Plant Poly(ADP-Ribose) Polymerase 1 Is a Potential Mediator of Cross-Talk between the Cajal Body Protein Coilin and Salicylic Acid-Mediated Antiviral Defence
Abstract
1. Introduction
2. Materials and Methods
2.1. Transgenic Plants and TRV-Based Constructs
2.2. RNA Extraction and Northern Blot Analysis
2.3. Real Time Quantitative RT-PCR (RT-qPCR)
2.4. Immunolabelling and Confocal Imaging Analysis
2.5. Co-Immunoprecipitation and Western Blotting
2.6. Immunological Detection of Poly ADP-Ribose (PAR)
2.7. 3-Aminobenzamide (3AB) Treatment
2.8. Expression and Purification of the Recombinant Coilin and Far-Western Blot Analysis
2.9. Virus-Induced Silencing of PARP1 Expression
2.10. Measurement of Endogenous SA
2.11. Foliar Application of SA
2.12. Callose Staining
3. Results
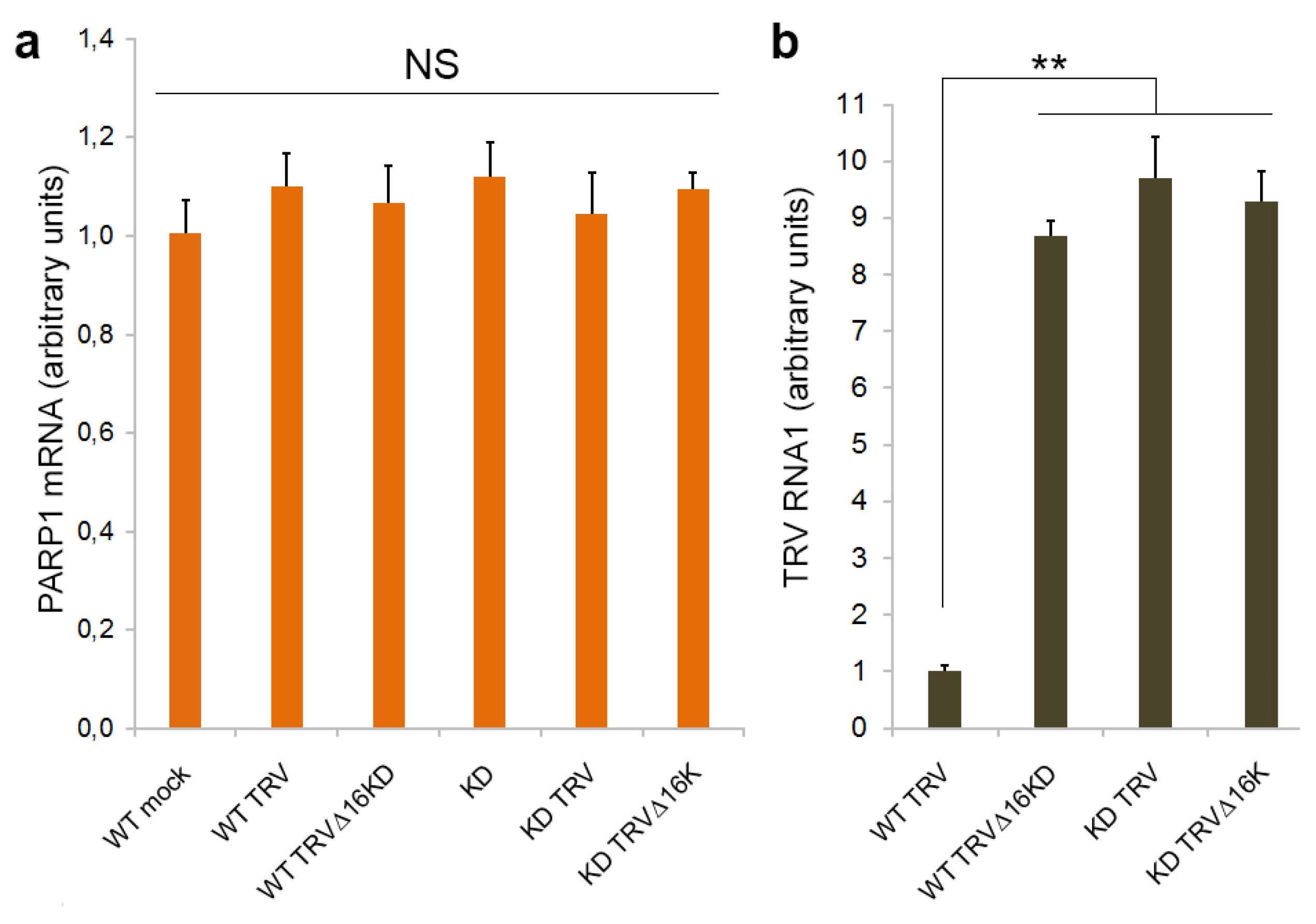
3.1. PARP1 Gene Expression Is Unaffected by TRV Infection
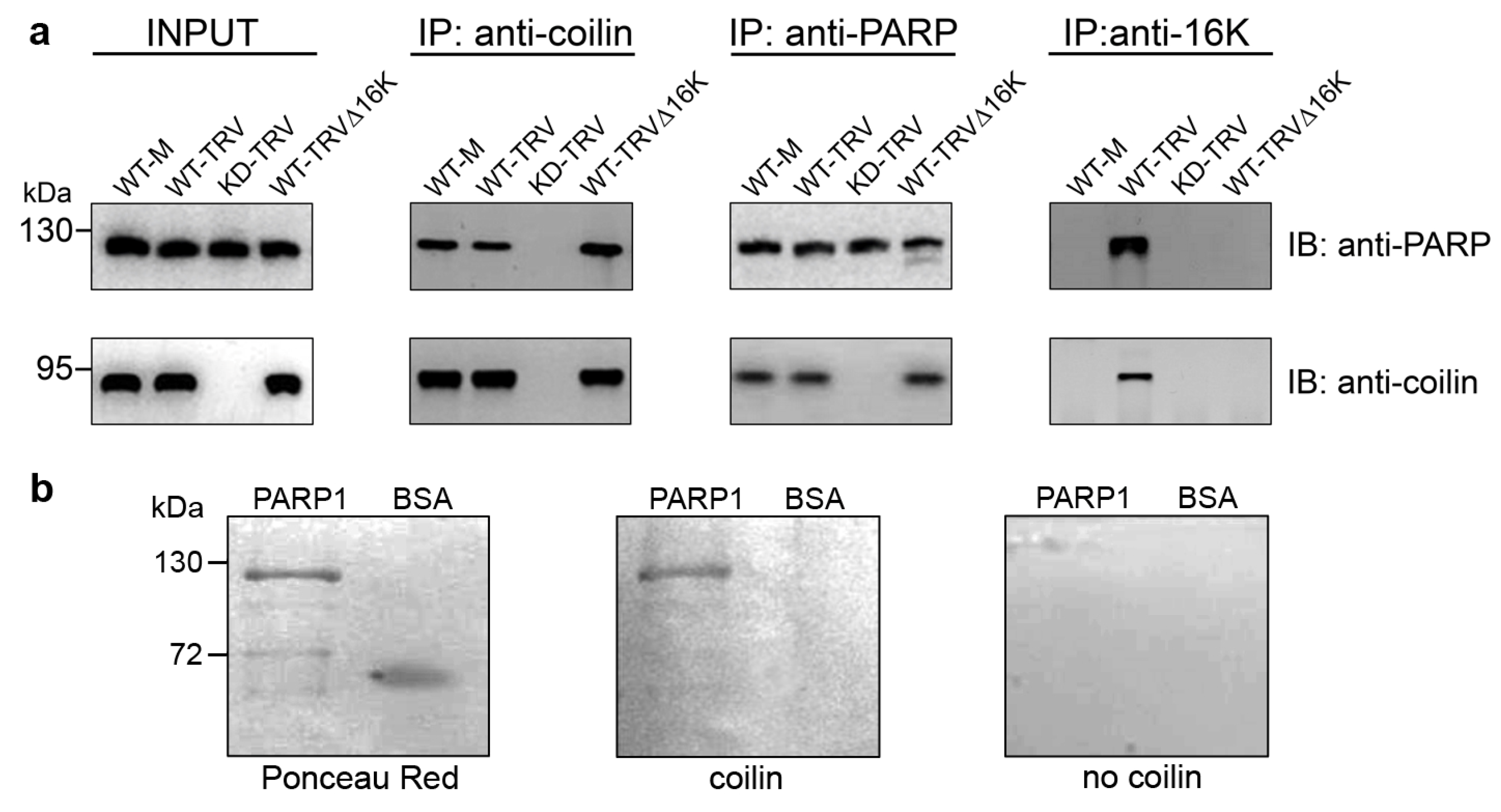
3.2. Interaction between Coilin and PARP1
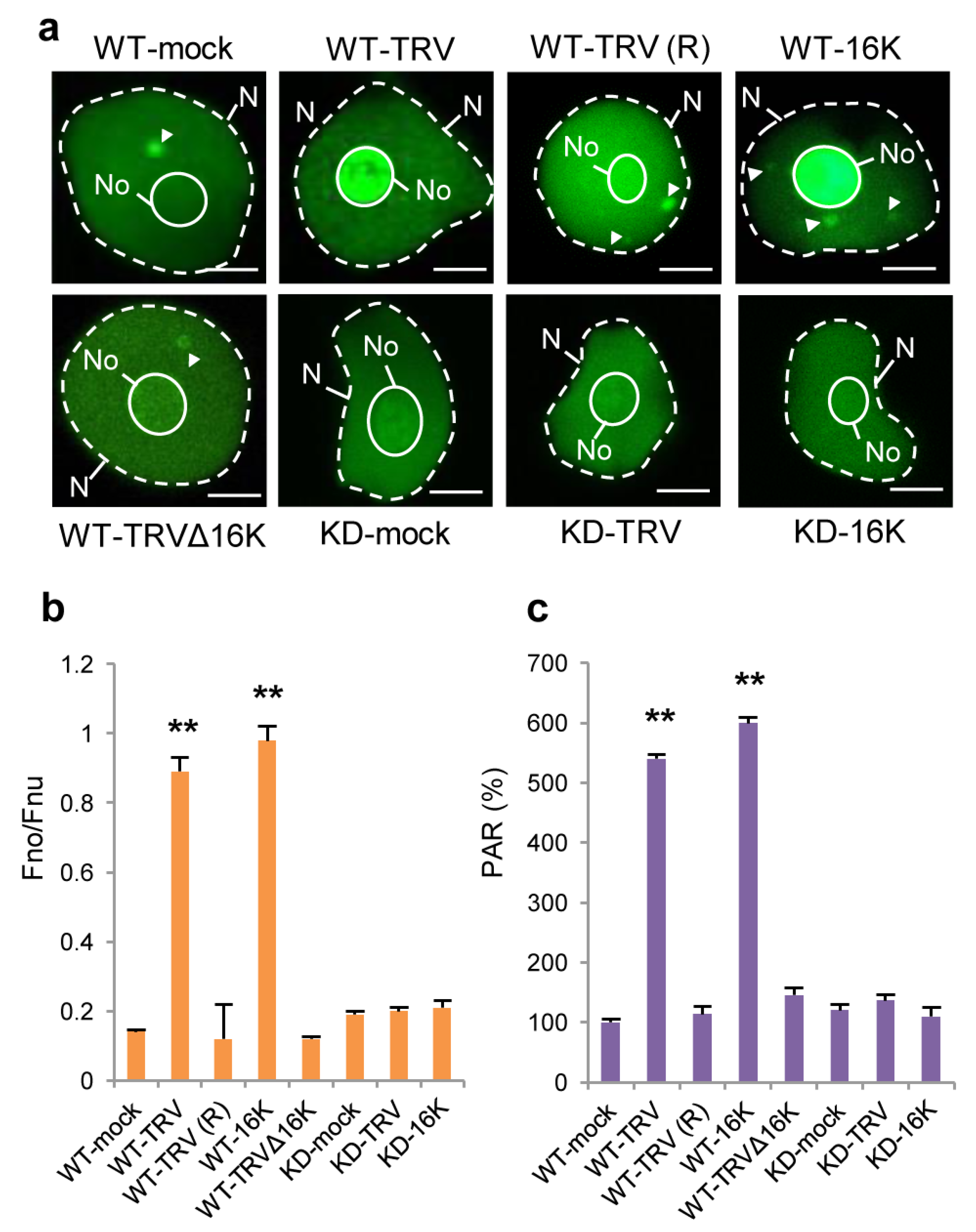
3.3. Effect of TRV Infection on the Intracellular Localisation of PARP1 and Accumulation of PARylated Proteins
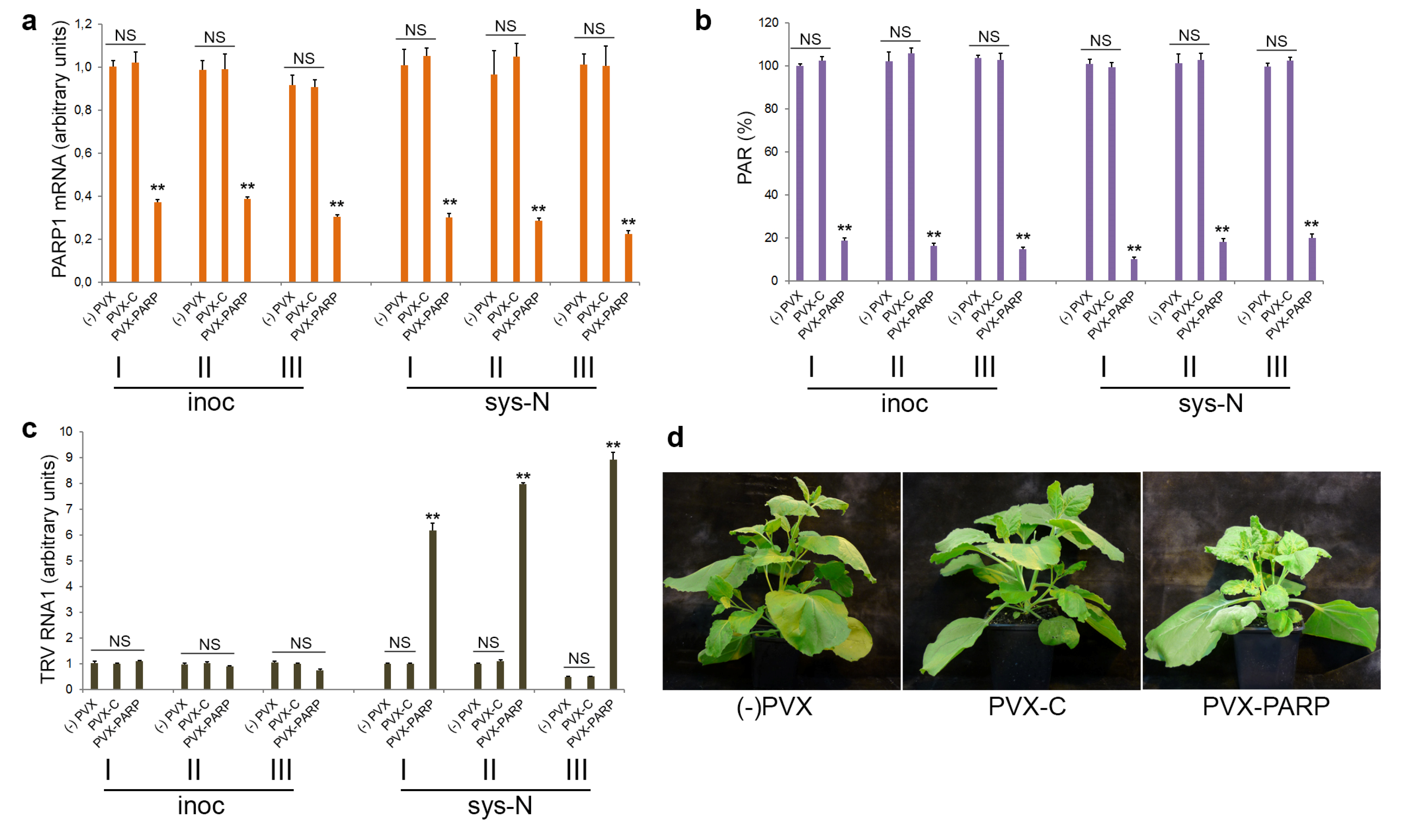
3.4. Effect of Deficiency in PARP Activity on TRV Infection
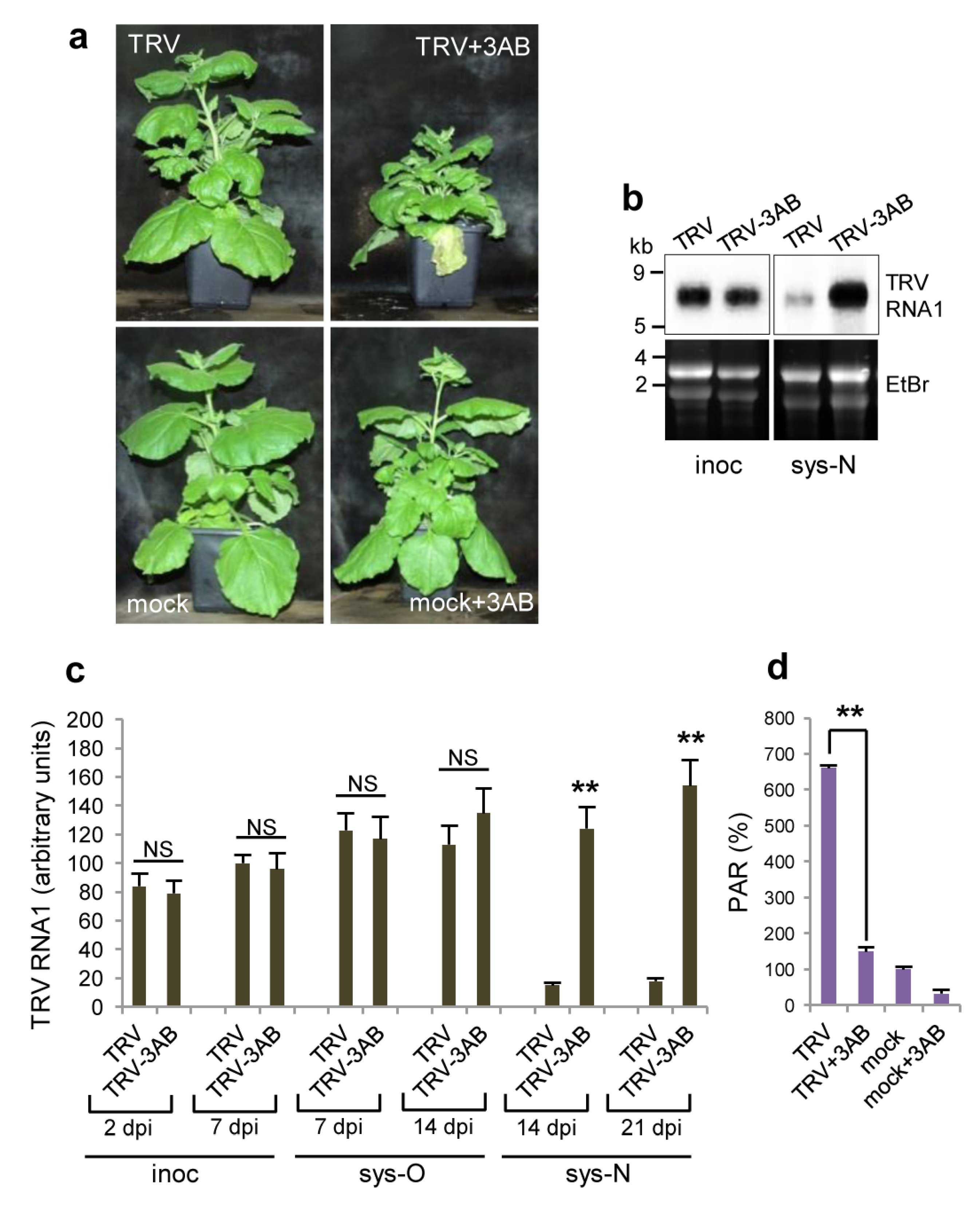
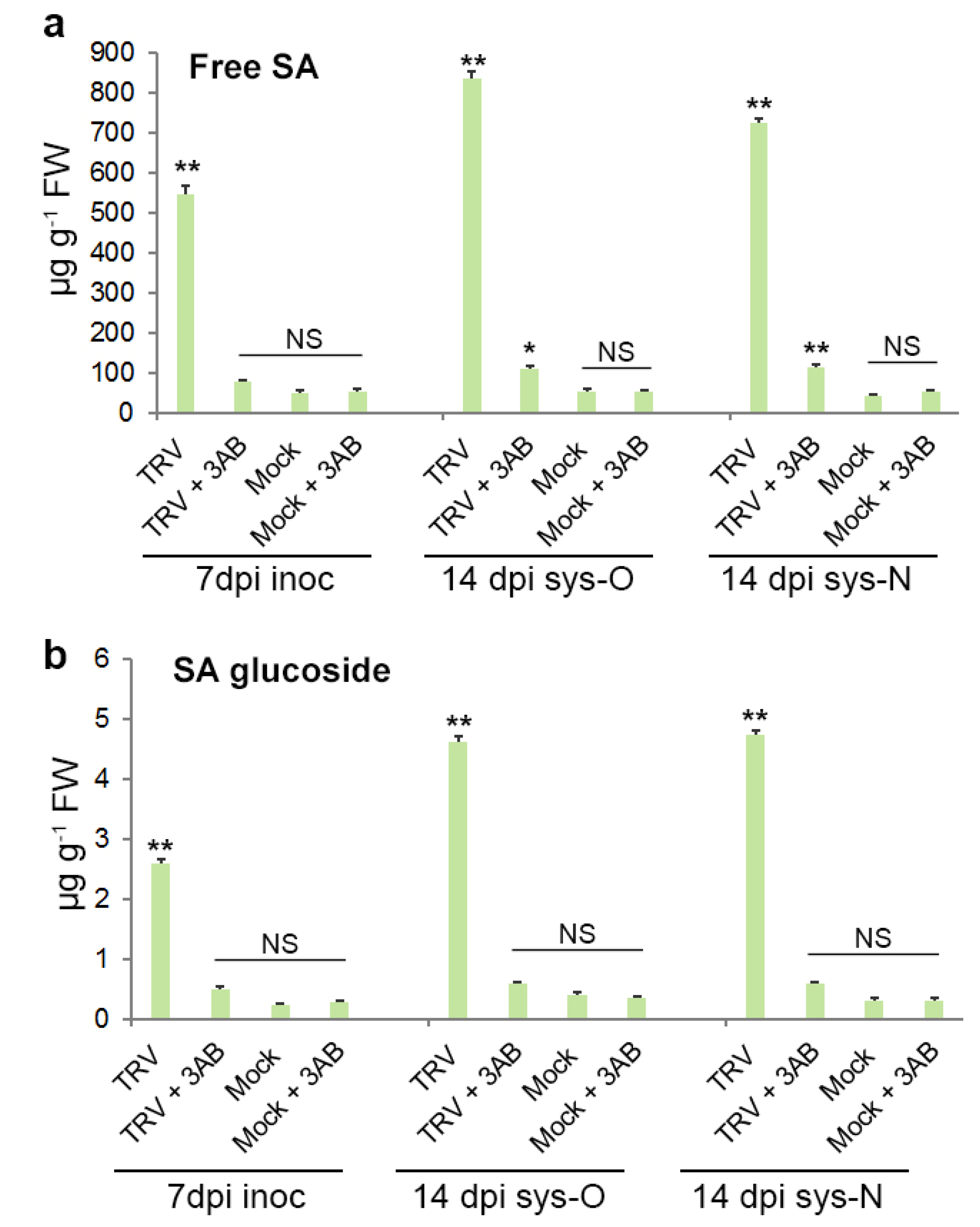
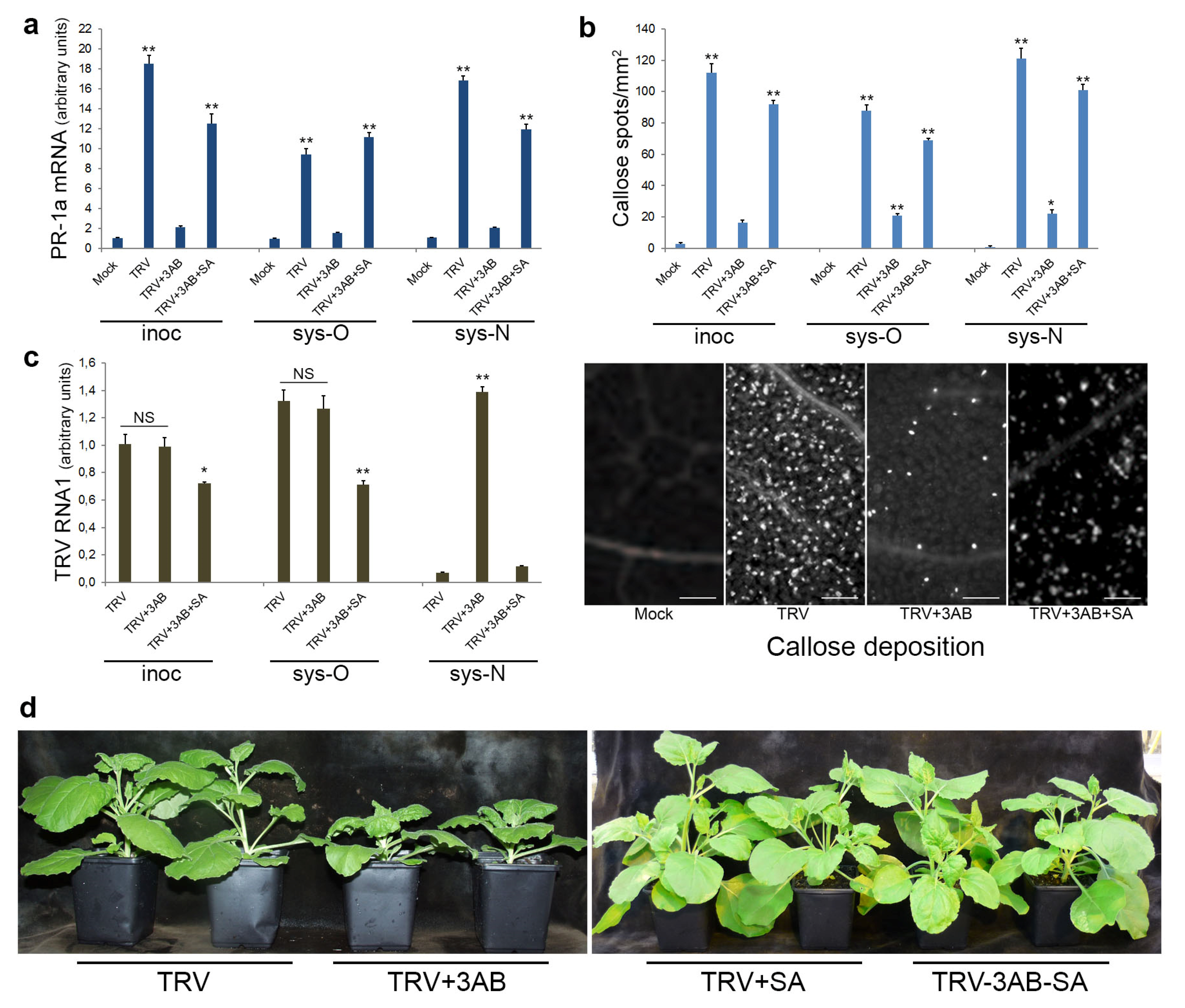
3.5. The Role of PARP1 in Plant SA-Mediated Defence Responses against TRV
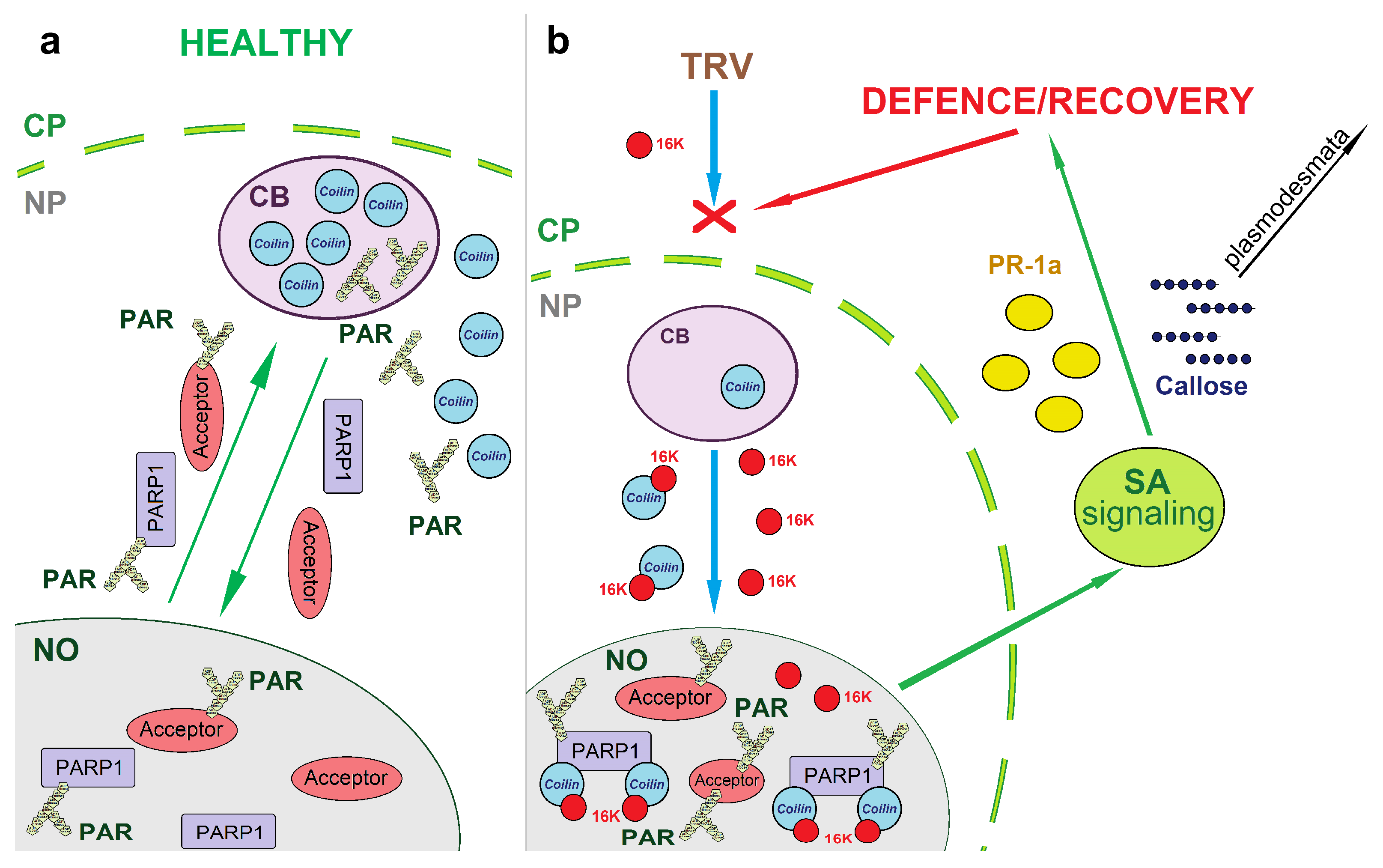
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rissel, D.; Peiter, E. Poly(ADP-Ribose) Polymerases in Plants and Their Human Counterparts: Parallels and Peculiarities. Int. J. Mol. Sci. 2019, 20, 1638. [Google Scholar] [CrossRef] [PubMed]
- Spechenkova, N.; Kalinina, N.O.; Zavriev, S.K.; Love, A.J.; Taliansky, M. ADP-Ribosylation and Antiviral Resistance in Plants. Viruses 2023, 15, 241. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Kraus, W.L. On PAR with PARP: Cellular Stress Signaling through Poly(ADP-Ribose) and PARP-1. Genes Dev. 2012, 26, 417–432. [Google Scholar] [CrossRef]
- Olson, M.O.J. Sensing Cellular Stress: Another New Function for the Nucleolus? Sci. STKE 2004, 2004, pe10. [Google Scholar] [CrossRef]
