and Meta-Analysis. Microb. Risk Anal. 2021, 17, 100129. [Google Scholar] [CrossRef]
- Aniţă, A.; Gorgan, L.; Aniţă, D.; Oşlobanu, L.; Pavio, N.; Savuţa, G. Evidence of Hepatitis E Infection in Swine and Humans in the East Region of Romania. Int. J. Infect. Dis. 2014, 29, 232–237. [Google Scholar] [CrossRef]
- Porea, D.; Anita, A.; Demange, A.; Raileanu, C.; Oslobanu Ludu, L.; Anita, D.; Savuta, G.; Pavio, N. Molecular Detection of Hepatitis E Virus in Wild Boar Population in Eastern Romania. Transbound. Emerg. Dis. 2018, 65, 527–533. [Google Scholar] [CrossRef]
- Porea, D.; Anita, A.; Vata, A.; Teodor, D.; Crivei, L.; Raileanu, C.; Gotu, V.; Ratoi, I.; Cozma, A.; Anita, D.; et al. Common European Origin of Hepatitis E Virus in Human Population From Eastern Romania. Front. Public Health 2020, 8, 578163. [Google Scholar] [CrossRef]
- Jothikumar, N.; Cromeans, T.L.; Robertson, B.H.; Meng, X.J.; Hill, V.R. A Broadly Reactive One-Step Real-Time RT-PCR Assay for Rapid and Sensitive Detection of Hepatitis E Virus. J. Virol. Methods 2006, 131, 65–71. [Google Scholar] [CrossRef]
- Barnaud, E.; Rogée, S.; Garry, P.; Rose, N.; Pavio, N. Thermal Inactivation of Infectious Hepatitis E Virus in Experimentally Contaminated Food. Appl. Env. Microbiol. 2012, 78, 5153–5159. [Google Scholar] [CrossRef]
- Cooper, K.; Huang, F.F.; Batista, L.; Rayo, C.D.; Bezanilla, J.C.; Toth, T.E.; Meng, X.J. Identification of Genotype 3 Hepatitis E Virus (HEV) in Serum and Fecal Samples from Pigs in Thailand and Mexico, Where Genotype 1 and 2 HEV Strains Are Prevalent in the Respective Human Populations. J. Clin. Microbiol. 2005, 43, 1684–1688. [Google Scholar] [CrossRef]
- Bouquet, J.; Tesse, S.; Lunazzi, A.; Eloit, M.; Rose, N.; Nicand, E.; Pavio, N. Close Similarity between Sequences of Hepatitis E Virus Recovered from Humans and Swine, France, 2008−2009. Emerg. Infect. Dis. 2011, 17, 2018. [Google Scholar] [CrossRef]
- Johne, R.; Plenge-Bonig, A.; Hess, M.; Ulrich, R.G.; Reetz, J.; Schielke, A. Detection of a Novel Hepatitis E-like Virus in Faeces of Wild Rats Using a Nested Broad-Spectrum RT-PCR. J. Gen. Virol. 2010, 91, 750–758. [Google Scholar] [CrossRef]
- Pavio, N.; Laval, M.; Maestrini, O.; Casabianca, F.; Charrier, F.; Jori, F. Possible Foodborne Transmission of Hepatitis E Virus from Domestic Pigs and Wild Boars from Corsica. Emerg. Infect. Dis. 2016, 22, 2197–2199. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the Number of Nucleotide Substitutions in the Control Region of Mitochondrial DNA in Humans and Chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Guan, D.; Su, J.; Takeda, N.; Wakita, T.; Li, T.-C.; Ke, C.W. High Prevalence of Rat Hepatitis E Virus in Wild Rats in China. Vet. Microbiol. 2013, 165, 275–280. [Google Scholar] [CrossRef]
- Parraud, D.; Lhomme, S.; Péron, J.M.; Da Silva, I.; Tavitian, S.; Kamar, N.; Izopet, J.; Abravanel, F. Rat Hepatitis E Virus: Presence in Humans in South-Western France? Front. Med. 2021, 8, 726363. [Google Scholar] [CrossRef]
- Faber, M.; Wenzel, J.J.; Erl, M.; Stark, K.; Schemmerer, M. No Evidence for Orthohepevirus C in Archived Human Samples in Germany, 2000–2020. Viruses 2022, 14, 742. [Google Scholar] [CrossRef]
- Johne, R.; Dremsek, P.; Kindler, E.; Schielke, A.; Plenge-Bönig, A.; Gregersen, H.; Wessels, U.; Schmidt, K.; Rietschel, W.; Groschup, M.H.; et al. Rat Hepatitis E Virus: Geographical Clustering within Germany and Serological Detection in Wild Norway Rats (Rattus Norvegicus). Infect. Genet. Evol. 2012, 12, 947–956. [Google Scholar] [CrossRef]