- Rubbi, C.P.; Milner, J. Disruption of the Nucleolus Mediates Stabilization of P53 in Response to DNA Damage and Other Stresses. EMBO J. 2003, 22, 6068–6077. [Google Scholar] [CrossRef] [PubMed]
- Sirri, V.; Urcuqui-Inchima, S.; Roussel, P.; Hernandez-Verdun, D. Nucleolus: The Fascinating Nuclear Body. Histochem. Cell Biol. 2008, 129, 13–31. [Google Scholar] [CrossRef] [PubMed]
- Pederson, T. The Nucleolus. Cold Spring Harb. Perspect. Biol. 2011, 3, a000638. [Google Scholar] [CrossRef]
- Gall, J.G. Cajal Bodies: The First 100 Years. Annu. Rev. Cell Dev. Biol. 2000, 16, 273–300. [Google Scholar] [CrossRef]
- Carmo-Fonseca, M. New Clues to the Function of the Cajal Body. EMBO Rep. 2002, 3, 726–727. [Google Scholar] [CrossRef]
- Ogg, S.C.; Lamond, A.I. Cajal Bodies and Coilin—Moving towards Function. J. Cell Biol. 2002, 159, 17–21. [Google Scholar] [CrossRef]
- Cioce, M.; Lamond, A.I. Cajal Bodies: A Long History of Discovery. Annu. Rev. Cell Dev. Biol. 2005, 21, 105–131. [Google Scholar] [CrossRef] [PubMed]
- Bassett, C.L. Cajal Bodies and Plant RNA Metabolism. Crit. Rev. Plant Sci. 2012, 31, 258–270. [Google Scholar] [CrossRef]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.-M.; Lamond, A.I. The Nucleolus under Stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Kotova, E.; Jarnik, M.; Tulin, A.V. Poly (ADP-Ribose) Polymerase 1 Is Required for Protein Localization to Cajal Body. PLoS Genet. 2009, 5, e1000387. [Google Scholar] [CrossRef]
- Love, A.J.; Yu, C.; Petukhova, N.V.; Kalinina, N.O.; Chen, J.; Taliansky, M.E. Cajal Bodies and Their Role in Plant Stress and Disease Responses. RNA Biol. 2017, 14, 779–790. [Google Scholar] [CrossRef]
- Kalinina, N.O.; Makarova, S.; Makhotenko, A.; Love, A.J.; Taliansky, M. The Multiple Functions of the Nucleolus in Plant Development, Disease and Stress Responses. Front. Plant Sci. 2018, 9, 132. [Google Scholar] [CrossRef]
- Vainonen, J.P.; Shapiguzov, A.; Vaattovaara, A.; Kangasjärvi, J. Plant PARPs, PARGs and PARP-like Proteins. Curr. Protein Pept. Sci. 2016, 17, 713–723. [Google Scholar] [CrossRef]
- Gu, Z.; Pan, W.; Chen, W.; Lian, Q.; Wu, Q.; Lv, Z.; Cheng, X.; Ge, X. New Perspectives on the Plant PARP Family: Arabidopsis PARP3 Is Inactive, and PARP1 Exhibits Predominant Poly (ADP-Ribose) Polymerase Activity in Response to DNA Damage. BMC Plant Biol. 2019, 19, 364. [Google Scholar] [CrossRef]
- Song, J.; Keppler, B.D.; Wise, R.R.; Bent, A.F. PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses. PLoS Genet. 2015, 11, e1005200. [Google Scholar] [CrossRef]
- Malgras, M.; Garcia, M.; Jousselin, C.; Bodet, C.; Lévêque, N. The Antiviral Activities of Poly-ADP-Ribose Polymerases. Viruses 2021, 13, 582. [Google Scholar] [CrossRef]
- Xia, C.; Wolf, J.J.; Sun, C.; Xu, M.; Studstill, C.J.; Chen, J.; Ngo, H.; Zhu, H.; Hahm, B. PARP1 Enhances Influenza A Virus Propagation by Facilitating Degradation of Host Type I Interferon Receptor. J. Virol. 2020, 94, e01572-19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Guo, J.; Chen, Q. Role of PARP-1 in Human Cytomegalovirus Infection and Functional Partners Encoded by This Virus. Viruses 2022, 14, 2049. [Google Scholar] [CrossRef] [PubMed]
- Sekine, K.-T.; Kawakami, S.; Hase, S.; Kubota, M.; Ichinose, Y.; Shah, J.; Kang, H.-G.; Klessig, D.F.; Takahashi, H. High Level Expression of a Virus Resistance Gene, RCY1, Confers Extreme Resistance to Cucumber Mosaic Virus in Arabidopsis Thaliana. Mol. Plant-Microbe Interact. 2008, 21, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Glushkevich, A.; Spechenkova, N.; Fesenko, I.; Knyazev, A.; Samarskaya, V.; Kalinina, N.O.; Taliansky, M.; Love, A.J. Transcriptomic Reprogramming, Alternative Splicing and RNA Methylation in Potato (Solanum tuberosum L.) Plants in Response to Potato Virus Y Infection. Plants 2022, 11, 635. [Google Scholar] [CrossRef] [PubMed]
- Samarskaya, V.O.; Spechenkova, N.; Markin, N.; Suprunova, T.P.; Zavriev, S.K.; Love, A.J.; Kalinina, N.O.; Taliansky, M. Impact of Exogenous Application of Potato Virus Y-Specific DsRNA on RNA Interference, Pattern-Triggered Immunity and Poly(ADP-Ribose) Metabolism. Int. J. Mol. Sci. 2022, 23, 7915. [Google Scholar] [CrossRef]
- Kim, S.H.; Ryabov, E.V.; Kalinina, N.O.; Rakitina, D.V.; Gillespie, T.; MacFarlane, S.; Haupt, S.; Brown, J.W.; Taliansky, M. Cajal Bodies and the Nucleolus Are Required for a Plant Virus Systemic Infection. EMBO J. 2007, 26, 2169–2179. [Google Scholar] [CrossRef]