- Mulyanto; Suparyatmo, J.B.; Andayani, I.G.A.S.; Khalid; Takahashi, M.; Ohnishi, H.; Jirintai, S.; Nagashima, S.; Nishizawa, T.; Okamoto, H. Marked Genomic Heterogeneity of Rat Hepatitis E Virus Strains in Indonesia Demonstrated on a Full-Length Genome Analysis. Virus Res. 2014, 179, 102–112. [Google Scholar] [CrossRef]
- Wolf, S.; Reetz, J.; Johne, R.; Heiberg, A.-C.; Petri, S.; Kanig, H.; Ulrich, R.G. The Simultaneous Occurrence of Human Norovirus and Hepatitis E Virus in a Norway Rat (Rattus Norvegicus). Arch. Virol. 2013, 158, 1575–1578. [Google Scholar] [CrossRef]
- Grierson, S.; Rabie, A.; Lambert, M.; Choudhury, B.; Smith, R.P. HEV Infection Not Evident in Rodents on English Pig Farms. Vet. Rec. 2018, 182, 81. [Google Scholar] [CrossRef]
- Camp, J.V.; Desvars-Larrive, A.; Nowotny, N.; Walzer, C. Monitoring Urban Zoonotic Virus Activity: Are City Rats a Promising Surveillance Tool for Emerging Viruses? Viruses 2022, 14, 1516. [Google Scholar] [CrossRef]
- Lhomme, S.; Top, S.; Bertagnoli, S.; Dubois, M.; Guerin, J.-L.; Izopet, J. Wildlife Reservoir for Hepatitis E Virus, Southwestern France. Emerg. Infect. Dis. 2015, 21, 1224–1226. [Google Scholar] [CrossRef]
| Species | Type of Samples | County | ||||
|---|---|---|---|---|---|---|
| Wild | Liver | Suceava | Iasi | Bacau | Tulcea | Vrancea |
| Rattus norvegicus | 3/16 (3/10 4) | 1/6 (1/1 5) | 2/16 (2/11 2) | 3/14 (3/12 1) | - | |
| Vulpes vulpes | - | - | - | - | 0/9 | |
| Mustela putorius | - | 0/1 | - | 0/3 | - | |
| Mus musculus | - | 0/1 | - | - | - | |
| Talpa europaea | 0/1 | - | - | - | - | |
| Myocastor coypus | - | - | - | 0/1 | - | |
| Ondatra zibethicus | - | - | - | 0/1 | - | |
| Domestic | ||||||
| Sus scrofa domesticus | Pool faecal | - | - | - | 0/10 | - |
| Sample Number | County | Accession Number | Best Sequence Identity (%) Country |
|---|---|---|---|
| SiD | Iasi | OQ601523 | MH729811 (87%) CN |
| StlF-1 | Tulcea | OQ601529 | KC294199 (89%) DK |
| StlF-5 | Tulcea | OQ601530 | |
| StlF-7 | Tulcea | OQ601531 | |
| Ssv4-1 | Suceava | OQ601526 | ON644869 (89%) NL OM037396 (89%) CN |
| Ssv4-2 | Suceava | OQ601527 | |
| Ssv4-8 | Suceava | OQ601528 | |
| SbcS-1 | Bacau | OQ601524 | KX774658 (87%) DE |
| SbcS-6 | Bacau | OQ601525 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porea, D.; Raileanu, C.; Crivei, L.A.; Gotu, V.; Savuta, G.; Pavio, N. First Detection of Hepatitis E Virus (Rocahepevirus ratti Genotype C1) in Synanthropic Norway Rats (Rattus norvegicus) in Romania. Viruses 2023, 15, 1337. https://doi.org/10.3390/v15061337
Porea D, Raileanu C, Crivei LA, Gotu V, Savuta G, Pavio N. First Detection of Hepatitis E Virus (Rocahepevirus ratti Genotype C1) in Synanthropic Norway Rats (Rattus norvegicus) in Romania. Viruses. 2023; 15(6):1337. https://doi.org/10.3390/v15061337
Chicago/Turabian StylePorea, Daniela, Cristian Raileanu, Luciana Alexandra Crivei, Vasilica Gotu, Gheorghe Savuta, and Nicole Pavio. 2023. "First Detection of Hepatitis E Virus (Rocahepevirus ratti Genotype C1) in Synanthropic Norway Rats (Rattus norvegicus) in Romania" Viruses 15, no. 6: 1337. https://doi.org/10.3390/v15061337
APA StylePorea, D., Raileanu, C., Crivei, L. A., Gotu, V., Savuta, G., & Pavio, N. (2023). First Detection of Hepatitis E Virus (Rocahepevirus ratti Genotype C1) in Synanthropic Norway Rats (Rattus norvegicus) in Romania. Viruses, 15(6), 1337. https://doi.org/10.3390/v15061337