- Kim, S.H.; MacFarlane, S.; Kalinina, N.O.; Rakitina, D.V.; Ryabov, E.V.; Gillespie, T.; Haupt, S.; Brown, J.W.S.; Taliansky, M. Interaction of a Plant Virus-Encoded Protein with the Major Nucleolar Protein Fibrillarin Is Required for Systemic Virus Infection. Proc. Natl. Acad. Sci. USA 2007, 104, 11115–11120. [Google Scholar] [CrossRef]
- Shaw, J.; Love, A.J.; Makarova, S.S.; Kalinina, N.O.; Harrison, B.D.; Taliansky, M.E. Coilin, the Signature Protein of Cajal Bodies, Differentially Modulates the Interactions of Plants with Viruses in Widely Different Taxa. Nucleus 2014, 5, 85–94. [Google Scholar] [CrossRef]
- Shaw, J.; Yu, C.; Makhotenko, A.V.; Makarova, S.S.; Love, A.J.; Kalinina, N.O.; MacFarlane, S.; Chen, J.; Taliansky, M.E. Interaction of a Plant Virus Protein with the Signature Cajal Body Protein Coilin Facilitates Salicylic Acid-Mediated Plant Defence Responses. New Phytol. 2019, 224, 439–453. [Google Scholar] [CrossRef]
- Ding, Y.; Lozano-Durán, R. The Cajal Body in Plant-Virus Interactions. Viruses 2020, 12, 250. [Google Scholar] [CrossRef]
- MacFarlane, S.A. Tobraviruses—Plant Pathogens and Tools for Biotechnology. Mol. Plant Pathol. 2010, 11, 577–583. [Google Scholar] [CrossRef]
- Ratcliff, F.; Harrison, B.D.; Baulcombe, D.C. A Similarity Between Viral Defense and Gene Silencing in Plants. Science 1997, 276, 1558–1560. [Google Scholar] [CrossRef] [PubMed]
- Ratcliff, F.; MacFarlane, S.; Baulcombe, D. Gene Silencing without DNA. Rna-Mediated Cross-Protection between Viruses. Plant Cell 1999, 11, 1207–1216. [Google Scholar] [CrossRef]
- Ghazala, W.; Waltermann, A.; Pilot, R.; Winter, S.; Varrelmann, M. Functional Characterization and Subcellular Localization of the 16K Cysteine-Rich Suppressor of Gene Silencing Protein of Tobacco Rattle Virus. J. Gen. Virol. 2008, 89, 1748–1758. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.H.; Robinson, D.J.; Duncan, G.H.; Harrison, B.D. Nuclear Location of the 16K Non-Structural Protein of Tobacco Rattle Virus. J. Gen. Virol. 1991, 72, 1811–1817. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, D.A.; Klessig, D.F. How Does the Multifaceted Plant Hormone Salicylic Acid Combat Disease in Plants and Are Similar Mechanisms Utilized in Humans? BMC Biol. 2017, 15, 23. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Dinesh-Kumar, S.P. Virus-Induced Gene Silencing in Tomato. Plant J. 2002, 31, 777–786. [Google Scholar] [CrossRef]
- Ghazala, W.; Varrelmann, M. Tobacco Rattle Virus 29K Movement Protein Is the Elicitor of Extreme and Hypersensitive-like Resistance in Two Cultivars of Solanum tuberosum. Mol. Plant-Microbe Interact. 2007, 20, 1396–1405. [Google Scholar] [CrossRef]
- Abramoff, M.D.; Magelhaes, P.J.; Ram, S.J. Image Processing with ImageJ. Biophotonics Int. 2004, 11, 36–42. [Google Scholar]
- Terzi, L.C.; Simpson, G.G. Arabidopsis RNA Immunoprecipitation. Plant J. 2009, 59, 163–168. [Google Scholar] [CrossRef]
- Block, M.D.; Verduyn, C.; Brouwer, D.D.; Cornelissen, M. Poly(ADP-Ribose) Polymerase in Plants Affects Energy Homeostasis, Cell Death and Stress Tolerance. Plant J. 2005, 41, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Affar, E.B.; Duriez, P.J.; Shah, R.G.; Winstall, E.; Germain, M.; Boucher, C.; Bourassa, S.; Kirkland, J.B.; Poirier, G.G. Immunological Determination and Size Characterization of Poly(ADP-Ribose) Synthesized in Vitro and in Vivo. Biochim. Biophys. Acta BBA—Gen. Subj. 1999, 1428, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Makarov, V.V.; Skurat, E.V.; Semenyuk, P.I.; Abashkin, D.A.; Kalinina, N.O.; Arutyunyan, A.M.; Solovyev, A.G.; Dobrov, E.N. Structural Lability of Barley Stripe Mosaic Virus Virions. PLoS ONE 2013, 8, e60942. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.; Hamilton, A.J.; Voinnet, O.; Thomas, C.L.; Maule, A.J.; Baulcombe, D.C. RNA-DNA Interactions and DNA Methylation in Post-Transcriptional Gene Silencing. Plant Cell 1999, 11, 2291–2301. [Google Scholar] [PubMed]
- Allasia, V.; Industri, B.; Ponchet, M.; Quentin, M.; Favery, B.; Keller, H. Quantification of Salicylic Acid (SA) and SA-Glucosides in Arabidopsis Thaliana. Bio Protoc. 2018, 8, e2844. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Shi, L.; Han, C.; Yu, J.; Li, D.; Zhang, Y. Validation of Reference Genes for Gene Expression Studies in Virus-Infected Nicotiana Benthamiana Using Quantitative Real-Time PCR. PLoS ONE 2012, 7, e46451. [Google Scholar] [CrossRef] [PubMed]
- Briggs, A.G.; Bent, A.F. Poly(ADP-Ribosyl)Ation in Plants. Trends Plant Sci. 2011, 16, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Briggs, A.G.; Adams-Phillips, L.C.; Keppler, B.D.; Zebell, S.G.; Arend, K.C.; Apfelbaum, A.A.; Smith, J.A.; Bent, A.F. A Transcriptomics Approach Uncovers Novel Roles for Poly(ADP-Ribosyl)Ation in the Basal Defense Response in Arabidopsis Thaliana. PLoS ONE 2017, 12, e0190268. [Google Scholar] [CrossRef]
- Faivre-Rampant, O.; Gilroy, E.M.; Hrubikova, K.; Hein, I.; Millam, S.; Loake, G.J.; Birch, P.; Taylor, M.; Lacomme, C. Potato Virus X-Induced Gene Silencing in Leaves and Tubers of Potato. Plant Physiol. 2004, 134, 1308–1316. [Google Scholar] [CrossRef]
- Lewsey, M.G.; Murphy, A.M.; MacLean, D.; Dalchau, N.; Westwood, J.H.; Macaulay, K.; Bennett, M.H.; Moulin, M.; Hanke, D.E.; Powell, G.; et al. Disruption of Two Defensive Signaling Pathways by a Viral RNA Silencing Suppressor. Mol. Plant-Microbe Interact. 2010, 23, 835–845. [Google Scholar] [CrossRef]
- Carr, J.P.; Donnelly, R.; Tungadi, T.; Murphy, A.M.; Jiang, S.; Bravo-Cazar, A.; Yoon, J.-Y.; Cunniffe, N.J.; Glover, B.J.; Gilligan, C.A. Chapter Seven—Viral Manipulation of Plant Stress Responses and Host Interactions with Insects. In Advances in Virus Research; Palukaitis, P., Roossinck, M.J., Eds.; Academic Press: Cambridge, MA, USA, 2018; Volume 102, pp. 177–197. [Google Scholar]
- Malamy, J.; Hennig, J.; Klessig, D.F. Temperature-Dependent Induction of Salicylic Acid and Its Conjugates during the Resistance Response to Tobacco Mosaic Virus Infection. Plant Cell 1992, 4, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Love, A.J.; Geri, C.; Laird, J.; Carr, C.; Yun, B.-W.; Loake, G.J.; Tada, Y.; Sadanandom, A.; Milner, J.J. Cauliflower Mosaic Virus Protein P6 Inhibits Signaling Responses to Salicylic Acid and Regulates Innate Immunity. PLoS ONE 2012, 7, e47535. [Google Scholar] [CrossRef] [PubMed]
- Whitham, S.A.; Quan, S.; Chang, H.-S.; Cooper, B.; Estes, B.; Zhu, T.; Wang, X.; Hou, Y.-M. Diverse RNA Viruses Elicit the Expression of Common Sets of Genes in Susceptible Arabidopsis Thaliana Plants. Plant J. 2003, 33, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Love, A.J.; Yun, B.W.; Laval, V.; Loake, G.J.; Milner, J.J. Cauliflower Mosaic Virus, a Compatible Pathogen of Arabidopsis, Engages Three Distinct Defense-Signaling Pathways and Activates Rapid Systemic Generation of Reactive Oxygen Species. Plant Physiol. 2005, 139, 935–948. [Google Scholar] [CrossRef]
- Ellinger, D.; Voigt, C.A. Callose Biosynthesis in Arabidopsis with a Focus on Pathogen Response: What We Have Learned within the Last Decade. Ann. Bot. 2014, 114, 1349–1358. [Google Scholar] [CrossRef]
- Wang, X.; Sager, R.; Cui, W.; Zhang, C.; Lu, H.; Lee, J.-Y. Salicylic Acid Regulates Plasmodesmata Closure during Innate Immune Responses in Arabidopsis. Plant Cell 2013, 25, 2315–2329. [Google Scholar] [CrossRef]
- Mahmoudi, S.; Henriksson, S.; Weibrecht, I.; Smith, S.; Söderberg, O.; Strömblad, S.; Wiman, K.G.; Farnebo, M. WRAP53 Is Essential for Cajal Body Formation and for Targeting the Survival of Motor Neuron Complex to Cajal Bodies. PLoS Biol. 2010, 8, e1000521. [Google Scholar] [CrossRef]
- Shpargel, K.B.; Matera, A.G. Gemin Proteins Are Required for Efficient Assembly of Sm-Class Ribonucleoproteins. Proc. Natl. Acad. Sci. USA 2005, 102, 17372–17377. [Google Scholar] [CrossRef]
- Carvalho, T.; Almeida, F.; Calapez, A.; Lafarga, M.; Berciano, M.T.; Carmo-Fonseca, M. The Spinal Muscular Atrophy Disease Gene Product, Smn: A Link between Snrnp Biogenesis and the Cajal (Coiled) Body. J. Cell Biol. 1999, 147, 715–728. [Google Scholar] [CrossRef]
- Kroiss, M.; Schultz, J.; Wiesner, J.; Chari, A.; Sickmann, A.; Fischer, U. Evolution of an RNP Assembly System: A Minimal SMN Complex Facilitates Formation of UsnRNPs in Drosophila Melanogaster. Proc. Natl. Acad. Sci. USA 2008, 105, 10045–10050. [Google Scholar] [CrossRef]
- Li, L.; Ji, G.; Ye, C.; Shu, C.; Zhang, J.; Liang, C. PlantOrDB: A Genome-Wide Ortholog Database for Land Plants and Green Algae. BMC Plant Biol. 2015, 15, 161. [Google Scholar] [CrossRef] [PubMed]
- Kørner, C.J.; Pitzalis, N.; Peña, E.J.; Erhardt, M.; Vazquez, F.; Heinlein, M. Crosstalk between PTGS and TGS Pathways in Natural Antiviral Immunity and Disease Recovery. Nat. Plants 2018, 4, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, B.; Sanfaçon, H. Symptom Recovery in Virus-Infected Plants: Revisiting the Role of RNA Silencing Mechanisms. Virology 2015, 479–480, 167–179. [Google Scholar] [CrossRef]
- Shimomura, T.; Dijkstra, J. The Occurrence of Callose during the Process of Local Lesion Formation. Neth. J. Plant Pathol. 1975, 81, 107–121. [Google Scholar] [CrossRef]
- Calil, I.P.; Fontes, E.P.B. Plant Immunity against Viruses: Antiviral Immune Receptors in Focus. Ann. Bot. 2017, 119, 711–723. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spechenkova, N.; Samarskaya, V.O.; Kalinina, N.O.; Zavriev, S.K.; MacFarlane, S.; Love, A.J.; Taliansky, M. Plant Poly(ADP-Ribose) Polymerase 1 Is a Potential Mediator of Cross-Talk between the Cajal Body Protein Coilin and Salicylic Acid-Mediated Antiviral Defence. Viruses 2023, 15, 1282. https://doi.org/10.3390/v15061282
Spechenkova N, Samarskaya VO, Kalinina NO, Zavriev SK, MacFarlane S, Love AJ, Taliansky M. Plant Poly(ADP-Ribose) Polymerase 1 Is a Potential Mediator of Cross-Talk between the Cajal Body Protein Coilin and Salicylic Acid-Mediated Antiviral Defence. Viruses. 2023; 15(6):1282. https://doi.org/10.3390/v15061282
Chicago/Turabian StyleSpechenkova, Nadezhda, Viktoriya O. Samarskaya, Natalya O. Kalinina, Sergey K. Zavriev, S. MacFarlane, Andrew J. Love, and Michael Taliansky. 2023. "Plant Poly(ADP-Ribose) Polymerase 1 Is a Potential Mediator of Cross-Talk between the Cajal Body Protein Coilin and Salicylic Acid-Mediated Antiviral Defence" Viruses 15, no. 6: 1282. https://doi.org/10.3390/v15061282
APA StyleSpechenkova, N., Samarskaya, V. O., Kalinina, N. O., Zavriev, S. K., MacFarlane, S., Love, A. J., & Taliansky, M. (2023). Plant Poly(ADP-Ribose) Polymerase 1 Is a Potential Mediator of Cross-Talk between the Cajal Body Protein Coilin and Salicylic Acid-Mediated Antiviral Defence. Viruses, 15(6), 1282. https://doi.org/10.3390/v15061282






